Key Points
Using genome-wide association study, we found the first replicated genetic association with acute chest syndrome in sickle cell disease patients.
The locus identified includes COMMD7, a gene highly expressed in the lung that interacts with NFκB to control inflammatory responses.
Abstract
Patients with sickle cell disease (SCD) present with a wide range of clinical complications. Understanding this clinical heterogeneity offers the prospects to tailor the right treatments to the right patients and also guide the development of novel therapies. Several environmental (eg, nutrition) and nonenvironmental (eg, fetal hemoglobin levels, α-thalassemia status) factors are known to modify SCD severity. To find new genetic modifiers of SCD severity, we performed a gene-centric association study in 1514 African American participants from the Cooperative Study of Sickle Cell Disease (CSSCD) for acute chest syndrome (ACS) and painful crisis. From the initial results, we selected 36 single nucleotide polymorphism (SNPs) and genotyped them for replication in 387 independent patients from the CSSCD, 318 SCD patients recruited at Georgia Health Sciences University, and 449 patients from the Duke SCD cohort. In the combined analysis, an association between ACS and rs6141803 reached array-wide significance (P = 4.1 × 10−7). This SNP is located 8.2 kilobases upstream of COMMD7, a gene highly expressed in the lung that interacts with nuclear factor-κB signaling. Our results provide new leads to gaining a better understanding of clinical variability in SCD, a “simple” monogenic disease.
Introduction
Sickle cell disease (SCD) is among the most common Mendelian diseases worldwide and is particularly prevalent in regions where malaria is endemic.1,2 SCD is caused by mutations in the β-globin gene that encodes 1 of the subunits of the oxygen carrier hemoglobin and is characterized by a wide spectrum of disease-specific complications. In the deoxygenated state, sickle hemoglobin forms long polymers that alter erythrocyte shape and flexibility, thus increasing hemolysis and the adherence between sickled red blood cells and the endothelium.3,4 Although hemolysis and cell adherence are the main causes of complications in SCD, little is known about the additional environmental and nonenvironmental factors that may modify disease severity and therefore explain the remarkable clinical heterogeneity observed in this otherwise simple monogenic disease.
Environmental variables such as nutrition and sufficient hydration are linked to clinical heterogeneity in SCD.4,5 Two nonenvironmental factors, high fetal hemoglobin (HbF) levels and concomitant α-thalassemia, also correlate with reduced morbidity and mortality in SCD.6 The identification of additional disease-severity modifiers may yield novel insights into SCD pathophysiology. A genetic association study may improve understanding of SCD clinical heterogeneity because it attempts to correlate DNA sequence variants with SCD-specific complications or relevant clinical variables (eg, HbF). Over the last 2 decades, several genetic associations in SCD have been published, but the results are questionable because of small sample size and lack of replication (reviewed in ref. 7). There are, however, 2 notable exceptions: robust associations between (1) 3 loci (BCL11A, HBS1L-MYB, and β-globin) and HbF levels8,9 and (2) the bilirubin levels–associated UGT1A1 locus and gallstones.10,11
To find novel genetic modifiers of SCD, we performed a gene-centric association study in 1514 participants from the Cooperative Study of Sickle Cell Disease (CSSCD) for acute chest syndrome (ACS) and painful crisis. For genotyping, we used the ITMAT-Broad-CARe (IBC) array, which covers genetic variation at ∼2100 genes important for heart, lung, and blood diseases.12 Although the CSSCD is one of the largest existing SCD cohorts, our discovery power is modest to detect variants of small effect on phenotypic variation. For this reason, we genotyped for replication 36 variants that reached P < 1.0 × 10−4 in the CSSCD discovery sample in the DNA of 387 independent SCD patients from the CSSCD. We also genotyped markers in 318 SCD patients recruited at Georgia Health Sciences University and 449 patients from Duke University. Our analysis identified 1 single nucleotide polymorphism (SNP; rs6141803) near COMMD7 that reached array-wide significance, defined as P < 2.0 × 10−6 after accounting for the number of independent SNPs present on the IBC array.13 Overall, our findings prioritize DNA sequence variants and genes for future genetic and functional follow-up experiments in order to better grasp patient-to-patient clinical variability in SCD.
Methods
Ethics statement
Informed consent was obtained for all participants in accordance with the Declaration of Helsinki. The Candidate-gene Association Resource (CARe) Study is approved by the ethics committees of the participating studies and of the Massachusetts Institute of Technology. This project was also reviewed and approved by the Montreal Heart Institute Ethics Committee, the Duke Institutional Review Board, and the different recruiting centers.
Samples and genotyping
The CSSCD is described in detail elsewhere.14 Briefly, the CSSCD was a multicenter prospective study of the natural history of SCD; participant enrollment into phase 1 of the CSSCD began in 1978. Participant entry ended in 1981 for all patients older than age 6 months; however, infants continued to be enrolled until 1988. Both mild and hospital-based SCD patients were recruited. A total of 4085 participants, mostly African Americans and ranging in age from newborns to adults, were enrolled in phase 1 from 23 centers across the United States. Data collection for phase 1 of the CSSCD ended in 1988 (see https://biolincc.nhlbi.nih.gov/studies/csscd/ for more information on the study design).
In the CSSCD, painful crisis and ACS events were defined as previously described by the CSSCD investigators and as reported in the CSSCD phase 1 clinical database.15-17 Briefly, a painful crisis episode was defined as an occurrence of pain lasting ≥2 hours in the extremities, back, abdomen, chest, or head that could not be explained by a mechanism other than SCD. Pain episodes within 14 days were treated as a single episode. An episode of ACS occurred when a participant developed a new infiltrate on chest radiograph and/or had a perfusion defect detected on a lung radioisotope scan. Painful crisis and ACS were analyzed as rates by dividing the number of events by the number of patient-years.
A total of 318 patients from the Adult Sickle Cell Clinic of Georgia Health Sciences University (GHSU) Sickle Cell Center were included in this study as a validation cohort; patient age ranged from 20 to 74 years and 169 women and 149 men were included in the cohort. In the GHSU cohort, ACS was defined as a new pulmonary infiltrate involving more than 1 lung segment with fever, chest pain, and hypoxia. This excludes patients who present with classic lobar pneumonia, although the distinction may not be 100%. The Duke SCD cohort included 449 adult patients (199 men and 250 women) and used the following questions to define ACS: Have you ever experienced acute chest syndrome or pneumonia requiring hospitalization? and to define painful crisis: In the past 12 months, have you had painful episodes requiring hospitalization?. Demographics for the 3 SCD cohorts used in this study are summarized in Table 1.
Description of the SCD cohorts
| Phenotype . | CSSCD discovery (N = 1514) . | CSSCD replication (N = 387) . | Georgia Health Sciences University (N = 318) . | Duke (N = 449) . | Statistical power (0.5% and 1% variance explained) . |
|---|---|---|---|---|---|
| Males/females | 740/764 | 177/210 | 149/169 | 199/250 | — |
| Age, y | 14.2 ± 11.9 | 11.2 ± 12.5 | NA | 33.7 ± 12.1 | — |
| Follow-up, y | 6.6 ± 1.6 | 6.1 ± 2.2 | NA | NA | — |
| α-thalassemia (N) | 27.1% (410) | 25.3% (98) | NA | NA | — |
| Acute chest syndrome (events/patient-year or affected/nonaffected) | 0.12 ± 0.24 | 0.16 ± 0.48 | 52/262 (4 missing) | 337/112 | 13%, 50% |
| Painful crisis (events/patient-year or any/none) | 0.80 ± 1.45 | 0.65 ± 1.17 | NA | 259/171 | 13%, 50% |
| Phenotype . | CSSCD discovery (N = 1514) . | CSSCD replication (N = 387) . | Georgia Health Sciences University (N = 318) . | Duke (N = 449) . | Statistical power (0.5% and 1% variance explained) . |
|---|---|---|---|---|---|
| Males/females | 740/764 | 177/210 | 149/169 | 199/250 | — |
| Age, y | 14.2 ± 11.9 | 11.2 ± 12.5 | NA | 33.7 ± 12.1 | — |
| Follow-up, y | 6.6 ± 1.6 | 6.1 ± 2.2 | NA | NA | — |
| α-thalassemia (N) | 27.1% (410) | 25.3% (98) | NA | NA | — |
| Acute chest syndrome (events/patient-year or affected/nonaffected) | 0.12 ± 0.24 | 0.16 ± 0.48 | 52/262 (4 missing) | 337/112 | 13%, 50% |
| Painful crisis (events/patient-year or any/none) | 0.80 ± 1.45 | 0.65 ± 1.17 | NA | 259/171 | 13%, 50% |
Analyses in this study were restricted to sickle cell anemia (HbSS) or HbSβ0 patients, with or without α-thalassemia. Means ± standard deviations are provided. Statistical power for the CSSCD discovery cohort was calculated using the following assumptions: minor allele frequency = 25%, effect size = 0.5% or 1% of the phenotypic variance explained, and α = 1 × 10−4.
NA; not available.
DNA genotyping on the Illumina IBC array was carried out at the Broad Institute as part of the National Heart, Lung and Blood Institute (NHLBI) CARe Project. The IBC array interrogates genotypes at ∼50 000 SNPs and captures genetic variation at ∼2100 genes relevant for heart, lung, and blood diseases.12 Data quality control and genotype imputation were performed as previously described.13 Imputation was performed using MACH 1.0.16.18 MACH requires phased reference haplotypes to perform imputation. For the African American CSSCD participants, a combined Northern European (CEU) + Western Africans (YRI) reference panel was created using HapMap phase 2 data.19 This panel includes SNPs segregating in both CEU and YRI, as well as SNPs segregating in 1 panel and monomorphic and nonmissing in the other. Imputation was performed in 2 steps: For the first step, 300 individuals were randomly extracted to generate recombination and error rate estimates. In the second step, these rates were used to impute all individuals across the entire reference panel. Imputation results were filtered at an rsq_hat threshold ≥0.6 and a minor allele frequency (MAF) threshold ≥1%. For imputed markers with strong statistical association, we directly genotyped an overlapping set of CSSCD DNA samples (N = 777) and found high concordance with the imputation results (mean Pearson correlation coefficient = 0.87; range, 0.65-1.0). The final CSSCD discovery dataset included 1514 DNA samples with a genotyping success rate >99.8% (47 092 genotyped SNPs and 190 551 imputed SNPs). Genotyping in the CSSCD and GHSU replication cohorts was performed using the mass spectrometry–based MassArray iPLEX platform from Sequenom, removing SNPs and DNA samples with a genotyping success rate <95% and <90%, respectively. The concordance rate, which was estimated from replicates, was >99.7%. Genotyping and quality-control filters for the Duke SCD cohort were described elsewhere.20 We used identity-by-descent methods to identify patients who overlap in the CSSCD and Duke cohorts; these patients (N = 23) were excluded from the analysis of the Duke cohort.
Statistical analysis
We performed our discovery experiment in the CSSCD,14,21 a large longitudinal study with hundreds of clinical variables available. Despite this wealth of phenotypic data, we focus initially on only 2 quantitative measures, ACS and painful crisis rates, for two reasons. First, statistical power to find genetic associations with quantitative phenotypes was higher than for dichotomous traits (despite remaining relatively modest), even in the large CSSCD. For instance, we estimated that our study design had 50% power to find an association between a quantitative trait and a SNP under the following assumptions: minor allele frequency = 25%, variance explained = 1%, and α = 1 × 10−4 (Table 1). In comparison, we only had 8% power to find an association with stroke (prevalence = 7%) under the same assumptions for a variant with an odds ratio (OR) = 1.5. Second, because the size of our replication cohorts is small, for low prevalence complications, it is likely that there would be too few affected SCD patients to robustly validate genetic associations observed in the CSSCD discovery cohort.
In the CSSCD discovery cohort, we tested associations between 237 643 genotyped (0, 1, 2) or imputed (0.0-2.0) common SNPs (MAF ≥1%) and phenotypes using Poisson regression (correction for overdispersion) for painful crisis and ACS rates.17 We implemented the analysis using custom scripts in the R 2.10.0 statistical package (www.r-project.org/). We used sex, age at baseline, and the first 10 principal components as covariates. Analyses were stratified based on α-thalassemia status, and association results were combined by inverse variance meta-analyses.22 Analyses of global and local ancestry were performed using, respectively, the EIGENSOFT and HAPMIX software with their default parameters.23-25
Analyses in the CSSCD replication cohort were performed as for the CSSCD discovery cohort, except that we used α-thalassemia status as a covariate because of the small sample size of the cohort and we did not have access to principal components. In the GHSU SCD cohort, age at baseline and painful crisis information were not available. We analyzed the association between genotypes and ACS (dichotomous) using logistic regression in PLINK26 and sex and year of birth as covariates. For the Duke SCD cohort, logistic regression was used to determine the effect of genotype on a binary definition of ACS using PLINK.26 To examine the impact of number of hospitalizations for painful crisis episodes, logistic regression was employed using SAS version 9.2 (SAS Systems, Cary, NC). In an attempt to reduce any population substructure that may exist, principal component analysis was performed using EIGENSOFT.23 All models were adjusted for sex, age, and the first 2 principal components.
To analyze the effect of heme on the expression of COMMD7 in pulmonary endothelial cells, we accessed the relevant gene expression dataset on the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi accession number = GSE25014).27 We performed the analyses separately for the pulmonary microvascular endothelial cells and the pulmonary artery endothelial cells using the GEO2R analytical module and default parameters (http://www.ncbi.nlm.nih.gov/geo/info/geo2r.html). We corrected P values using the Benjamini–Hochberg false-discovery rate method.
Results
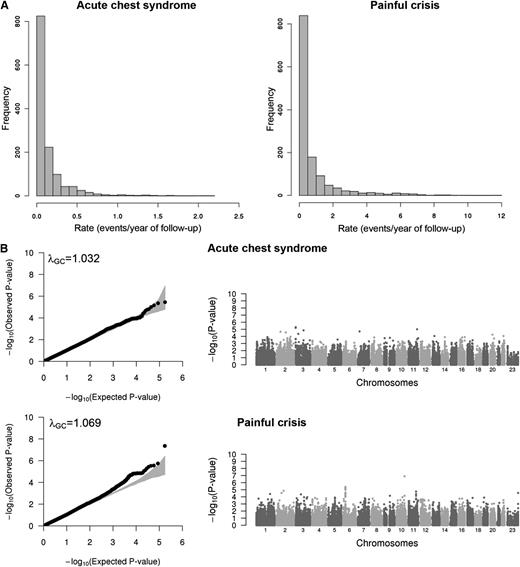
Several complications are observed in SCD and are linked to the quality of life and life expectancy of patients with this hemoglobinopathy.15,16,28-31 Our goal in this study was to identify genetic variants associated with 2 of these complications, ACS and painful crisis, in order to better understand clinical heterogeneity in SCD. We modeled and analyzed ACS and painful crisis rates as previously described using Poisson regression.15-17 Although for ACS the distribution of the observed test statistics does not show major departure from the expected null distribution (λGC = 1.032), there was a slight inflation for the painful crisis association results (λGC = 1.069; Figure 1).32 For this reason, we corrected both ACS and painful crisis association results using the genomic control (GC) approach. However, overall the limited inflation that was observed indicates that our analysis was appropriate and accounted for the main possible confounders. To declare statistical significance on the IBC array, we selected a threshold of α = 2 × 10−6 that is sufficient to account for the number of independent tests performed.13 Using this criterion, we identified a single locus that met array-wide significance: an association between rs11817401 in the SORCS1 gene and painful crisis rate (P = 1.2 × 10−7; Figure 1 and Table 2).
Association results for acute chest syndrome and painful crisis. (A) Histograms showing the distribution of the acute chest syndrome (left) and painful crisis (right) rates in the CSSCD participants analyzed in this study (N = 1514). (B) Quantile–quantile (QQ; left column) and Manhattan (right column) plots of association results for acute chest syndrome (top) and painful crisis (bottom) analyzed in participants from the CSSCD. In the QQ plots, the gray area corresponds to the 90% CI. Results are corrected using the genomic control method.
Association results for acute chest syndrome and painful crisis. (A) Histograms showing the distribution of the acute chest syndrome (left) and painful crisis (right) rates in the CSSCD participants analyzed in this study (N = 1514). (B) Quantile–quantile (QQ; left column) and Manhattan (right column) plots of association results for acute chest syndrome (top) and painful crisis (bottom) analyzed in participants from the CSSCD. In the QQ plots, the gray area corresponds to the 90% CI. Results are corrected using the genomic control method.
Painful crisis association results
| SNP . | Chromosome (position) . | Effect allele/other allele . | Effect allele frequency . | CSSCD discovery . | CSSCD replication . | Combined CSSCD . | Duke . | Gene (annotation) . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) . | P value . | β (SE) . | P value . | β (SE) . | P value . | OR (95% CI) . | P value . | |||||
| rs12720497 | 1 (185097043) | T/C | 0.10 | 0.529 (0.125) | 3.9 × 10−5 | 0.336 (0.194) | .08 | 0.470 (0.107) | 1.2 × 10−5 | NA | NA | PLA2G4A (intron) |
| rs540006 | 2 (70636930) | T/C | 0.98 | −0.723 (0.167) | 2.9 × 10−5 | −0.310 (0.338) | .36 | −0.637 (0.154) | 3.5 × 10−5 | 0.94 [0.08, 10.65] | .96 | Intergenic |
| rs3917296 | 2 (102151265) | A/G | 0.98 | −0.773 (0.172) | 1.5 × 10−5 | 1.393 (1.013) | .17 | −0.708 (0.176) | 5.5 × 10−5 | 8.5 × 10−8 [-inf, 0] | 1.00 | IL1R1 (intron) |
| rs324035 | 3 (115351544) | A/C | 0.74 | −0.303 (0.076) | 1.2 × 10−4 | 0.061 (0.170) | .72 | −0.239 (0.071) | 8.3 × 10−4 | NA | NA | DRD3 (intron) |
| rs10513478 | 3 (156723359) | T/C | 0.02 | 0.836 (0.209) | 1.1 × 10−4 | −1.779 (2.450) | .47 | 0.816 (0.216) | 1.5 × 10−4 | 0.59 [0.13, 2.72] | .50 | PLCH1 (intron) |
| rs11732673 | 4 (2652853) | A/G | 0.77 | −0.338 (0.089) | 2.2 × 10−4 | −0.374 (0.154) | .02 | −0.347 (0.079) | 9.9 × 10−6 | NA | NA | FAM193A (intron) |
| rs6858735 | 4 (37592622) | T/C | 0.12 | 0.375 (0.095) | 1.4 × 10−4 | −0.165 (0.218) | .45 | 0.283 (0.090) | 1.6 × 10−3 | NA | NA | TBC1D1 (intron) |
| rs13113915 | 4 (187702958) | A/G | 0.40 | 0.261 (0.067) | 1.6 × 10−4 | 0.245 (0.131) | .06 | 0.258 (0.061) | 2.5 × 10−5 | NA | NA | MTNR1A (intron) |
| rs10942625 | 5 (90446448) | A/G | 0.25 | −0.362 (0.091) | 1.2 × 10−4 | −0.329 (0.178) | .07 | −0.355 (0.083) | 2.0 × 10−5 | 0.93 [0.68, 1.28] | .66 | GPR98 (intron) |
| rs1851426 | 7 (99220872) | A/G | 0.68 | −0.290 (0.074) | 1.6 × 10−4 | 0.269 (0.145) | .07 | −0.168 (0.068) | 0.01 | NA | NA | CYP3A4 (intergenic) |
| rs10107231 | 8 (18098691) | T/C | 0.83 | −0.375 (0.096) | 1.7 × 10−4 | −0.105 (0.203) | .61 | −0.322 (0.089) | 3.1 × 10−4 | NA | NA | NAT1 (intron) |
| rs7034457 | 9 (77807918) | A/G | 0.90 | −0.418 (0.102) | 7.1 × 10−5 | 0.045 (0.249) | .86 | −0.348 (0.097) | 3.3 × 10−4 | NA | NA | PCSK5 (intron) |
| rs7899453 | 10 (79469965) | A/C | 0.22 | 0.317 (0.079) | 1.1 × 10−4 | 0.179 (0.149) | .23 | 0.285 (0.072) | 7.3 × 10−5 | NA | NA | RPS24 (missense) |
| rs11817401 | 10 (108823893) | T/C | 0.08 | 0.602 (0.110) | 1.2 × 10−7 | −0.176 (0.273) | .52 | 0.487 (0.105) | 3.4 × 10−6 | NA | NA | SORCS1 (intron) |
| rs17101814 | 11 (103367886) | T/C | 0.09 | 0.444 (0.100) | 1.8 × 10−5 | 0.128 (0.196) | .51 | 0.375 (0.091) | 4.1 × 10−5 | 0.82 [0.50, 1.35] | .44 | PDGFD (intron) |
| rs9933611 | 16 (52331386) | A/G | 0.93 | −0.449 (0.116) | 1.8 × 10−4 | −0.245 (0.272) | .37 | −0.415 (0.110) | 1.5 × 10−4 | NA | NA | FTO (intron) |
| rs445683 | 17 (71913117) | T/C | 0.67 | 0.311 (0.079) | 1.5 × 10−4 | 0.089 (0.142) | .53 | 0.256 (0.071) | 3.2 × 10−4 | 0.91 [0.68, 1.23] | .56 | UBE2O (intron) |
| rs7507634 | 19 (10297562) | T/C | 0.02 | 0.899 (0.226) | 1.2 × 10−4 | −0.821 (0.674) | .22 | 0.714 (0.221) | 1.2 × 10−3 | NA | NA | RAVER1 (intron) |
| rs2872817 | 23 (153211612) | A/G | 0.70 | −0.249 (0.058) | 3.0 × 10−5 | −0.158 (0.121) | .19 | −0.231 (0.053) | 1.5 × 10−5 | NA | NA | TKTL1 (3′UTR) |
| SNP . | Chromosome (position) . | Effect allele/other allele . | Effect allele frequency . | CSSCD discovery . | CSSCD replication . | Combined CSSCD . | Duke . | Gene (annotation) . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) . | P value . | β (SE) . | P value . | β (SE) . | P value . | OR (95% CI) . | P value . | |||||
| rs12720497 | 1 (185097043) | T/C | 0.10 | 0.529 (0.125) | 3.9 × 10−5 | 0.336 (0.194) | .08 | 0.470 (0.107) | 1.2 × 10−5 | NA | NA | PLA2G4A (intron) |
| rs540006 | 2 (70636930) | T/C | 0.98 | −0.723 (0.167) | 2.9 × 10−5 | −0.310 (0.338) | .36 | −0.637 (0.154) | 3.5 × 10−5 | 0.94 [0.08, 10.65] | .96 | Intergenic |
| rs3917296 | 2 (102151265) | A/G | 0.98 | −0.773 (0.172) | 1.5 × 10−5 | 1.393 (1.013) | .17 | −0.708 (0.176) | 5.5 × 10−5 | 8.5 × 10−8 [-inf, 0] | 1.00 | IL1R1 (intron) |
| rs324035 | 3 (115351544) | A/C | 0.74 | −0.303 (0.076) | 1.2 × 10−4 | 0.061 (0.170) | .72 | −0.239 (0.071) | 8.3 × 10−4 | NA | NA | DRD3 (intron) |
| rs10513478 | 3 (156723359) | T/C | 0.02 | 0.836 (0.209) | 1.1 × 10−4 | −1.779 (2.450) | .47 | 0.816 (0.216) | 1.5 × 10−4 | 0.59 [0.13, 2.72] | .50 | PLCH1 (intron) |
| rs11732673 | 4 (2652853) | A/G | 0.77 | −0.338 (0.089) | 2.2 × 10−4 | −0.374 (0.154) | .02 | −0.347 (0.079) | 9.9 × 10−6 | NA | NA | FAM193A (intron) |
| rs6858735 | 4 (37592622) | T/C | 0.12 | 0.375 (0.095) | 1.4 × 10−4 | −0.165 (0.218) | .45 | 0.283 (0.090) | 1.6 × 10−3 | NA | NA | TBC1D1 (intron) |
| rs13113915 | 4 (187702958) | A/G | 0.40 | 0.261 (0.067) | 1.6 × 10−4 | 0.245 (0.131) | .06 | 0.258 (0.061) | 2.5 × 10−5 | NA | NA | MTNR1A (intron) |
| rs10942625 | 5 (90446448) | A/G | 0.25 | −0.362 (0.091) | 1.2 × 10−4 | −0.329 (0.178) | .07 | −0.355 (0.083) | 2.0 × 10−5 | 0.93 [0.68, 1.28] | .66 | GPR98 (intron) |
| rs1851426 | 7 (99220872) | A/G | 0.68 | −0.290 (0.074) | 1.6 × 10−4 | 0.269 (0.145) | .07 | −0.168 (0.068) | 0.01 | NA | NA | CYP3A4 (intergenic) |
| rs10107231 | 8 (18098691) | T/C | 0.83 | −0.375 (0.096) | 1.7 × 10−4 | −0.105 (0.203) | .61 | −0.322 (0.089) | 3.1 × 10−4 | NA | NA | NAT1 (intron) |
| rs7034457 | 9 (77807918) | A/G | 0.90 | −0.418 (0.102) | 7.1 × 10−5 | 0.045 (0.249) | .86 | −0.348 (0.097) | 3.3 × 10−4 | NA | NA | PCSK5 (intron) |
| rs7899453 | 10 (79469965) | A/C | 0.22 | 0.317 (0.079) | 1.1 × 10−4 | 0.179 (0.149) | .23 | 0.285 (0.072) | 7.3 × 10−5 | NA | NA | RPS24 (missense) |
| rs11817401 | 10 (108823893) | T/C | 0.08 | 0.602 (0.110) | 1.2 × 10−7 | −0.176 (0.273) | .52 | 0.487 (0.105) | 3.4 × 10−6 | NA | NA | SORCS1 (intron) |
| rs17101814 | 11 (103367886) | T/C | 0.09 | 0.444 (0.100) | 1.8 × 10−5 | 0.128 (0.196) | .51 | 0.375 (0.091) | 4.1 × 10−5 | 0.82 [0.50, 1.35] | .44 | PDGFD (intron) |
| rs9933611 | 16 (52331386) | A/G | 0.93 | −0.449 (0.116) | 1.8 × 10−4 | −0.245 (0.272) | .37 | −0.415 (0.110) | 1.5 × 10−4 | NA | NA | FTO (intron) |
| rs445683 | 17 (71913117) | T/C | 0.67 | 0.311 (0.079) | 1.5 × 10−4 | 0.089 (0.142) | .53 | 0.256 (0.071) | 3.2 × 10−4 | 0.91 [0.68, 1.23] | .56 | UBE2O (intron) |
| rs7507634 | 19 (10297562) | T/C | 0.02 | 0.899 (0.226) | 1.2 × 10−4 | −0.821 (0.674) | .22 | 0.714 (0.221) | 1.2 × 10−3 | NA | NA | RAVER1 (intron) |
| rs2872817 | 23 (153211612) | A/G | 0.70 | −0.249 (0.058) | 3.0 × 10−5 | −0.158 (0.121) | .19 | −0.231 (0.053) | 1.5 × 10−5 | NA | NA | TKTL1 (3′UTR) |
We genotyped 19 SNPs with P < 1 × 10−4 in the CSSCD Discovery dataset (before GC correction) in an independent subset of the CSSCD. We combined results from the discovery and replication cohorts using an inverse variance meta-analysis approach. Only 1 SNP (rs11732673, in bold) replicated at nominal significance, but no SNPs reached array-wide significance (P < 2 × 10−6) in the combined analysis. We also report results for the analysis of painful crisis as a dichotomous trait in the Duke cohort. P values for the CSSCD Discovery cohort are G -corrected. Genomic positions are on NCBI build 37.1.
NA, not available; SE, standard error.
To confirm this association and also identify additional loci that appeared promising but did not reach statistical significance in the CSSCD discovery cohort, we selected all SNPs with discovery P < 1 × 10−4 (before GC correction; 19 SNPs for painful crisis and 17 SNPs for ACS) and genotyped them in 387 independent CSSCD participants. Replication and combined association results are presented in Table 2 and Table 3 for painful crisis and ACS, respectively. The association between SORCS1-rs11817401 and painful crisis did not replicate (replication P = .52, opposite direction of effect). Overall, we replicated in this small CSSCD cohort 2 associations at nominal level (P < .05): an association between FAM193A-rs11732673 and painful crisis (replication P = .02, combined P = 9.9 × 10−6) and an association between rs6141803 and ACS (replication P = .003, combined P = 5.2 × 10−7), the latter reaching array-wide significance when combining the CSSCD discovery and replication results (Table 3). FAM193A encodes a protein with no clear biological functions. The rs6141803 SNP is located between the COMMD7 and DNMT3B genes on chromosome 20. DNMT3B encodes a DNA methyltransferase, which is important for development, and COMMD7, a gene highly expressed in the lung, codes for an adaptor protein that interacts with subunits of the nuclear factor (NF)-κB complex.33 Importantly, treating human pulmonary endothelial cells with free heme, a model that recapitulates some of the cellular responses observed when ACS is induced in a SCD mouse model,34 significantly modulates the expression of COMMD7 (differential expression in pulmonary microvascular endothelial cells [P = 5 × 10−4] and in pulmonary artery endothelial cells [P = 3 × 10−5]).27 This result, together with a role in NF-κB signaling and inflammation, adds additional evidence supporting a role for COMMD7 in ACS.
Acute chest syndrome association results
| SNP . | Chromosome (position) . | Effect allele/other allele . | Effect allele frequency . | CSSCD discovery . | CSSCD replication . | Combined CSSCD . | Georgia Health Sciences University . | Duke . | Gene (annotation) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) . | P value . | β (SE) . | P value . | β (SE) . | P value . | OR (95% CI) . | P value . | OR [95% CI] . | P value . | |||||
| rs10399947 | 1 (149128584) | A/G | 0.57 | −0.319 (0.082) | 1.2 × 10−4 | 0.339 (0.156) | .03 | −0.174 (0.073) | .02 | 0.98 [0.64, 1.50] | .93 | 0.89 [0.66, 1.21] | .47 | Intergenic |
| rs34661029 | 2 (61002492) | C/G | 0.98 | −0.822 (0.191) | 2.3 × 10−5 | −0.103 (0.450) | .82 | −0.709 (0.178) | 7.1 × 10−5 | 0.87 [0.24, 3.19] | .83 | 0.75 [0.16, 3.61] | .72 | REL (missense) |
| rs17749316 | 2 (191548954) | C/G | 0.99 | −0.891 (0.228) | 1.2 × 10−4 | 0.518 (1.013) | .61 | −0.821 (0.226) | 2.7 × 10−4 | 3.2 × 108 [0, inf] | 1.00 | 5.5 × 108 [0, inf] | 1.00 | STAT1 (intron) |
| rs13021001 | 2 (128125889) | A/G | 0.96 | −0.631 (0.149) | 3.0 × 10−5 | 0.450 (0.482) | .35 | −0.534 (0.144) | 2.1 × 10−4 | 0.55 [0.14, 2.19] | .39 | 0.84 [0.3, 2.34] | .73 | GPR17 (3′UTR) |
| rs6778854 | 3 (563448) | A/G | 0.61 | 0.374 (0.081) | 5.0 × 10−6 | 0.057 (0.161) | .72 | 0.309 (0.073) | 2.4 × 10−5 | 0.98 [0.64, 1.50] | .92 | NA | NA | Intergenic |
| rs13315133 | 3 (38624908) | A/C | 0.79 | −0.339 (0.082) | 4.3 × 10−5 | 0.031 (0.197) | .88 | −0.284 (0.076) | 2.0 × 10−4 | 0.90 [0.53, 1.54] | .71 | NA | NA | SCN5A (intron) |
| rs3910551 | 3 (106659325) | C/G | 0.02 | 0.793 (0.180) | 1.4 × 10−5 | −0.333 (0.532) | .53 | 0.675 (0.172) | 9.1 × 10−5 | 1.13 [0.38, 3.32] | .83 | 0.25 [0.08, 0.81] | .02 | ALCAM (intron) |
| rs5030094 | 3 (187943852) | T/C | 0.99 | −0.801 (0.221) | 3.5 × 10−4 | −0.727 (0.424) | .09 | −0.785 (0.198) | 7.5 × 10−5 | 0.54 [0.14, 2,15] | .38 | 0.73 [0.11, 4.71] | .74 | KNG1 (intron) |
| rs10478813 | 5 (127859164) | C/G | 0.16 | 0.400 (0.109) | 2.8 × 10−4 | −0.002 (0.240) | .99 | 0.330 (0.100) | 9.9 × 10−4 | 0.81 [0.38, 1.75] | .60 | NA | NA | FBN2 (intron) |
| rs5576 | 7 (24297876) | A/G | 0.97 | −0.655 (0.152) | 2.1 × 10−5 | 0.078 (0.477) | .87 | −0.586 (0.146) | 6.3 × 10−5 | 1.66 [0.37, 7.55] | .51 | 1.05 [0.37, 3.03] | .92 | NPY (3′UTR) |
| rs1176758 | 11 (113275565) | A/G | 0.92 | −0.528 (0.117) | 9.7 × 10−6 | 0.110 (0.275) | .69 | −0.427 (0.109) | 9.7 × 10−5 | 1.61 [0.49, 5.33] | .43 | 0.68 [0.33, 1.38] | .29 | Intergenic |
| rs6309 | 13 (46368601) | A/G | 0.04 | 0.631 (0.159) | 9.0 × 10−5 | −0.391 (0.477) | .41 | 0.526 (0.153) | 5.6 × 10−4 | 1.43 [0.45, 4.56] | .55 | 0.68 [0.34, 1.34] | .26 | HTR2A (intron) |
| rs9927848 | 16 (23740572) | A/C | 0.44 | −0.354 (0.099) | 4.3 × 10−4 | 0.317 (0.147) | .03 | −0.141 (0.083) | .09 | 1.14 [0.74, 1.75] | .55 | 1.03 [0.75, 1.40] | .87 | Intergenic |
| rs12447481 | 16 (52306470) | T/G | 0.98 | −0.794 (0.206) | 1.5 × 10−4 | −0.830 (0.450) | .07 | −0.800 (0.190) | 2.5 × 10−5 | 3.3 × 108 [0, inf] | 1 | 1.70 [0.31, 9.43] | .54 | FTO (intron) |
| rs6141803 | 20 (30804017) | T/C | 0.98 | −0.701 (0.171) | 5.4 × 10−5 | −0.797 (0.266) | .003 | −0.730 (0.145) | 5.2 × 10−7 | 0.41 [0.07, 2.34] | .32 | 0.16 [0.02, 1.23] | .08 | Intergenic |
| rs17728960 | 20 (49563123) | T/C | 0.98 | −0.789 (0.208) | 1.9 × 10−4 | −0.053 (0.354) | .88 | −0.595 (0.182) | 1.1 × 10−3 | 0.35 [0.12, 0.99] | .05 | 0.28 [0.04, 2.19] | .22 | NFATC2 (intron) |
| rs16998437 | 22 (36418341) | T/G | 0.92 | −0.483 (0.1216) | 9.3 × 10−5 | −0.061 (0.233) | .79 | 0.390 (0.109) | 3.5 × 10−4 | NA | NA | 1.07 [0.52, 2.18] | .86 | NOL12 (3′UTR) |
| SNP . | Chromosome (position) . | Effect allele/other allele . | Effect allele frequency . | CSSCD discovery . | CSSCD replication . | Combined CSSCD . | Georgia Health Sciences University . | Duke . | Gene (annotation) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) . | P value . | β (SE) . | P value . | β (SE) . | P value . | OR (95% CI) . | P value . | OR [95% CI] . | P value . | |||||
| rs10399947 | 1 (149128584) | A/G | 0.57 | −0.319 (0.082) | 1.2 × 10−4 | 0.339 (0.156) | .03 | −0.174 (0.073) | .02 | 0.98 [0.64, 1.50] | .93 | 0.89 [0.66, 1.21] | .47 | Intergenic |
| rs34661029 | 2 (61002492) | C/G | 0.98 | −0.822 (0.191) | 2.3 × 10−5 | −0.103 (0.450) | .82 | −0.709 (0.178) | 7.1 × 10−5 | 0.87 [0.24, 3.19] | .83 | 0.75 [0.16, 3.61] | .72 | REL (missense) |
| rs17749316 | 2 (191548954) | C/G | 0.99 | −0.891 (0.228) | 1.2 × 10−4 | 0.518 (1.013) | .61 | −0.821 (0.226) | 2.7 × 10−4 | 3.2 × 108 [0, inf] | 1.00 | 5.5 × 108 [0, inf] | 1.00 | STAT1 (intron) |
| rs13021001 | 2 (128125889) | A/G | 0.96 | −0.631 (0.149) | 3.0 × 10−5 | 0.450 (0.482) | .35 | −0.534 (0.144) | 2.1 × 10−4 | 0.55 [0.14, 2.19] | .39 | 0.84 [0.3, 2.34] | .73 | GPR17 (3′UTR) |
| rs6778854 | 3 (563448) | A/G | 0.61 | 0.374 (0.081) | 5.0 × 10−6 | 0.057 (0.161) | .72 | 0.309 (0.073) | 2.4 × 10−5 | 0.98 [0.64, 1.50] | .92 | NA | NA | Intergenic |
| rs13315133 | 3 (38624908) | A/C | 0.79 | −0.339 (0.082) | 4.3 × 10−5 | 0.031 (0.197) | .88 | −0.284 (0.076) | 2.0 × 10−4 | 0.90 [0.53, 1.54] | .71 | NA | NA | SCN5A (intron) |
| rs3910551 | 3 (106659325) | C/G | 0.02 | 0.793 (0.180) | 1.4 × 10−5 | −0.333 (0.532) | .53 | 0.675 (0.172) | 9.1 × 10−5 | 1.13 [0.38, 3.32] | .83 | 0.25 [0.08, 0.81] | .02 | ALCAM (intron) |
| rs5030094 | 3 (187943852) | T/C | 0.99 | −0.801 (0.221) | 3.5 × 10−4 | −0.727 (0.424) | .09 | −0.785 (0.198) | 7.5 × 10−5 | 0.54 [0.14, 2,15] | .38 | 0.73 [0.11, 4.71] | .74 | KNG1 (intron) |
| rs10478813 | 5 (127859164) | C/G | 0.16 | 0.400 (0.109) | 2.8 × 10−4 | −0.002 (0.240) | .99 | 0.330 (0.100) | 9.9 × 10−4 | 0.81 [0.38, 1.75] | .60 | NA | NA | FBN2 (intron) |
| rs5576 | 7 (24297876) | A/G | 0.97 | −0.655 (0.152) | 2.1 × 10−5 | 0.078 (0.477) | .87 | −0.586 (0.146) | 6.3 × 10−5 | 1.66 [0.37, 7.55] | .51 | 1.05 [0.37, 3.03] | .92 | NPY (3′UTR) |
| rs1176758 | 11 (113275565) | A/G | 0.92 | −0.528 (0.117) | 9.7 × 10−6 | 0.110 (0.275) | .69 | −0.427 (0.109) | 9.7 × 10−5 | 1.61 [0.49, 5.33] | .43 | 0.68 [0.33, 1.38] | .29 | Intergenic |
| rs6309 | 13 (46368601) | A/G | 0.04 | 0.631 (0.159) | 9.0 × 10−5 | −0.391 (0.477) | .41 | 0.526 (0.153) | 5.6 × 10−4 | 1.43 [0.45, 4.56] | .55 | 0.68 [0.34, 1.34] | .26 | HTR2A (intron) |
| rs9927848 | 16 (23740572) | A/C | 0.44 | −0.354 (0.099) | 4.3 × 10−4 | 0.317 (0.147) | .03 | −0.141 (0.083) | .09 | 1.14 [0.74, 1.75] | .55 | 1.03 [0.75, 1.40] | .87 | Intergenic |
| rs12447481 | 16 (52306470) | T/G | 0.98 | −0.794 (0.206) | 1.5 × 10−4 | −0.830 (0.450) | .07 | −0.800 (0.190) | 2.5 × 10−5 | 3.3 × 108 [0, inf] | 1 | 1.70 [0.31, 9.43] | .54 | FTO (intron) |
| rs6141803 | 20 (30804017) | T/C | 0.98 | −0.701 (0.171) | 5.4 × 10−5 | −0.797 (0.266) | .003 | −0.730 (0.145) | 5.2 × 10−7 | 0.41 [0.07, 2.34] | .32 | 0.16 [0.02, 1.23] | .08 | Intergenic |
| rs17728960 | 20 (49563123) | T/C | 0.98 | −0.789 (0.208) | 1.9 × 10−4 | −0.053 (0.354) | .88 | −0.595 (0.182) | 1.1 × 10−3 | 0.35 [0.12, 0.99] | .05 | 0.28 [0.04, 2.19] | .22 | NFATC2 (intron) |
| rs16998437 | 22 (36418341) | T/G | 0.92 | −0.483 (0.1216) | 9.3 × 10−5 | −0.061 (0.233) | .79 | 0.390 (0.109) | 3.5 × 10−4 | NA | NA | 1.07 [0.52, 2.18] | .86 | NOL12 (3′UTR) |
We genotyped 17 SNPs with P < 1 × 10−4 in the CSSCD Discovery dataset (before GC correction) in an independent subset of the CSSCD. We combined results from the discovery and replication CSSCD cohorts using an inverse variance meta-analysis approach. Only 1 SNP (rs6141803, in bold) replicated at nominal significance and reached array-wide significance (P < 2 × 10−6) in the combined analysis. We also report results for the analysis of acute chest syndrome as a dichotomous trait in the GHSU and Duke cohorts. For rs6141803, the association was not significant but trended in the right direction. P values for the CSSCD Discovery cohort are GC corrected. Genomic positions are on NCBI build 37.1.
NA, not available; SE, standard error.
Our group had access to 2 additional SCD replication cohorts: 318 patients recruited at GHSU and 449 SCD patients from Duke University (Table 1). Painful crisis information was not available for the GHSU cohort and only available as categories for the Duke cohort. ACS information was available for both cohorts but only in the form of a binary presence/absence phenotype. In many situations, dichotomizing a quantitative trait can lead to substantial loss in statistical power as individuals with 1 or several ACS events are all labeled as affected.35 For replication in the GSHU SCD cohort, we genotyped the top 17 SNPs associated with ACS (Table 3). A single variant, rs17728960 in the NFATC2 gene, was nominally significant (P = .05), but the combined association result was not significant. The association between ACS and COMMD7-rs6141803 was not significant (P = .32) but trended in the right direction (OR = 0.41; Table 3). Genome-wide genotype data were available for the Duke SCD cohort. After quality-control steps and genotype imputation (see Materials and Methods section), 6 painful crisis and 14 ACS SNPs were available for association testing. The association between ACS and rs6141803 near COMMD7 in the Duke cohort showed a consistent direction of effect (OR = 0.16, P = .08; Table 3). When we combine at the P value level results from the CSSCD discovery, CSSCD replication, and GHSU and Duke cohorts using a Z-score method weighted based on sample size, the association between ACS and rs6141803 is array-wide significant (weighted P = 4.1 × 10−7).
We noted that COMMD7-rs6141803 is an ancestry informative marker: the C allele has a frequency of 17% and 0% in the HapMap individuals of Northern European (CEU) and African (YRI) ancestry, respectively. This observation raises the possibility that the association between ACS and COMMD7-rs6141803 is a false-positive result owing to admixture. However, this is unlikely because the ACS rate is not correlated with the first principal component, which captures European vs African admixture (Spearman ρ = −0.0188, P = .47), and we used the first 10 principal components in our analysis to account for global admixture. Although the association between ACS rate and genotypes at rs6141803 is not spurious because of admixture, we tried to use local ancestry to fine-map the causal variant. We inferred local European vs African ancestry at the locus and used this estimate as a covariate in our regression model.24 The strength of the genetic association between ACS and rs6141803 was reduced when controlling for local ancestry but remained significant (P = 5.4 × 10−5 and P = .001 without and with local ancestry as covariate), suggesting that rs6141803 is unlikely to be the causal variant at the locus. rs6141803 is intergenic and not in linkage disequilibrium (LD) with any nonsynonymous DNA sequence variants identified by the 1000 Genomes Project.36 Interestingly, however, it is in weak LD (r2∼0.2–0.3) with cis-eQTL SNPs associated with COMMD7 transcript levels in human liver37 and monocytes.38
Discussion
Painful crisis and ACS are, respectively, the first and second most frequent causes of hospital admissions in patients with SCD.39 Although they share common causes (eg, vaso-occlusion), it is also clear that some of the triggering factors are different (eg, the role of infections in ACS). We performed one of the largest genetic association experiments to date in order to identify DNA sequence variants that modify SCD clinical severity through these 2 measures of morbidity.
Our experimental design identified a single SNP, rs6141803, that reached array-wide significance (P = 5.2 × 10−7 in the CSSCD discovery + replication, P = 4.1 × 10−7 if we add the GHSU and Duke SCD samples). Analyzing ACS as a dichotomous phenotype in the GHSU and Duke cohorts, as we did in this study, could account for a loss of statistical power. One additional difference between the CSSCD and the other 2 cohorts is the age of the enrolled patients. The GHSU and Duke cohorts are essentially adult cohorts, whereas the CSSCD includes a large number of children (Table 1). Age is a known predictor of ACS events16 and, for this reason, was included as a covariate in our statistical model. We performed a sensitivity analysis in the CSSCD discovery cohort that clearly shows that the association signal between ACS and rs6141803 is observed most often in children: the association was strong in patients recruited before they were aged 5 years (N = 335, P = 4 × 10−6) but not significant in older patients (N = 978, P = .80). Thus, this age effect might explain why we could replicate this association in the CSSCD replication cohort (which also includes children) but not in the adult GHSU and Duke cohorts. It is also possible that this association is a false-positive report. Despite allele frequency differences between the ancestral populations at rs6141803, we performed analyses that suggest that admixture is unlikely to confound this result. Additional replication attempts in large SCD cohorts with quantitative measures of ACS are needed before drawing a final conclusion on the ACS-rs6141803 genetic association.
rs6141803 is an intergenic SNP located between DNMT3B and COMMD7. DNMT3B encodes for a DNA methyltransferase that is involved in maintenance DNA methylation. Mutations in DNMT3B cause immunodeficiency-centromeric instability-facial anomalies syndrome-1 (ICF-1; MIM #242860), a very rare syndrome that has not been linked to lung-related complications. COMMD7 encodes a poorly known protein that contains a copper metabolism gene MURR1 (COMM) domain. The gene is abundantly expressed in the lung33 and is overexpressed in hepatocellular carcinoma.40 The knockdown of COMMD7 using short-hairpin RNA increases apoptosis and cell cycle arrest, in part, by interfering with NF-κB signaling.33,40 NF-κB is a master regulator of acute inflammation; upon stimulation, it transcriptionally activates interleukins, interferon, tumor necrosis factor-α, and adhesion molecules. The expression of COMMD7 in pulmonary endothelial cells is also affected upon heme treatment27 ; this is promising as free heme can induce ACS in a SCD mouse model.34 Although more work is needed to clarify the possible role of COMMD7 in ACS, these are potentially interesting observations given the importance of inflammation and free radical production in aggravating ACS episodes.41
A recent large candidate gene study in 942 SCD children identified an association between a microsatellite in the heme oxygenase-1 (HMOX1) promoter and ACS: longer alleles were associated with increased rate of hospitalization for ACS.42 Results from this initial study were not replicated in an independent cohort. Therefore, we queried our own results to test if HMOX1 SNPs were associated with ACS rate in the CSSCD. Although the HMOX1 gene was targeted for genotyping on the IBC array (28 genotyped or imputed nearby SNPs), the microsatellite was not directly tested. Of the HMOX1 SNPs that are accessible on the IBC array, 1 SNP in the 3′ untranslated region of HMOX1 is associated with ACS at nominal significance (rs12160039; P = .02). We would need to directly genotype the promoter microsatellite to determine if this SNP captures the association signal with ACS through LD. Similarly, although an intronic sequence repeat polymorphism in the NOS1 gene has been proposed to influence ACS risk,43,44 none of the NOS1 SNPs tested in our study showed significant association results with ACS.
The second most interesting association identified in our experiment is between painful crisis rate and rs12720497, an intronic SNP in the PLA2G4A gene. The association is not array-wide significant, but the directions of effect are consistent between the discovery and replication CSSCD panels (replication P = .08, combined P = 1.2 × 10−5; Table 2). PLA2G4A encodes a cytosolic phospholipase A2, an enzyme implicated in the production of proinflammatory molecules (prostaglandins and leukotrienes) that has previously been implicated in increased sensitivity to pain (hyperalgesia) in humans.45,46 Enzymes in the phospholipase A2 family can be divided into 4 groups (cytosolic, secreted, calcium-independent, and lipoprotein-associated), and high levels of secreted phospholipase A2 have been suggested to be predictive of future ACS events.47-49 The link between cytosolic and secreted phospholipase A2 and their role in SCD complications is intriguing, especially because severe ACS often occurs in the course of vaso-occlusive painful crisis. In our data, however, rs12720497 in PLA2G4A (coding for cytosolic phospholipase A2) is not associated with ACS rate (P = .67).
We performed our discovery search in 1514 participants from the CSSCD whose DNA was genotyped on gene-centric genotyping arrays.12 With the caveat that this genotyping platform only captures genetic variation at a subset (∼10%) of the predicted human genes, we did not identify loci with moderate to strong effect on phenotype, which is consistent with most reported genome-wide association study results.50 Our results highlight promising variants for further replication in independent SCD cohorts and biologically plausible candidate genes (eg, COMMD7, PLA2G4A) to test functionally, for instance, in SCD mouse models. They are also indicative of the importance of combining genome-wide association study results through meta-analyses between SCD cohorts to gain sufficient statistical power to identify genetic associations of weak phenotypic effect.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the contribution of Mélissa Beaudoin for DNA genotyping and Cameron D. Palmer for genotype imputation and thank all the patients who contributed to this study.
CSSCD is supported in part by the National Institutes of Health, National Heart, Lung, and Blood Institute (N01-HB-47110). CARe is supported by the National Heart, Lung, and Blood Institute (HHSN268200625226C). A full listing of the grants and contracts that have supported CARe is provided at http://www.nhlbi.nih.gov/resources/geneticsgenomics/programs/care.htm. The work in the Lettre Laboratory is supported by a Innovation in Clinical Research Award grant from the Doris Duke Charitable Foundation (2009089), the Canada Research Chair Program, the Canadian Institute of Health Research (123382), and the Fonds de Recherche Santé Québec. The work at Duke University was funded in part by the National Heart, Lung, and Blood Institute (RO1 HL079915 and RC2-HL101212).
Authorship
Contribution: G.G., S.C., G.L., and G.J.P. conceived and designed the experiment; G.G., S.C., M.E.G., N.J., K.S., and G.L. performed experiments; G.G., S.C., M.E.G., N.J., M.P., D.P., K.S., A.G., A.E.A.-K., M.J.T., A.K., G.L., and G.J.P. analyzed the results; and G.G. and G.L. wrote the manuscript with contributions from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guillaume Lettre, Montreal Heart Institute, 5000 Belanger St, Montreal, Quebec, Canada, H1T 1C8; e-mail: guillaume.lettre@umontreal.ca; and George J. Papanicolaou, National Heart, Lung, and Blood Institute, 6701 Rockledge Dr, Bethesda, MD 20892; e-mail: gjp@mail.nih.gov.
References
Author notes
G.L. and G.J.P codirected the study.