In this issue of Blood, Johnson et al1 report their findings of the hepatic cytochrome P450 (CYP) enzyme gene polymorphisms in patients with chronic lymphocytic leukemia (CLL) treated with fludarabine and cyclophosphamide (FC) within a clinical trial. Patients with at least one *6 allele had significantly lower rates of complete response and also less toxicity than patients with *1/*1 alleles, as well as a trend of shorter progression-free survival (PFS). Albeit indirect, this is the first indication in a controlled clinical trial of a significant impact of cyclophosphamide (CPA) bioactivation in CLL and a step toward tailored therapy of cancer.
CPA needs bioactivation in the liver to 4OH-CPA by CYP enzymes, particularly CYP2B6, which occurs mainly with *1/*1 alleles (wild type). As a result of SNPs, variant alleles occur, including the *6 variant associated with reduced levels of the CYP2B6 enzyme. In CLL patients treated with FC within a controlled clinical trial, Johnson et al1 found that patients with at least one *6 allele had lower complete response rates, less toxicity, and a trend of shorter PFS than *1/*1 subjects, indicating lower CPA bioactivation by the CYP2B6 *6 enzyme variant. In contrast, no differences in outcome were detected in patients treated with chlorambucil or fludarabine. Professional illustration by Paulette Dennis.
CPA needs bioactivation in the liver to 4OH-CPA by CYP enzymes, particularly CYP2B6, which occurs mainly with *1/*1 alleles (wild type). As a result of SNPs, variant alleles occur, including the *6 variant associated with reduced levels of the CYP2B6 enzyme. In CLL patients treated with FC within a controlled clinical trial, Johnson et al1 found that patients with at least one *6 allele had lower complete response rates, less toxicity, and a trend of shorter PFS than *1/*1 subjects, indicating lower CPA bioactivation by the CYP2B6 *6 enzyme variant. In contrast, no differences in outcome were detected in patients treated with chlorambucil or fludarabine. Professional illustration by Paulette Dennis.
When combined, FC exerts synergism owing to inhibition by fludarabine of the repair of CPA-induced point mutations.2 The British LFR CLL4 trial,3 which compared frontline therapy of CLL with chlorambucil, fludarabine, or FC, is one of the 3 important controlled trials3-5 that established FC as the standard chemotherapy in fit CLL patients and led to the CLL8 trial, which subsequently established rituximab-FC as the standard immuno-chemotherapy6 and was the first shown to prolong survival in CLL.
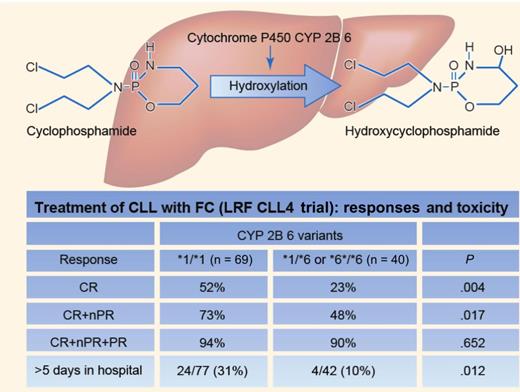
CPA is a pro-drug that requires bioactivation by hydroxylation to 4-hydroxy-CPA (4OH-CPA) by hepatic CYP enzymes, in particular CYP2B6 (see figure). 4OH-CPA is in equilibrium with its tautomer aldophosphamide, which, by off-cleavage of the toxic metabolite acrolein, turns into phosphoramide mustard, the active compound. CYP2B6 mainly expresses *1/*1 alleles (wild type), but is quite polymorphic, with several known single nucleotide polymorphisms (SNPs) that lead to variant alleles, particularly the *6 allele, which occurs in approximately one-third of whites. In such individuals, the enzyme levels are reduced but may be more active in CPA hydroxylation than the *1/*1 allele enzyme.7,8
Johnson and colleagues1 performed real-time polymerase chain reaction for the CYP2B6 SNP genotypes in 455 (roughly all with available DNA) of the 777 patients in the CLL4 study. The 428 patients with the *1/*1 (265), *1/*6 (134) or *6/*6 (29) genotypes were selected for further investigation, of whom 206 had been assigned to chlorambucil, 103 to fludarabine, and 119 to FC (77 *1/*1 and 42 with at least one *6 allele, respectively). Clinical or other genomic characteristics did not differ among these treatment- or enzyme-variant subgroups, or among those with and without available DNA, and the main trial findings were the same in this cohort as in the original entire CLL4 cohort.
Among the patients assigned to FC, the patients expressing at least one *6 allele (*6 carriers) achieved a complete remission (CR) or a CR + nodal partial remission significantly less often than *1/*1 carriers, also confirmed by analysis of specific response indicators like white blood cell count and lymphadenopathy. In addition, a trend was found of shorter PFS among the *6 carriers than among the *1/*1 carriers (P = .055). Of note, in multivariate analysis of biological variables and response, only TP53 deletion/mutation and CYP2B6 variant had an independent influence on the likelihood of achieving CR. Furthermore, some typical FC-related adverse events and in-hospital days were also less frequent among *6 carriers. None of these differences were observed among the 309 patients assigned to either CLB or F. These findings clearly indicate a negative impact of the *6 allele on conversion of CPA to its active form.
That the response to CPA may vary with CYP2B variants is not a new observation, but has been suggested in lymphoma, myeloma, breast cancer, and stem cell transplantation, but often in retrospective studies with divergent methodologies and results (summarized by Raccor et al and Zanger and Klein).7,8 However, the present study is the first based on a prospective, controlled clinical trial and is the first to indicate an impact of host pharmacogenetics in CLL. Given the extensive use of CPA for malignancies in general, the observation is relevant for the whole field of cancer chemotherapy and may contribute to progress toward tailored therapy.
But what will this observation mean for the use of CPA-containing regimens in the future? Although 4OH-CPA can be measured by routine methods (high-performance liquid chromatography), it is cumbersome7 and hardly feasible in larger clinical trials. Instead, a pharmacogenetic approach like the present study may make it possible to intervene in future trials, for example by CPA dose adjustment in individuals or groups with variant CYP enzymes including CYP2B6.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal