Key Points
Runx1 is necessary for survival and development of B cell–specified progenitors and also the transition through the pre-B-cell stage.
Genomewide expression and Runx1 occupancy analyses identified critical target genes and collaborating transcription partners.
Abstract
The t(12;21) chromosomal translocation, targeting the gene encoding the RUNX1 transcription factor, is observed in 25% of pediatric acute lymphoblastic leukemia (ALL) and is an initiating event in the disease. To elucidate the mechanism by which RUNX1 disruption initiates leukemogenesis, we investigated its normal role in murine B-cell development. This study revealed 2 critical functions of Runx1: (1) to promote survival and development of progenitors specified to the B-cell lineage, a function that can be substituted by ectopic Bcl2 expression, and (2) to enable the developmental transition through the pre-B stage triggered by the pre-B-cell antigen receptor (pre-BCR). Gene expression analysis and genomewide Runx1 occupancy studies support the hypothesis that Runx1 reinforces the transcription factor network governing early B-cell survival and development and specifically regulates genes encoding members of the Lyn kinase subfamily (key integrators of interleukin-7 and pre-BCR signaling) and the stage-specific transcription factors SpiB and Aiolos (critical downstream effectors of pre-BCR signaling). Interrogation of expression databases of 257 ALL samples demonstrated the specific down-regulation of the SPIB and IKZF3 genes (the latter encoding AIOLOS) in t(12;21) ALL, providing novel insight into the mechanism by which the translocation blocks B-cell development and promotes leukemia.
Introduction
Disruptions of pivotal transcription factors that govern lineage-specific differentiation of lympho-hematopioetic cells are common events in acute lymphoblastic leukemia (ALL) and acute myelogenous leukemia.1,2 In B-cell precursor (BCP)-ALL, the most common form of leukemia in children, specific genetic mutations affecting the IKZF1, EBF1, TCF3, or PAX5 genes occur at overall incidences ranging from ∼1% (for EBF1) to 15% (for PAX5).1,3-5 All 4 genes encode transcription factors (IKAROS, EBF1, E2A, and PAX5, respectively) that are key components of a hierarchical regulatory circuit that coordinate B-cell specification, commitment, and/or development.6-9
Significantly, the most frequent genetic aberration in pediatric BCP-ALL, occurring in close to 25% of all cases, is the chromosomal translocation t(12;21) that juxtapositions the ETV6 and RUNX1 genes to form a fusion protein. Insightful studies of twins with discordant incidence and/or latency of BCP-ALL, coupled with molecular genetics, cell biology, and animal models, have provided strong evidence that ETV6-RUNX1 is the initiating lesion in t(12;21) BCP-ALL, often occurring in utero and leading to the expansion and persistence of a preleukemic cell pool.10 However, the mechanism by which the ETV6/RUNX1 fusion protein establishes a preleukemic condition and thereby initiates leukemogenesis has not been resolved. The fusion protein retains the N-terminal PNT domain of ETV6, which is thought to deregulate the normal activity of RUNX1 by promoting multimerization and recruitment of transcriptional corepressors, leading to avid DNA binding and repression of its gene targets.11 Understanding the role of RUNX1 in B-cell development is thus essential to delineate the molecular mechanism by which ETV6/RUNX1 initiates leukemogenesis.
RUNX1 encodes a transcription factor belonging to the highly conserved family of DNA-binding proteins that contain a Runt homology domain. Despite a growing understanding of Runx1 function in several lympho-hematopoietic lineages12-14 and its major impact in BCP-ALL, little is known with regard to its role in B-cell development. The aim of this work was to extend the work of previous studies demonstrating a loss of B cells in Runx1-deficient mice15-18 by determining the precise developmental stage(s) at which early B-cell development is blocked and to identify critical Runx1 target genes in B-cell development and ALL.
Methods
Experimental animals
The B6.129-Runx1fl/fl strain was established after backcrossing 129-Runx1fl/fl mice17 onto a C57Bl/6 background for over ten generations. The B6.129-Tg(vav-Cre) and B6.129-Cd79ahCre/+ strains were provided by T. Graf (CGI, Barcelona)19 and M. Reth (Max-Planck-Institute, Freiburg),20 respectively. The B6-Tg(Bcl2)36Wehi/J and B6.129 × 1-Gt(ROSA)26Sortm1(EYFP)Cos/J strains were obtained from Jackson Laboratories. All animal strains were maintained at the Heinrich-Pette-Institute animal facility. All animal studies were approved by the Hamburg commission for animal experiments.
Antibodies and flow cytometry
Cells for fluorescence-activated cell sorter (FACS) analysis and sorting were prepared from blood, bone marrow (BM), and spleen from 6- to 12-week-old mice as described.21 Single cell suspensions were stained with fluorophore- or biotin-conjugated antibodies (supplemental Table 1 on the Blood website) and analyzed or sorted on a FACS Canto-I or Aria-I, respectively (BD Biosciences). The EasySep Mouse Hematopoietic Progenitor Enrichment Kit (StemCell Technologies) aided analysis of early progenitor compartments.
Cell culture assays
For proliferation and apoptosis assays, sorted pro-B/pre-B cells (1 × 106 cell/mL) were stained with carboxyfluorescein diacetate succinimidyl ester (1.65 µM; Sigma) in phosphate-buffered saline and 0.1% fetal calf serum (FCS; PAN Biotech) at 20°C for 10 minutes and then incubated with 10% FCS at 37°C for 15 minutes. Cells were plated either with or without preseeded OP9 cells (2 × 103) in α-modified minimum essential medium (PAA Laboratories) supplemented with 10% FCS, 10 to 20 ng interleukin-7 (IL7)/mL (Peprotech), and 50 µM β-mercaptoethanol. FACS analysis was evaluated using the FlowJo software (Tree Star, Inc.). Apoptosis was assessed with the AnnexinV Apoptosis Detection Kit 1 (BD Pharmingen). Common lymphoid progenitors (CLPs) were sorted at various cell densities into wells containing 2 × 103 irradiated OP9 cells (13.5 Gy for 35 minutes) supplemented with α-modified minimum essential medium, 10% FCS, 40 µM β-mercaptoethanol, 10 ng/mL FLT3-ligand, 10 ng/mL stem cell factor, and 20 ng/mL IL7 (Peprotech). Pre-B-cell colony assays were performed with Methocult M3630 (Stem Cell Technologies) in triplicate and scored after 1 week.
Gene expression analysis
For loss-of-function gene expression analysis, B-cell progenitors (CD19+,Lin2neg,B220med,CD93+) were isolated from the BM of either Runx1fl/flCd79ahCre/+ or Runx1+/+Cd79ahCre/+. For gain-of-function experiments, BMiFLT3(15-3) pro-B cells isolated from a murine model for BCP-ALL and expressing low levels of Runx1 (B.N., A. Werk, N.K., and C.S., unpublished results) were transduced with retroviral vectors expressing RUNX1-ERt2 or ERt2 coexpressing Venus fluorescent protein and sorted. Cells were treated with 0.2 µM 4-hydroxytamoxifen (Sigma) for 24 hours, and viable cells were sorted for Venus/B220med expression. RNA was extracted (NucleoSpin RNA II; Agilent Technologies), labeled with Cy3, and hybridized to a Whole Mouse Genome Microarray (4 × 44K or 4 × 44K v.2; Agilent Technologies) by Miltenyi Biotec. The Agilent Feature Extraction Software was used to read out and process the microarray image files.
ChIP-coupled deep sequencing
Chromatin immunoprecipitation (ChIP) using anti-Runx1 or anti-IgG antibodies was performed using the EZ-ChIP Kit (Upstate/Millipore) on murine BMiFLT3(15-3) cells induced to activate Runx1. Extracted DNA fragments were isolated, sequenced on a Genome Analyzer IIx (Illumina), and mapped to the mm9 assembly. Details for ChIP-coupled deep sequencing (ChIP-Seq), sequence analysis, and associated references are available in supplemental Methods.
Accession numbers
Expression array and ChIP-Seq data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE45425).
Results
Runx1 is essential for survival and development of cells specified to the B-cell lineage
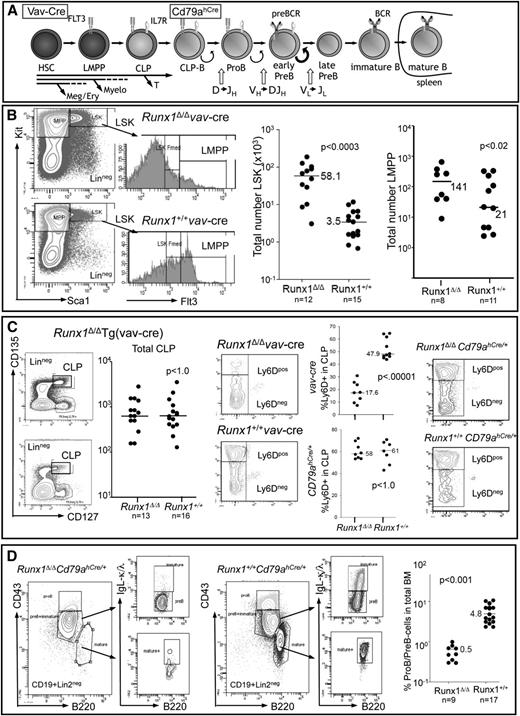
To determine the hematopoietic stage at which Runx1 becomes critical for B-cell development, we compared progenitor compartments of mice in which Runx1 was excised either in the hematopoietic stem cell (HSC) compartment [Tg(vav-Cre)] or in the B cell–specified compartment (Cd79ahCre/+) with control mice (Figure 1A). Excision of Runx1 at the level of the HSC confirmed the importance of Runx1 in the development of several different lineages (supplemental Figure 1) and for negatively regulating the size of the Lin-Sca1+Kithi-stem cell and myeloid multipotent progenitor compartments, but also showed that the number of lymphoid-primed multipotent progenitors is only moderately affected by loss of Runx1 (Figure 1B). CLPs were also found at similar absolute cell numbers in the BM of Runx1-deficient mice compared with Runx1+/+ littermates, but a significant reduction in the proportion of Ly6D+ CLPs marking the B cell–specified CLPs (CLP-B)22,23 was observed (Figure 1C). In contrast, Runx1fl/flCd79ahCre/+ mice showed no changes in the CLP compartment, with the Ly6D+ fraction (CLP-B) unchanged with respect to control Runx1+/+Cd79ahCre/+ littermates (Figure 1C). Control experiments demonstrated that Cd79ahCre-mediated excision occurs within the CLP-B compartment (supplemental Figure 2A). Despite normal levels of CLP-B cells in Runx1fl/flCd79ahCre/+ mice in contrast to Runx1fl/flTg(vav-Cre) mice, both Runx1-deficient strains had markedly reduced numbers of pro-B/pre-B-cells (Figure 1D; supplemental Figure 1) and no detectable immature/mature B cells in the BM, spleen, or lymph nodes (supplemental Figures 1 and 2). Several assays were performed to confirm that the residual pro-B/pre-B cells were not due to incomplete excision (supplemental Figure 3).
Functional Runx1 deletion by targeted excision by Tg(vav-Cre) or Cd79ahCre/+ expression leads to profound defects in B-cell development. (A) Schematic representation of B-cell development starting from the HSC compartment. B-cell specification occurs at the CLP-B stage (Ly6D+), before the expression of the classical B-cell antigens CD19 and B220. The stage in B-cell development at which Tg(vav-Cre) and Cd79ahCre/+ mediate excision is indicated (see also supplemental Figure 2). Receptors necessary for the proliferative expansion of the indicated cell types are depicted. (B) FACS analysis of lineage-depleted BM cells to detect HSC (LinnegSca1+Kithi), myeloid multipotent progenitor (Lin−Sca1−Kithi), and lymphoid-primed multipotent progenitors (LMPP) (LinnegScal+KithiFlt3hi) compartments in vav-Cre mediated excision. The total cell number determined for each analyzed mouse (2 femora) is depicted by a dot, and the median for the indicated number of mice is shown. (C) FACS strategy for detection of total CLP (LinnegIl7r+Flt3hiKitmedSca1med) and the percentage therein specified for B-cell development (Ly6D+) from lineage-depleted BM cells. Each dot represents the total number of CLPs per Tg(vav-Cre) mouse (2 femora). The percentage of Ly6D+ cells within the CLP fraction of either (upper graph) Tg(vav-Cre) or (lower graph) Cd79ahCre/+ mice is indicated. Representative FACS analysis of total CLPs analyzed for Ly6d expression is shown on the right. (D) FACS analysis of CD19+Lin2neg cells of the BM, detecting pro-B (B220medCD43hiIgLneg), pre-B cells (B220medCD43med/loIgLneg), immature B cells (B220medCD43med/loIgL+), and mature (recirculating) B cells (B220hiCD43negIgL+) of Cd79ahCre/+ mice. Graph depicts the percentage of pro-B/pre-B cells within the total BM of mice with the indicated genotypes. P values were calculated by a nonpaired Student t test.
Functional Runx1 deletion by targeted excision by Tg(vav-Cre) or Cd79ahCre/+ expression leads to profound defects in B-cell development. (A) Schematic representation of B-cell development starting from the HSC compartment. B-cell specification occurs at the CLP-B stage (Ly6D+), before the expression of the classical B-cell antigens CD19 and B220. The stage in B-cell development at which Tg(vav-Cre) and Cd79ahCre/+ mediate excision is indicated (see also supplemental Figure 2). Receptors necessary for the proliferative expansion of the indicated cell types are depicted. (B) FACS analysis of lineage-depleted BM cells to detect HSC (LinnegSca1+Kithi), myeloid multipotent progenitor (Lin−Sca1−Kithi), and lymphoid-primed multipotent progenitors (LMPP) (LinnegScal+KithiFlt3hi) compartments in vav-Cre mediated excision. The total cell number determined for each analyzed mouse (2 femora) is depicted by a dot, and the median for the indicated number of mice is shown. (C) FACS strategy for detection of total CLP (LinnegIl7r+Flt3hiKitmedSca1med) and the percentage therein specified for B-cell development (Ly6D+) from lineage-depleted BM cells. Each dot represents the total number of CLPs per Tg(vav-Cre) mouse (2 femora). The percentage of Ly6D+ cells within the CLP fraction of either (upper graph) Tg(vav-Cre) or (lower graph) Cd79ahCre/+ mice is indicated. Representative FACS analysis of total CLPs analyzed for Ly6d expression is shown on the right. (D) FACS analysis of CD19+Lin2neg cells of the BM, detecting pro-B (B220medCD43hiIgLneg), pre-B cells (B220medCD43med/loIgLneg), immature B cells (B220medCD43med/loIgL+), and mature (recirculating) B cells (B220hiCD43negIgL+) of Cd79ahCre/+ mice. Graph depicts the percentage of pro-B/pre-B cells within the total BM of mice with the indicated genotypes. P values were calculated by a nonpaired Student t test.
To rigorously determine whether Runx1 is necessary for B-cell specification itself and not just the expansion of early B-cell progenitors, we sorted CLPs from Runx1fl/fl Tg(vav-Cre) and Runx1+/+ Tg(vav-Cre) mice and performed limiting dilution assays under conditions that stimulate B-cell differentiation. Assessed after 1 week of culture, cloning efficiencies between Runx1-deficient and normal CLPs were not significantly different (Figure 2A). However, there was a striking contrast in both the number of cells per well (reflecting proliferation and/or survival rates) and the number of wells that showed initial growth (colonies with >50 cells) but that died, as determined by light refraction (Figure 2B). FACS analysis of cells pooled from positive wells demonstrated that, despite poor proliferation and survival rates, most cells arising from Runx1fl/fl Tg(vav-Cre) CLPs were positive for B-cell markers (CD19 and B220; Figure 2C). These results support the hypothesis that Runx1 is not required for B-cell specification but rather their survival or continued development. To address the discrepancy between these results and the low incidence of CLP-B (Ly6d+) observed in vav-Cre Runx1-deficient mice, we examined the level of Ly6d in committed progenitors lacking Runx1 (Figure 2D). The severely reduced Ly6d protein levels suggest that Ly6d is a direct target gene for Runx1, an observation confirmed in subsequent analysis. Thus, in the absence of Runx1, Ly6D cannot be used to assay CLP-B.
Runx1 is critical for the survival of B cell–specified CLP progeny. (A) End point dilution of CLPs under B-cell differentiation conditions shows that Runx1-deficient CLPs form colonies at the same efficiency as Runx1+/+ CLPs. CLPs from either Runx1Δ/Δ [Runx1fl/fl-Tg(vav-Cre)] or Runx1+/+ [Runx1+/+-Tg(vav-Cre)] were sorted, and the indicated cell number was plated on OP9 stroma cells supplemented with cytokines. Positive wells were scored on day 7 by microscopic visualization. (B) CLP progeny from Runx1-deficient mice showed a high frequency of cell death and low proliferation rates. The number of wells exhibiting cell growth was determined at both days 7 and 12 in 2 independent limiting dilution assays. The difference in the number of positive clones is shown as the percentage of surviving clones for each genotype. To estimate the proliferation rate of CLP progeny, 200 CLPs were sorted on OP9 stroma, and the total number of viable cells was determined on day 7. Shown are the results from 4 independent wells seeded in 2 independent sorts. Runx1+/+, solid line; Runx1Δ/Δ, stippled line. (C) FACS analysis of CLP progeny demonstrates the predominant expression of B-cell (CD19+/B220+) but not myeloid (CD11b/Gr1) antigens. For Runx1+/+ CLPs, cells from single wells analyzed by FACS after 1 week; for Runx1Δ/Δ CLPs, cells from 6 positive wells were pooled before analysis. (D) FACS analysis for Ly6D expression levels on pro-B/pre-B cells (CD19+Lin2negB220med) isolated from either Runx1fl/fl Tg(vav-Cre) or Runx1+/+ Tg(vav-Cre) mice.
Runx1 is critical for the survival of B cell–specified CLP progeny. (A) End point dilution of CLPs under B-cell differentiation conditions shows that Runx1-deficient CLPs form colonies at the same efficiency as Runx1+/+ CLPs. CLPs from either Runx1Δ/Δ [Runx1fl/fl-Tg(vav-Cre)] or Runx1+/+ [Runx1+/+-Tg(vav-Cre)] were sorted, and the indicated cell number was plated on OP9 stroma cells supplemented with cytokines. Positive wells were scored on day 7 by microscopic visualization. (B) CLP progeny from Runx1-deficient mice showed a high frequency of cell death and low proliferation rates. The number of wells exhibiting cell growth was determined at both days 7 and 12 in 2 independent limiting dilution assays. The difference in the number of positive clones is shown as the percentage of surviving clones for each genotype. To estimate the proliferation rate of CLP progeny, 200 CLPs were sorted on OP9 stroma, and the total number of viable cells was determined on day 7. Shown are the results from 4 independent wells seeded in 2 independent sorts. Runx1+/+, solid line; Runx1Δ/Δ, stippled line. (C) FACS analysis of CLP progeny demonstrates the predominant expression of B-cell (CD19+/B220+) but not myeloid (CD11b/Gr1) antigens. For Runx1+/+ CLPs, cells from single wells analyzed by FACS after 1 week; for Runx1Δ/Δ CLPs, cells from 6 positive wells were pooled before analysis. (D) FACS analysis for Ly6D expression levels on pro-B/pre-B cells (CD19+Lin2negB220med) isolated from either Runx1fl/fl Tg(vav-Cre) or Runx1+/+ Tg(vav-Cre) mice.
Together these results support the conclusion that Runx1 is not required for the specification of CLPs to the B-cell lineage but is required for the survival and/or proliferation of early progenitors.
Proapoptotic Bcl2 protein rescues the survival of Runx1-deficient early B-cell progenitors
To provide further evidence that the low levels of pro-B/pre-B cells in Runx1-deficient mice are not due to specification but rather a defect in proliferation or survival signals, we crossed Runx1-deficient mice [both Tg(vav-Cre) and Cd79ahCre/+ strains] with Tg(Eµ-bcl2) mice that express antiapoptotic Bcl2 in both T cells and B cells. Strikingly, the levels of B cells in Runx1-deficient mice expressing Bcl2 approached that observed in Runx1+/+ controls (Figure 3A). The number of B cells in control Runx1+/+ Tg(Eµ-bcl2) was also increased compared with wild-type mice; however, this observed 2.5-fold increase was significantly lower than the close to 10-fold increase observed in Runx1-deficient cohorts. The largest increase in cell numbers was observed at the early pre-B stage (CD43lo) in Runx1-deficient Bcl2+ (sixfold; n = 5 per cohort), whereas only a twofold increase at this stage was observed in Runx1+/+ Bcl2+ mice (n = 4 per cohort; Figure 3B-C). Immature cells were also detected in the BM of most (7/11) Bcl2+ Runx1-deficient mice, but their number was ∼10% of that found in wild-type mice. Notably, examination of spleen tissue revealed a small but easily detectable subset of CD19+/B220+ cells expressing IgM and/or IgD in all mice (Figure 3D), but at a significantly lower frequency than in wild-type mice (median of 8% vs 55% of total splenic cells). Small numbers of B cells could also be detected in the blood of animals lacking Runx1 but expressing Bcl2 (Figure 3D).
Bcl2 expression promotes the expansion of Runx1Δ/Δ pre-/pro-B cells, resulting in a limited number of mature B cells. (A) Dot blot showing the percentage of CD19+Lin2neg in total BM from Runx1fl/fl Tg(vav-Cre) or Runx1+/+ Tg(vav-Cre) mice with or without Tg-Bcl2. Each dot represents an independent mouse. (B) Bar graph demonstrates that Tg-Bcl2 leads to a larger increase in pro-/pre-B cells in Runx1Δ/Δ mice compared with Runx1+/+ controls. Immature cells (as detected with IgL-κ/λ antibody) were observed in 4 of 5 Bcl2+Runx1Δ/Δ mice analyzed. The level of mature (transient) B cells in the BM of Runx1Δ/Δ mice was below detection in most mice. Values are compiled from 5 mice per cohort using the gating strategy shown in C. A minimum of 106 events was registered per analysis. (C) Representative FACS analysis of B cells isolated from BM of Runx1fl/flCd79ahCre/+ Tg(Eµ-Bcl2) or Runx1+/+Cd79ahCre/+ Tg(Eµ-Bcl2) mice. The CD43 gate discriminating pro-B- and pre-B-cell populations was determined by analysis of pro-B cells from Igα-deficient mice (Cd79ahCre/hCre), which do not express a functional pre-BCR. (D) Mature B cells (CD19+B220+IgM+/IgD+) are detectable in blood and spleen of Bcl2 mice, albeit at reduced levels. The percentage of B cells (CD19+B220+) found in total blood (inverted triangles) or spleen (black circles) of mice with the indicated genotype is depicted. Histograms show analysis of splenic CD19+ cells and expression of IgM/IgD of representative mice for each genotype. Similar results were obtained with both vav-Cre and Cd79ahCre/+ strains. (E) Colony numbers obtained from 5 × 104 BM cells isolated from Runx1fl/flCd79ahCre/+ (KO), Runx1fl/flCd79ahCre/+ Tg(Eµ-Bcl2) (KO+Bcl2), and Runx1+/+Cd79ahCre/+ (WT) mice and plated in methylcellulose in the presence of IL7. Shown is the mean of 2 independent experiments performed in triplicate. Colonies with similar morphology were picked, and cells were pooled (2-4 colonies per analysis) and analyzed for B-cell markers by FACS.
Bcl2 expression promotes the expansion of Runx1Δ/Δ pre-/pro-B cells, resulting in a limited number of mature B cells. (A) Dot blot showing the percentage of CD19+Lin2neg in total BM from Runx1fl/fl Tg(vav-Cre) or Runx1+/+ Tg(vav-Cre) mice with or without Tg-Bcl2. Each dot represents an independent mouse. (B) Bar graph demonstrates that Tg-Bcl2 leads to a larger increase in pro-/pre-B cells in Runx1Δ/Δ mice compared with Runx1+/+ controls. Immature cells (as detected with IgL-κ/λ antibody) were observed in 4 of 5 Bcl2+Runx1Δ/Δ mice analyzed. The level of mature (transient) B cells in the BM of Runx1Δ/Δ mice was below detection in most mice. Values are compiled from 5 mice per cohort using the gating strategy shown in C. A minimum of 106 events was registered per analysis. (C) Representative FACS analysis of B cells isolated from BM of Runx1fl/flCd79ahCre/+ Tg(Eµ-Bcl2) or Runx1+/+Cd79ahCre/+ Tg(Eµ-Bcl2) mice. The CD43 gate discriminating pro-B- and pre-B-cell populations was determined by analysis of pro-B cells from Igα-deficient mice (Cd79ahCre/hCre), which do not express a functional pre-BCR. (D) Mature B cells (CD19+B220+IgM+/IgD+) are detectable in blood and spleen of Bcl2 mice, albeit at reduced levels. The percentage of B cells (CD19+B220+) found in total blood (inverted triangles) or spleen (black circles) of mice with the indicated genotype is depicted. Histograms show analysis of splenic CD19+ cells and expression of IgM/IgD of representative mice for each genotype. Similar results were obtained with both vav-Cre and Cd79ahCre/+ strains. (E) Colony numbers obtained from 5 × 104 BM cells isolated from Runx1fl/flCd79ahCre/+ (KO), Runx1fl/flCd79ahCre/+ Tg(Eµ-Bcl2) (KO+Bcl2), and Runx1+/+Cd79ahCre/+ (WT) mice and plated in methylcellulose in the presence of IL7. Shown is the mean of 2 independent experiments performed in triplicate. Colonies with similar morphology were picked, and cells were pooled (2-4 colonies per analysis) and analyzed for B-cell markers by FACS.
To determine whether Bcl2 expression was able to rescue early B-cell development in vitro, colony assays of total BM cells were performed in the presence of IL7. As expected, no or only limited small colonies were observed in cells isolated from Runx1fl/flCd79ahCre/+ mice, whereas Runx1-deficient Bcl2+ BM gave rise to colony numbers approaching those from wild-type mice. Notably, the majority of Bcl2+ colonies had a typical B-cell morphology (densely packed, small cells) with a similar size to that of control colonies and was composed of C19+B220+ cells (Figure 3E). However, a minor but reproducible fraction (10%) of the Runx1-deficient, Bcl2+, IL7-responsive cells did not express B-cell markers, suggesting abortive differentiation.
Together, these results demonstrate that the ectopic expression of a survival signal (Bcl2) bypasses a critical barrier for the accumulation of Runx1-deficient B-cell progenitors in vitro and in vivo. Nevertheless, a second block to B-cell development was unveiled in the mouse that impinges at the pre-B stage. A small fraction of cells was able to overcome this block, as evidenced by the accumulation of mature B cells residing in the spleen and circulating in blood.
Runx1 deficiency leads to a block at the early pre-B-cell stage of development
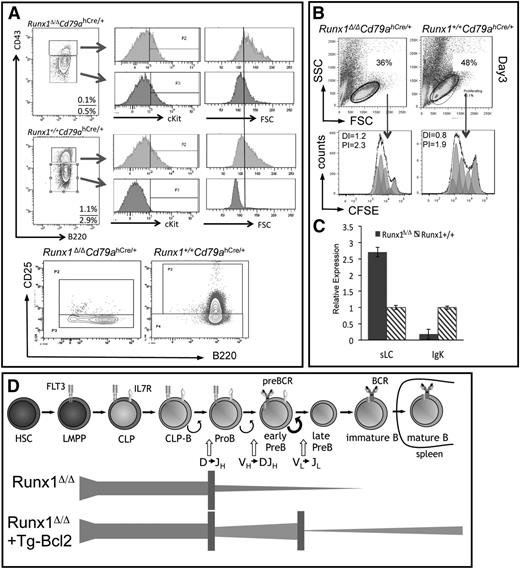
A number of assays were performed to define the second B-cell development block in Runx1-deficient mice. FACS analysis showed that the majority of the Runx1-deficient cells have an aberrant pre-B-cell phenotype, with low CD43 expression, but with aberrant cKit expression (a marker of late pro-B cells) and the absence of CD25 expression (marking stimulated pre-B cells) (Figure 4A). Furthermore, Runx1-deficient CD43lo cells continued to produce large, proliferating cells (as ascertained by forward scatter in FACS analysis), in contrast to those in wild-type mice, which are uniformly small in size, reflecting the cell cycle attenuation that follows the proliferative stimulation of the precursor (pre)B-cell antigen receptor (BCR; Figure 4A). In vitro assays demonstrated that, although Runx1-deficient pro-/pre-B-cell cultures stimulated with IL7 showed a greater proportion of dead cells, the responding fraction proliferated at similar frequencies as control cells, but no accumulation of nonproliferating cells was observed (Figure 4B).
Characterization of Runx1-deficient B-cell progenitors. (A) Shown is the analysis of pre-B- and pro-B-cells (gated on CD19+Lin2negIgLneg) isolated from BM of mice with the indicated genotype and examined for expression of cKit, CD25 (IL2Rα), or for size (FSC). Gates for pro-B cells were set using cells isolated from Cd79ahCre/hCre mice. (B) FACS analysis of sorted pro-/pre-B-cells isolated from mice with the indicated genotypes. Sorted cells were plated on OP9 cells supplemented with IL7 and analyzed after 3 days. Half of the sorted cells were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE) before plating. The proliferation index (PI; based on proliferating cells only) and the division index (DI; all viable cells) were determined by calculation of the fluorescent intensity of the CFSE-stained cells. Results shown are representative of 3 independent experiments. To obtain sufficient cells for sorting, staining, and consequent culture, 3 Runx1-deficient mice were used per experiment. (C) Expression analysis of sLC and LC transcripts (λ5 encoded by Igll1 and rearranged VJ-κ transcripts, respectively) was performed on sorted pro-B/pre-B cells from mice with the given genotypes. cDNA levels were normalized to Hprt expression levels. (D) Schematic representation of the observed blocks in B-cell development in Runx1-deficient mice and the impact of Bcl2 on the cell survival of the pro-B- and pre-B-cell compartments, but their low frequency of transition to the immature stage.
Characterization of Runx1-deficient B-cell progenitors. (A) Shown is the analysis of pre-B- and pro-B-cells (gated on CD19+Lin2negIgLneg) isolated from BM of mice with the indicated genotype and examined for expression of cKit, CD25 (IL2Rα), or for size (FSC). Gates for pro-B cells were set using cells isolated from Cd79ahCre/hCre mice. (B) FACS analysis of sorted pro-/pre-B-cells isolated from mice with the indicated genotypes. Sorted cells were plated on OP9 cells supplemented with IL7 and analyzed after 3 days. Half of the sorted cells were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE) before plating. The proliferation index (PI; based on proliferating cells only) and the division index (DI; all viable cells) were determined by calculation of the fluorescent intensity of the CFSE-stained cells. Results shown are representative of 3 independent experiments. To obtain sufficient cells for sorting, staining, and consequent culture, 3 Runx1-deficient mice were used per experiment. (C) Expression analysis of sLC and LC transcripts (λ5 encoded by Igll1 and rearranged VJ-κ transcripts, respectively) was performed on sorted pro-B/pre-B cells from mice with the given genotypes. cDNA levels were normalized to Hprt expression levels. (D) Schematic representation of the observed blocks in B-cell development in Runx1-deficient mice and the impact of Bcl2 on the cell survival of the pro-B- and pre-B-cell compartments, but their low frequency of transition to the immature stage.
Thus, in addition to increased apoptotic frequencies, Runx1-deficient CD19+ cells do not progress to the quiescent phase of pre-B-cell development, a defect that could be attributed to structural or signaling defects in the pre-BCR. No obvious disruption in the expression or rearrangement of the immunoglobulin Igh locus, encoding the heavy chain components of the pre-BCR, was observed in Runx1-deficient B-cell progenitors (supplemental Figure 4). Similarly, normal gene expression levels of Ig signaling proteins, the coreceptor CD19, and downstream effectors (Syk tyrosine kinases and adapter proteins) were observed in RunxΔ/ΔCd79ahCre/+ progenitors (supplemental Figure 5). Notably, normal to high expression levels of the four genes encoding surrogate light chains (sLCs) were observed, but LC transcripts encoded by the Igl-κ locus were reduced by 1 order of magnitude in Runx1-deficient pro-B/pre-B cells (Figure 4C). Analysis of Igl-κ gene rearrangements revealed no significant difference in the gross level of V-Jκ rearrangements in Runx1-deficient cells (supplemental Figure 5C).
We conclude that Runx1 deficiency leads to 2 blocks in B-cell development (Figure 4D). The first impacts at the earliest stages of B-cell development and can be rescued by Bcl2, leading to the accumulation of early progenitors. Runx1-deficient progenitors represent a continuum of pro-B- to early pre-B cells that do not or very inefficiently progress to the late stage of pre-B-cell development, which is accompanied by down-regulation of Igll1, cell cycle attenuation, and functional production of LC-κ. This block is not due to a deficiency in either Igh or Igl-κ rearrangements or expression of key pre-BCR components, but rather more likely reflect a defect in the stage-specific regulatory circuits initiated by pre-BCR signaling.
Identification of critical Runx1 target genes at the early pre-B-cell stage
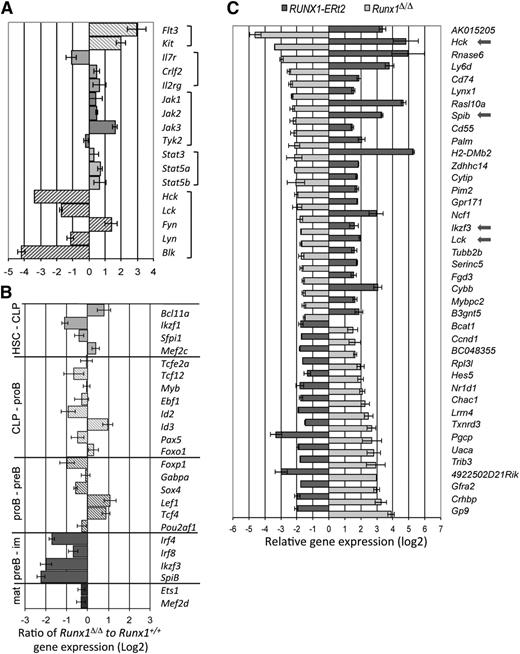
To identify the Runx1 target genes that specifically regulate pro-B-/pre-B-cell survival and development, gene expression profiles of wild-type and Runx1-deficient cells were interrogated. When stringent criteria for differential expression are used, 209 genes were found to be down-regulated and 378 genes up-regulated in Runx1-deficient pro-B/pre-B cells compared with controls. A functional annotation tool identified several functional gene clusters enriched for Runx1-deregulated genes (supplemental Table 2), primarily related to extra- and intracellular signaling (ie, membrane proteins and proteins with SH2 domains or that bind GTPase, respectively), but also enriched for genes in proapoptotic pathways. Included in this list was the proapoptotic Bok gene, whose expression level was increased by 8.8-fold (±2.4) in the Runx1-deficient B-cell progenitors. Up-regulation of this gene would sensitize the cells to apoptosis by disrupting the balance of proapoptotic and antiapoptotic Bcl2 proteins, in agreement with the ability of proapoptotic Bcl2 to partially rescue the Runx1-deficient phenotype. Receptor signaling is known to be a critical mediator of survival in B-cell development, but pivotal cytokine receptor or JAK-STAT effector genes were not notably down-regulated in the absence of Runx1 (Figure 5A). However, significantly reduced transcript levels of all genes encoding the 4 members of the Lyn subfamily of src family kinases (Lyn-SFK: Hck, Blk, Lck, and Lyn) were found, ranging from a 50% (for Lyn) to 90% reduction (for Blk) (Figure 5A). The gene expression pattern for most known B-cell transcription factors was only moderately altered in Runx1-deficient progenitors compared with controls (Figure 5B; supplemental Figure 6). However, three genes encoding transcription factors critical for pre-B-cell development (Irf4, SpiB, and Aiolos) showed greatly reduced expression levels in Runx1-deficient cells.
Potential target genes of Runx1 in B-cell development revealed by expression analysis. (A) Differentially regulated genes in pro-B/pre-B cells of Runx1-deficient mice known to encode receptor proteins or their signal effectors that are critical regulators of B-cell development. Qualitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis confirmed the levels for the Hck, Blk, and Lck genes. (B) Relative expression levels of genes encoding transcription factors from Runx1fl/flCd79ahCre/+ pro-B/pre-B cells were compared with that isolated from Runx1+/+Cd79ahCre/+ mice. Shown are genes encoding transcription factors that are critical for the development of multipotent, lymphoid-restricted, or specific B-cell developmental stages, as indicated. Qualitative RT-PCR analysis confirmed the levels of genes encoding key regulators. (C) Depicted is the relative expression ratio of 40 genes differentially regulated in Runx1-deficient pro-B/pre-B cells compared with controls and reciprocally regulated in a pro-B-cell line that expresses an inducible RUNX1 protein.
Potential target genes of Runx1 in B-cell development revealed by expression analysis. (A) Differentially regulated genes in pro-B/pre-B cells of Runx1-deficient mice known to encode receptor proteins or their signal effectors that are critical regulators of B-cell development. Qualitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis confirmed the levels for the Hck, Blk, and Lck genes. (B) Relative expression levels of genes encoding transcription factors from Runx1fl/flCd79ahCre/+ pro-B/pre-B cells were compared with that isolated from Runx1+/+Cd79ahCre/+ mice. Shown are genes encoding transcription factors that are critical for the development of multipotent, lymphoid-restricted, or specific B-cell developmental stages, as indicated. Qualitative RT-PCR analysis confirmed the levels of genes encoding key regulators. (C) Depicted is the relative expression ratio of 40 genes differentially regulated in Runx1-deficient pro-B/pre-B cells compared with controls and reciprocally regulated in a pro-B-cell line that expresses an inducible RUNX1 protein.
To investigate whether the identified genes were direct target genes of Runx1, we used a pro-B-cell line to conditionally express Runx1. Gene expression arrays were compared with results from differential expression analysis of Runx1-deficient pro-B/pre-B cells. Using a strict threefold change of normalized expression values, 40 gene loci were identified with reciprocal expression patterns between the 2 approaches (Figure 5C). In this short list of Runx1 target genes was the Ly6d gene, consistent with protein data presented in Figure 2D, but also the Lyn family src genes Lck and Hck. Lyn but not Blk was also up-regulated in the overexpression assay, but at levels below our threefold threshold. Finally, whereas Irf4 appears not to be a direct target of Runx1, both the Spib and Ikzf3 genes (encoding SpiB and Aiolos, respectively) were dramatically up-regulated on Runx1 activation.
Runx1 occupies enhancer regions of genes critical for pre-B-cell transition
To ascertain what genes identified in our microarray analysis are direct targets of Runx1, we assessed global Runx1 occupancy in pro-B cells by performing ChIP-Seq, identifying 6792 Runx1-occupied sites that mapped to 4327 gene loci (supplemental Table 3). To determine whether the set of Runx1-deregulated genes were preferentially found in the vicinity of Runx1 occupancy sites, we determined the closest Runx1 peak for all annotated gene transcripts and determined its location relative to the gene (Figure 6A). In 39% of the putative Runx1-target genes (n = 1436), a Runx1-bound site was found within −10 kb of the transcriptional start site to +10 kb of the transcriptional end site. In contrast, in a set of control genes not targeted by Runx1, but with similar expression levels in pro-B cells (n = 8703), less than 25% of the genes contained a Runx1-occupied region within these borders.
Genomewide analysis of Runx1 occupancy in pro-B cells confirms binding to enhancer regions and colocalization with SpiB/Pu.1 and Ebf1 binding sites. (A) Distribution of Runx1-bound regions in relation to potential Runx1 target genes demonstrates an enrichment of Runx1 occupancy within close proximity to the regulated genes. The nearest peak to genes deregulated in either gain- or loss-of-function expression or to control genes, exhibiting similar expression levels but whose expression remains constant, was determined. Identified peaks were classified based on their relative position to their associated gene as (1) intragenic (between or coincident with gene boundaries); (2) proximal (within 10 kb upstream or downstream); (3) distal (between 10 and 100 kb upstream or downstream); or (4) >100 kb from gene boundaries. (B) Sequence logos corresponding to enriched sequence elements identified by de novo motif analysis of Runx1-binding sites. The frequency of the enriched motif near Runx1 summits (±100 bp) is indicated (n = 500), as is the calculated E-value. (C) The frequency of Runx1 peaks with a summit mapping at the indicated distance from the summits of E2a, Pax5, or Ebf1 peaks or H3K4me marks in pro-B cells6,24 is shown. (D) Snapshot of relevant gene loci demonstrating association of Runx1-bound regions with E2a-binding sites and regions with mono- and trimethylated H3K4 mapped in pro-B cells6 and DNaseI hypersensitive sites mapped in B cells.50 The University of California - Santa Cruz Genome Browser was used to visualize binding patterns. Vertical rectangles mark regions with proposed enhancer (black) or promoter (green) functions.
Genomewide analysis of Runx1 occupancy in pro-B cells confirms binding to enhancer regions and colocalization with SpiB/Pu.1 and Ebf1 binding sites. (A) Distribution of Runx1-bound regions in relation to potential Runx1 target genes demonstrates an enrichment of Runx1 occupancy within close proximity to the regulated genes. The nearest peak to genes deregulated in either gain- or loss-of-function expression or to control genes, exhibiting similar expression levels but whose expression remains constant, was determined. Identified peaks were classified based on their relative position to their associated gene as (1) intragenic (between or coincident with gene boundaries); (2) proximal (within 10 kb upstream or downstream); (3) distal (between 10 and 100 kb upstream or downstream); or (4) >100 kb from gene boundaries. (B) Sequence logos corresponding to enriched sequence elements identified by de novo motif analysis of Runx1-binding sites. The frequency of the enriched motif near Runx1 summits (±100 bp) is indicated (n = 500), as is the calculated E-value. (C) The frequency of Runx1 peaks with a summit mapping at the indicated distance from the summits of E2a, Pax5, or Ebf1 peaks or H3K4me marks in pro-B cells6,24 is shown. (D) Snapshot of relevant gene loci demonstrating association of Runx1-bound regions with E2a-binding sites and regions with mono- and trimethylated H3K4 mapped in pro-B cells6 and DNaseI hypersensitive sites mapped in B cells.50 The University of California - Santa Cruz Genome Browser was used to visualize binding patterns. Vertical rectangles mark regions with proposed enhancer (black) or promoter (green) functions.
To determine what other B-cell transcription factors cooperate with Runx1 in B-cell gene regulation, sequences flanking the calculated Runx1-binding peaks were used to search for common de novo motifs within 100 bp of the peak summit. In 99.9% of all random peaks analyzed (n = 500), a motif was identified that matches the known consensus binding sequence for Runx1, validating the robustness of the analysis (Figure 6B). Furthermore, motifs containing the consensus sequence for either the SpiB/Pu.1 subfamily of ETS transcription factors or the Ebf1 transcription factor were found at frequencies of 20.1% and 4.2%, respectively, suggesting cotranscriptional regulation. Although no other motifs were identified with significant statistics by this de novo motif discovery approach, searches for known motifs identified several other consensus-binding sites, including those for the E2a, AP1, and CTCF transcription factors. Conserved spacing between the Runx1 summit and the identified motif in a subset of these occupied regions were highly significant and provide evidence that these transcription factors collaborate in Runx1 gene regulation (supplemental Table 4). We analyzed previously published E2a, Pax5, and Ebf1 occupancy sites6,24 and confirmed close proximity (<300 bp) to the summits of 16%, 12%, and 1% of the Runx1 peaks, respectively (Figure 6C). We also determined a correlation between histone marks and Runx1 occupancy; ∼25% of Runx1 peaks mapped within 1 kb of chromosome domains occupied in pro-B cells by histone H3 proteins with monomethylated lysine-4 (H3K4me1)6 (Figure 6C).
Closer scrutiny of Lck, Hck, and Blk gene loci, as well as Spib and Ikzf3, confirmed Runx1 occupancy (Figure 6D). Notably, each of these gene loci contained ≥1 Runx1-occupied site that was (1) coincident with the E2A binding site; (2) flanked by H3K4me1; and (3) coincided with a DNaseI hypersensitive site, suggestive of enhancer function. A similar pattern was observed for the Ly6d and Lyn loci (supplemental Figure 7). In addition, the Lck and Blk loci also contained Runx1-binding sites within proximal promoter regions (<3 kb upstream of the transcriptional start site) and were in close vicinity to H3 marked by trimethylated K4 (H3K4me3), frequently found at active promoter elements.
RUNX1-target genes IKZF3 and SPIB are differentially regulated during pre-B-cell transition and specifically dysregulated in t(12;21) ALL
The expression pattern of the identified Runx1 target genes was assessed during B-cell development. A striking 10- to 15-fold up-regulation of Spib, Ikzf3, and Hck was observed at the transition from pro-B- to pre-B cells, but Hck expression increased again by more than 4-fold from the pre-B to immature B stage (Figure 7A). In contrast, Lck expression levels remained relatively constant during development, as did that of Runx1. These results demonstrate the dynamic expression patterns of this Runx1-targeted gene set, which are linked to specific B-cell development stages. To gain insight into whether the deregulation of these potential target genes of Runx1 are also deregulated in ETV6/RUNX1-induced leukemia, we screened a large, public database for expression profiles in different subclasses of BCP-ALL using the Leukemia Gene Atlas website. BCP-ALL subgroups were defined by the presence of translocation involving t(1;19) (TCF3/PBX1); t(9;22) (BCR/ABL); t(12;21) (ETV6/RUNX1); or chormosome hyperdiploidy. Although expression levels of the Lyn-SFK genes were variably deregulated between these different subgroups, SPIB and IKZF3 showed significantly lower expression in t(12;21) samples (Figure 7B).
Pivotal Runx1 target genes during pre-B-cell progression and ALL induction. (A) RNA was isolated from B-cell progenitors isolated using the FACS scheme depicted in Figure 1E. To confirm the development stage of the isolated cell compartments, Igll gene transcript levels (encoding λ5 sLCs) and Vκ-Jκ rearranged transcripts of the Igl-κ (encoding LCs) locus were determined by qualitative reverse transcriptase-polymerase chain reaction. Subsequently, the relative gene expression levels of Lck, Blk, and Hck were determined, using Hprt transcript levels as control. (B) Plotted are the gene expression data for IKZF3 and SPIB for BCP-ALL patient samples [hyperdiploid (HD; n = 40), t(1;19) (E2A/PBX1; n = 36), t(12;21) (ETV6/RUNX1; n = 58), and t(9;22) (BCR/ABL, n = 122)]51 evaluated by the Leukemia Gene Atlas (www.leukemia-gene-atlas.org). Statistical significance was calculated with the Welch’s t test; ***P < .001; *P < .05. (C) Schematic model of key transcription events regulated by Runx1 during pre-B-cell progression. Green arrows denote interactions leading to up-regulation of gene transcription by Runx1 or products of the indicated gene, whereas yellow lines denote interactions leading to transcription repression of the indicated genes. Gray arrow denotes a possible feedback mechanism in which the product of the indicated gene may alter Runx1 activity by phosphorylation (P). ETV6-RUNX1 expression would repress SPIB and IKZF3 transcription, thereby blocking pre-B-cell progression, an important event in leukemogenesis.
Pivotal Runx1 target genes during pre-B-cell progression and ALL induction. (A) RNA was isolated from B-cell progenitors isolated using the FACS scheme depicted in Figure 1E. To confirm the development stage of the isolated cell compartments, Igll gene transcript levels (encoding λ5 sLCs) and Vκ-Jκ rearranged transcripts of the Igl-κ (encoding LCs) locus were determined by qualitative reverse transcriptase-polymerase chain reaction. Subsequently, the relative gene expression levels of Lck, Blk, and Hck were determined, using Hprt transcript levels as control. (B) Plotted are the gene expression data for IKZF3 and SPIB for BCP-ALL patient samples [hyperdiploid (HD; n = 40), t(1;19) (E2A/PBX1; n = 36), t(12;21) (ETV6/RUNX1; n = 58), and t(9;22) (BCR/ABL, n = 122)]51 evaluated by the Leukemia Gene Atlas (www.leukemia-gene-atlas.org). Statistical significance was calculated with the Welch’s t test; ***P < .001; *P < .05. (C) Schematic model of key transcription events regulated by Runx1 during pre-B-cell progression. Green arrows denote interactions leading to up-regulation of gene transcription by Runx1 or products of the indicated gene, whereas yellow lines denote interactions leading to transcription repression of the indicated genes. Gray arrow denotes a possible feedback mechanism in which the product of the indicated gene may alter Runx1 activity by phosphorylation (P). ETV6-RUNX1 expression would repress SPIB and IKZF3 transcription, thereby blocking pre-B-cell progression, an important event in leukemogenesis.
Discussion
The pivotal function of Runx1 in both myelopoiesis and B-cell lymphopoiesis is reflected in the frequency at which it is disrupted in both acute myelogenous leukemia and BCP-ALL. Indeed, disruption of the RUNX1 gene by t(12;21) is the most frequent event in pediatric BCP-ALL. Thus, the strict functional requirement of Runx1 during early stages of B-cell development and the Runx1-targeted genes identified in this study has not only significant implications for understanding the complex regulatory circuits that modulate B-cell development, but also for the function of the t(12;21) product ETV6/RUNX1 in initiating leukemogenesis.
Runx1 in early B-cell development
An important conclusion of our study is that Runx1 is not required for the specification of the B-cell compartment per se, but rather for the survival and the development of early B-cell progenitors. This is based on a number of observations. First, we found no changes in the absolute numbers of CLPs when Runx1 was inactivated in the HSC compartment, a finding in agreement with 2 of 3 previous studies.15,16,25 Second, although our analysis identified Ly6d as a Runx1 target and thus its expression cannot be used to identify CLP-Bs, we could show that CLPs from Runx1fl/fl Tg(vav-Cre) mice cloned at the same frequency under B-cell conditions as wild-type controls. Finally, the drastic reduction of pro-B/pre-B cells in Runx1-deficient mice could be reversed by introducing Tg(Eμ-bcl2).
How does the Runx1 transcription factor mediate survival and developmental signals in early B-cell progenitors? Although our results do not support an instructive role of Runx1 in B-cell specification, they are consistent with the hypothesis that Runx1 is instrumental in reinforcing a B-cell developmental program. IL7-mediated activation of Ebf1 is thought to be the essential step in B-cell specification,26,27 but an interplay of feedback loops, reciprocal activation, and cross-antagonism of several transcription factors has been postulated to be essential for stabilization of B-cell specification and commitment.6,7,28,29 What is the evidence that Runx1 is also a player in this network? First of all, analysis of Runx1 occupancy sites in pro-B cells showed coincident binding of Runx1 with E2a, Pax5, and Ebf1 binding sites. These results are concordant with previous reports showing an enrichment of Runx1 consensus sites in E2a and Pax5 occupied loci6,24 and functional synergy between Runx1 and Ebf1 or Pax5 in gene regulation.30-32 A recent report has implicated Runx1 as key regulator of Ebf1 gene expression and has shown that Ebf1 can rescue in vitro B-cell development of Runx1Δ/ΔCd79ahCre/+ progenitors.18 Although no significant Runx1-occupied sites were called near the Ebf1 gene in our analysis nor was deregulated Ebf1 expression in Runx1-deficient proB-cells observed, we can confirm the ability of Ebf1-mediated rescue of Runx1-deficient B cells in vitro (U.B. and C.S., unpublished results). We thus favor the hypothesis that the key function of Runx1 is reinforcing the complex transcriptional network initiated by Ebf1 during B-cell development. Increasing the life span (via Bcl2) of B cell–specified progenitors may allow alternative or parallel regulatory mechanisms to intervene, leading to the necessary transcription rates of key differentiation, proliferation, or survival genes that may be impaired in the absence of Runx1. Ebf1 has been shown to regulate key survival signal in early B-cell development via an IL7 receptor–independent pathway; however, Bcl2 is insufficient to rescue pro-B-cell survival in vitro.33 Thus, the survival pathway mediated by Runx1 may only partially overlap with that maintained by Ebf1. Interestingly, many Runx1 target genes encode membrane-bound or signaling proteins implicated in relaying extracellular signals to cells. In the absence of these interactions, Bok expression may be triggered, leading to the observed apoptosis.
Critical role of Runx1 in pre-B-cell development
Our second finding was the essential role of Runx1 for the transition to the late pre-B-cell stage of development, normally characterized by cell cycle attenuation, reduced levels of sLC, and increased rearrangement and expression of the Igl-κ.34,35 We identified 2 sets of Runx1 target genes that encode downstream effectors of the IL7 and pre-BCR signaling pathway and that likely contribute to the pre-B-cell development block.
The first set of critical Runx1 target genes is composed of all 4 Lyn-SFK members, all of which were deregulated in Runx1-deficient pro-B cells. Our results predict that the dynamic regulation of Lyn-SFK genes during B-cell development may be orchestrated by Runx1 and interaction partners such as ETS transcription factors.36 Interestingly, an important feedback mechanism may be provided by the ability of SFKs to directly regulate Runx1 activity.37 It is well established that Lyn-SFK are important mediators of IL7 and pre-BCR stimulation, but the exact mechanism is unknown.38 Lyn-SFKs have been linked to Stat5 activation,39 and IL7-STAT5 signaling pathways have been shown to be critical for repressing Igl-κ rearrangements at the pro-B stage,40,41 which could account for relatively high Igl-κ rearrangements observed in the Runx1-deficient pro-/pre-B cells. Furthermore, the critical role of Lyn-SFK in pre-BCR signaling has also been demonstrated in mice with either a triple knockout (Blk, Lyn, Fyn) or that are transgenic for a constitutively active form of Blk.42,43
The second set of target genes encodes transcription factors that are sharply up-regulated after pre-BCR signaling: specifically, the Ikaros family member Aiolos and the ETS-related factor SpiB. Low levels of the Ikzf3 gene product Aiolos, which is a critical negative modulator of Igll1 (encoding the λ5 subunit of sLC) and Myc gene expression,44,45 likely contribute to the high sLC transcript levels and the absence of cell cycle arrest observed in Runx1-deficient cells. Similarly, SpiB and its close homolog Pu.1 have also been implicated in pre-BCR signaling, although the critical target genes are not well defined.46,47 Significantly, SpiB and Runx1 likely share several target genes in this process, as evidenced by the high incidence (20%) of SpiB/Pu.1 consensus sites within Runx1-occupied loci. These results support a scenario in which Runx1 and SpiB constitute an interdependent feed-forward loop: transcriptional activation of Spib by Runx1 leads to the production of SpiB, which together with Runx1 activates (or inactivates) downstream genes necessary for pre-B-cell transition. A similar interplay between Runx1 and Pu.1 has recently been revealed in the myeloid compartment48 and thus may represent a conserved mechanism for driving differentiation.
Runx1 and BCP-ALL
In t(12;21) BCP-ALL, the ETV6/RUNX1 fusion protein is thought to antagonize the function of wild-type RUNX1.10,11 Our study is the first to identify a critical role of Runx1 in regulating the transition to a late pre-B stage, a block that is a defining feature of BCP-ALL. Two RUNX1 target genes (IKZF3 and SPIB) are regulators of this transition and are significantly down-regulated in t(12;21) BCP-ALL. Thus, targeting of ETV6/RUNX1 to these loci most likely inhibits their up-regulation during pre-B-cell development, impairing further development (Figure 7C). Paradoxically, our study also demonstrated that Runx1 is necessary for survival functions of early B-cell progenitors. This apparent contradiction, however, can be explained by the fact that many target genes are actually repressed and not activated by Runx1 and that ETV6/RUNX1 can be recruited to unique genes that may overcome this block.49 Nevertheless, it is interesting to speculate that the relative good response of t(12;21) BCP-ALL to chemotherapy is due to an inherent susceptibility to apoptosis incurred by inhibiting RUNX1 function.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the many scientists who provided valuable mouse strains and Y. Groner and D. Levanon (Weizmann Institute, Rehovot) for supplying the Runx1 antibody.
This work was funded by the Deutsche Krebshilfe e.V. and the Else Kröner-Fresenius-Stiftung. The Heinrich-Pette-Institute, Leibniz Institute for Experimental Virology, is supported by the Bundesministerium für Gesundheit and the Freie und Hansestadt Hamburg.
Authorship
Contribution: B.N., N.K., M.F., and K.B. designed and performed experiments, analyzed results, and helped write the manuscript; T.G., M.A., and A.G. evaluated ChIP-Seq data; U.B., U.M., S.R., and M.Z. performed experiments; F.B. developed the mouse strain and discussed results; and C.S. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carol Stocking, Heinrich-Pette-Institute, Leibniz Institute of Experimental Virology, Martinstrasse 52, 20251 Hamburg, Germany; e-mail: carol.stocking@hpi.uni-hamburg.de.

![Figure 2. Runx1 is critical for the survival of B cell–specified CLP progeny. (A) End point dilution of CLPs under B-cell differentiation conditions shows that Runx1-deficient CLPs form colonies at the same efficiency as Runx1+/+ CLPs. CLPs from either Runx1Δ/Δ [Runx1fl/fl-Tg(vav-Cre)] or Runx1+/+ [Runx1+/+-Tg(vav-Cre)] were sorted, and the indicated cell number was plated on OP9 stroma cells supplemented with cytokines. Positive wells were scored on day 7 by microscopic visualization. (B) CLP progeny from Runx1-deficient mice showed a high frequency of cell death and low proliferation rates. The number of wells exhibiting cell growth was determined at both days 7 and 12 in 2 independent limiting dilution assays. The difference in the number of positive clones is shown as the percentage of surviving clones for each genotype. To estimate the proliferation rate of CLP progeny, 200 CLPs were sorted on OP9 stroma, and the total number of viable cells was determined on day 7. Shown are the results from 4 independent wells seeded in 2 independent sorts. Runx1+/+, solid line; Runx1Δ/Δ, stippled line. (C) FACS analysis of CLP progeny demonstrates the predominant expression of B-cell (CD19+/B220+) but not myeloid (CD11b/Gr1) antigens. For Runx1+/+ CLPs, cells from single wells analyzed by FACS after 1 week; for Runx1Δ/Δ CLPs, cells from 6 positive wells were pooled before analysis. (D) FACS analysis for Ly6D expression levels on pro-B/pre-B cells (CD19+Lin2negB220med) isolated from either Runx1fl/fl Tg(vav-Cre) or Runx1+/+ Tg(vav-Cre) mice.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/3/10.1182_blood-2013-01-480244/4/m_413f2.jpeg?Expires=1767723493&Signature=YxdcIfSaujtyMMqvJf52HOwCDHm19-D4EjVFWeq5r4Qoolye5f5Kmbbjhis1Ls~~dHRIymE9CSd5TZgPh6pV7NLaIv3MX5IhqkLoTqA07kU8z2DDny92DAn-~K9~wQuNxg6dlB7Xbcifj5uENjEvCjJg--yezd25FSp0Rq6IBz6fR2yyFvAtc4A4xxZ2gU8auPSXMdRUFvCKWiKQIX1rj29k664Nqi3GVHQU7eJ8GmSf0b3pOemJrfqYxhMHte3wad8KFU3GiwQYClE9eCJxLUq1b~hJKT3H3WR3uqUC4h8ykO0gXmXJvQmRRX8u74CUvQxunhlohxUsU2aaoe41AA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 7. Pivotal Runx1 target genes during pre-B-cell progression and ALL induction. (A) RNA was isolated from B-cell progenitors isolated using the FACS scheme depicted in Figure 1E. To confirm the development stage of the isolated cell compartments, Igll gene transcript levels (encoding λ5 sLCs) and Vκ-Jκ rearranged transcripts of the Igl-κ (encoding LCs) locus were determined by qualitative reverse transcriptase-polymerase chain reaction. Subsequently, the relative gene expression levels of Lck, Blk, and Hck were determined, using Hprt transcript levels as control. (B) Plotted are the gene expression data for IKZF3 and SPIB for BCP-ALL patient samples [hyperdiploid (HD; n = 40), t(1;19) (E2A/PBX1; n = 36), t(12;21) (ETV6/RUNX1; n = 58), and t(9;22) (BCR/ABL, n = 122)]51 evaluated by the Leukemia Gene Atlas (www.leukemia-gene-atlas.org). Statistical significance was calculated with the Welch’s t test; ***P < .001; *P < .05. (C) Schematic model of key transcription events regulated by Runx1 during pre-B-cell progression. Green arrows denote interactions leading to up-regulation of gene transcription by Runx1 or products of the indicated gene, whereas yellow lines denote interactions leading to transcription repression of the indicated genes. Gray arrow denotes a possible feedback mechanism in which the product of the indicated gene may alter Runx1 activity by phosphorylation (P). ETV6-RUNX1 expression would repress SPIB and IKZF3 transcription, thereby blocking pre-B-cell progression, an important event in leukemogenesis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/3/10.1182_blood-2013-01-480244/4/m_413f7.jpeg?Expires=1767723493&Signature=iZtyDHvfucyfK5LAxW~dfG3oRA9pOlVQ0pgzeFooTsG83bqvQCtDcSCrCaVRPSloh~Ckpi~I60LYRu-IQDkXWCh9fMjpDU2UdaHG7gdwUZ6Q2dO5GgP~dDpZBUQeV2KAfWOSfpih4POH0fW1Y1T7rwcG52ENYAyDjq2fllhbpPFN5Ro~eaauTU1cICtk8Ro2xqfYm61bhM3FVRGKtkoCcKH~iMyQm82GDOQYM~F~xpF84G0FaIonEnk0sz6rykDzgiGgOu33Mo-5xv2ckj8lxFfjujeetks3Pog75QsfRBKJ5V9ObLB78L0Q6trGzlVr2TWdKX44oHX-g0nDHxaTQg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal