Key Points
DS TMD shows no DNA rearrangements and a low rate of mutations other than GATA1.
DS AMKL always has rearrangements and mutations in genes known for leukemic progression; affected pathways share upregulation of MYC.
Abstract
Some neonates with Down syndrome (DS) are diagnosed with self-regressing transient myeloproliferative disorder (TMD), and 20% to 30% of those progress to acute megakaryoblastic leukemia (AMKL). We performed exome sequencing in 7 TMD/AMKL cases and copy-number analysis in these and 10 additional cases. All TMD/AMKL samples contained GATA1 mutations. No exome-sequenced TMD/AMKL sample had other recurrently mutated genes. However, 2 of 5 TMD cases, and all AMKL cases, showed mutations/deletions other than GATA1, in genes proven as transformation drivers in non-DS leukemia (EZH2, APC, FLT3, JAK1, PARK2-PACRG, EXT1, DLEC1, and SMC3). One patient at the TMD stage revealed 2 clonal expansions with different GATA1 mutations, of which 1 clone had an additional driver mutation. Interestingly, it was the other clone that gave rise to AMKL after accumulating mutations in 7 other genes. Data suggest that GATA1 mutations alone are sufficient for clonal expansions, and additional driver mutations at the TMD stage do not necessarily predict AMKL progression. Later in infancy, leukemic progression requires “third-hit driver” mutations/somatic copy-number alterations found in non-DS leukemias. Putative driver mutations affecting WNT (wingless-related integration site), JAK-STAT (Janus kinase/signal transducer and activator of transcription), or MAPK/PI3K (mitogen-activated kinase/phosphatidylinositol-3 kinase) pathways were found in all cases, aberrant activation of which converges on overexpression of MYC.
Introduction
Children with Down syndrome (DS) have a 10- to 50-fold higher incidence of leukemias than euploid children, including most types of acute myeloid leukemia (AML) and acute lymphocytic leukemia (ALL).1-3 Acute megakaryoblastic leukemia (AMKL), which is preceded by transient myeloproliferative disorder (TMD), has a 500-fold increased incidence relative to euploid children.4 DS TMD/AMKL is a unique condition of DS that enables the study of stages of leukemogenesis, beginning in the fetus, as precancerous hyperproliferation that manifests itself as TMD in the first weeks postnatally and thereafter regresses spontaneously to complete remission.5,6 In 20% to 30% of cases of regressed DS TMD, the dormant clonal TMD is thought to accumulate additional changes and, 1 to 4 years later, cause AMKL,5 in 70% of cases preceded by a myelodysplastic syndrome (MDS)-like stage.7 The exact nature of changes driving the progression of this malignancy are only partially understood. Trisomy 21 (T21) and in utero–acquired truncating mutations in the transcription factor GATA1 are always observed in cell expansions of DS TMD and DS AMKL.5,8,9 These mutations eliminate the full-length GATA1 protein and leave only the shorter, truncated form called GATA1s. These typical GATA1 mutations have never been observed in cells lacking T21. However, as T21 in the absence of GATA1 mutations can cause rampant hyperproliferative megakaryopoiesis leading to perinatal death,10 and as the presence or type of GATA1 mutations does not predict the outcome of TMD (spontaneous regression or progression to AMKL),5,9 it remains unclear whether GATA1 mutations represent the first initiating event sufficient for the onset of TMD in T21 cells, or whether other acquired changes precede or accompany it, such as mutations in JAK2, JAK3, TP53, and FLT35,11,12 that have been observed together with GATA1 mutations, at various stages of this disease.
It has been suggested that GATA1 controls proliferation of hematopoietic progenitors through repression of a MYC transcription factor.13,14 It has been shown in megakaryocyte progenitors that GATA1s fails to properly downregulate MYC, which results in substantially elevated (≤10-fold) levels of MYC expression.13,15 Nevertheless, despite substantial efforts in recent years, the nature of affected pathways that drive the progression of DS TMD to DS AMKL, and their association with the loss of full-length GATA1 protein, remain elusive.6 To address this question, we have analyzed mutational profiles of DS TMDs and DS AMKLs using exome sequencing, as well as somatic copy-number alterations (SCNAs) restricted to DS AMKL samples using genome-wide single nucleotide polymorphism (SNP) microarrays. This study reveals driver mutations and SCNAs common to other types of non-DS leukemia that potentially trigger progression of TMD to AMKL. Our data also suggest that T21 and the mutation in GATA1 are sufficient events for DS TMD. The analysis of the spectrum of likely “third-hit driver” mutations suggests that their converging downstream effect may be partly mediated by overexpression of MYC.
Materials and methods
Patient samples
Patient samples consisted of surplus clinical or archived clinical material collected by the tissue bank of the Italian National Association for Pediatric Haematology-Oncology. In accordance with the Declaration of Helsinki, informed written consent was obtained by the tissue bank for all subjects. Samples were processed and stored in the tissue bank at the Blizard Institute, which is licensed for tissue storage and monitored by the United Kingdom Human Tissue Authority.
Exome sequencing
The overall methodology was as previously described.16,17 DNA was extracted from the frozen samples using the QIAamp DNA Mini Kit (Qiagen), exome capture was conducted using the SureSelect Human Exon v3 50Mb (Agilent Technologies) kit, and sequencing was conducted on an Illumina HiSeq2000 instrument with paired-end 105-nt reads. Burrows–Wheeler Aligner (BWA) software was used to align the sequence reads to the human reference genome National Center for Biotechnology Information build (GRCh37/hg19). SAMtools was used to remove polymerase chain reaction duplicates and call single-nucleotide variants (SNVs). Detection of small insertions and deletions (smINDEL) was conducted with Pindel 0.2.2 software. The search for somatic mutations was restricted to the regions that were covered at least 10-fold. The average sequencing coverage for the target region was >100× (supplemental Table 1, available on the Blood Web site).
The remission samples for patients 15 and 16 were considered germline controls; however, no paired normal tissue for the other patients was available. The initial list of SNVs was filtered against the common (>5%) germline polymorphisms present in build 135 of the dbSNP and 1000 Genomes databases. SNVs present in the normal tissue sample from the same patient at a frequency of >2% were also filtered out. Qscore 500 for indel calling and 100 for SNV calling were selected as thresholds. All the variants above those thresholds and that had at least 1 read with the mutation on the forward and reverse strands were subjected to validation with Sanger sequencing. This revealed false-positive rates of 22% for indels and <5% for SNVs. For both SNVs and smINDELs, we focused on the mutations that map to the protein coding sequences and splice sites, as the untranslated exonic regions are less well covered by commercially available exome capture kits. All SNVs and indels were subjected to validation by Sanger sequencing.
Genome-wide SNP microarray analysis
In all cases (10 DS TMDs, 7 DS AMKLs, and 2 remission controls of DS AMKLs) the Genome-Wide Human SNP Array 6.0 (Affymetrix) was used to screen for SCNAs. Genome-wide SNP-array hybridization was performed with 500 ng genomic DNA following the manufacturer’s instructions. Genome-wide copy number analysis was performed using the Affymetrix Genotyping Console version 4.0, the Affymetrix Genotyping Console Browser version 1.10.12, and a reference set of 47 in-house control DNAs isolated from blood of healthy donors, none of whom presented with any type of leukemia.
Results
We performed exome sequencing of 10 samples from 7 ML-DS patients, of which 5 samples were DS TMD, 3 were DS AMKL (including 1 paired TMD-AMKL sample), and 2 were complete remission controls of the other 2 DS AMKLs (Table 1; supplemental Table 2). In addition, we performed a high-resolution analysis for SCNAs on 10 DS TMD and 7 DS AMKL samples (including the exome-sequenced series) using Affymetrix 6.0 arrays that combine 1.9 million SNP and copy number probes genome-wide. For patient 4, both TMD and AMKL stages of the disease were exome-sequenced (TMD4→AMKL4). The samples and experiments are summarized in Table 1.
Overview of DS TMD and DS AMKL samples analyzed by exome sequencing and SNP microarray
| Patient . | Diagnosis . | Chr21 . | GATA1 sequence . | SNP microarray . | SCNA gain . | SCNA loss . | Exome sequence . | Putative third-hit driver event? . |
|---|---|---|---|---|---|---|---|---|
| TMD1 | TMD | Trisomy | p.S12Fs | Yes | No | |||
| TMD2 | TMD | Trisomy | p.50_61ins | Yes | No | |||
| TMD3 | TMD | Trisomy | p.F34Fs; P.51_56ins | Yes | Yes | No | ||
| TMD4 [= AMKL4] | TMD | Trisomy | p.58_64ins; p.71_74del | Yes | Yes | Yes (PIK3C2A) | ||
| TMD5 | TMD | Trisomy | p.V77Fs | Yes | No | |||
| TMD8 | TMD | Trisomy | p.G31Fs | Yes | No | |||
| TMD10 | TMD | Trisomy | p.E13× | Yes | Yes | No | ||
| TMD11 | TMD | Trisomy | p.G7Fs | Yes | No | |||
| TMD12 | TMD | Trisomy | splicing 5′ exon 2 | Yes | Yes | Yes (FLT3) | ||
| TMD14 | TMD | Trisomy | P.V74I (exon2 3′ splice site) | Yes | Yes | No | ||
| AMKL4 [= TMD4] | AML-M7 | Trisomy | p.71_74del | Yes | Chr5 1.3-3.7, 87-122 | Yes | Yes (EZH2, SMC3, DHX29, APC) | |
| AMKL15 | AML-M7 | Tetrasomy | p.Y62Fs (dupl) | Yes | Chr14 tetrasomy 19.5-106.3 | Chr8 113.1-119.3 | Yes | Yes (DLEC1, EXT1) |
| AMKL15rem | Remission | Trisomy | wt | Yes | Yes | No | ||
| AMKL16 | AML-M7 | Trisomy | p.52_58ins | Yes | Chr7 q-arm | Chr3 60.2-60.9, 153.4-153.6, 176.2-176.8, Chr6 161.9-162.9, Chr7 p-arm | Yes | Yes (JAK1, POLE, PARK2-PACRG) |
| AMKL16rem | Remission | Trisomy | wt | Yes | Yes | No | ||
| AMKL20 | AML-M7 | Trisomy | p.P50Fs | Yes | Chr1 174-247, Chr15 62-99.5 | Chr7 0-10.1 | (SCNAs seen in leukemias) | |
| AMKL21 | AML-M7 | Trisomy | p.F33Fs | Yes | Chr1 200-247 | Chr17 0-7.7, 75.9-77.3 | (SCNAs seen in leukemias) | |
| AMKL24 | AML-M7 | Trisomy | p.P50Fs | Yes | Chr1 222-247 | (SCNAs seen in leukemias) | ||
| AMKL25 | AML-M7 | Trisomy | p.T52Fs (dupl) | Yes | t(X;16)(q26;q23) (associated with multiple myeloma) |
| Patient . | Diagnosis . | Chr21 . | GATA1 sequence . | SNP microarray . | SCNA gain . | SCNA loss . | Exome sequence . | Putative third-hit driver event? . |
|---|---|---|---|---|---|---|---|---|
| TMD1 | TMD | Trisomy | p.S12Fs | Yes | No | |||
| TMD2 | TMD | Trisomy | p.50_61ins | Yes | No | |||
| TMD3 | TMD | Trisomy | p.F34Fs; P.51_56ins | Yes | Yes | No | ||
| TMD4 [= AMKL4] | TMD | Trisomy | p.58_64ins; p.71_74del | Yes | Yes | Yes (PIK3C2A) | ||
| TMD5 | TMD | Trisomy | p.V77Fs | Yes | No | |||
| TMD8 | TMD | Trisomy | p.G31Fs | Yes | No | |||
| TMD10 | TMD | Trisomy | p.E13× | Yes | Yes | No | ||
| TMD11 | TMD | Trisomy | p.G7Fs | Yes | No | |||
| TMD12 | TMD | Trisomy | splicing 5′ exon 2 | Yes | Yes | Yes (FLT3) | ||
| TMD14 | TMD | Trisomy | P.V74I (exon2 3′ splice site) | Yes | Yes | No | ||
| AMKL4 [= TMD4] | AML-M7 | Trisomy | p.71_74del | Yes | Chr5 1.3-3.7, 87-122 | Yes | Yes (EZH2, SMC3, DHX29, APC) | |
| AMKL15 | AML-M7 | Tetrasomy | p.Y62Fs (dupl) | Yes | Chr14 tetrasomy 19.5-106.3 | Chr8 113.1-119.3 | Yes | Yes (DLEC1, EXT1) |
| AMKL15rem | Remission | Trisomy | wt | Yes | Yes | No | ||
| AMKL16 | AML-M7 | Trisomy | p.52_58ins | Yes | Chr7 q-arm | Chr3 60.2-60.9, 153.4-153.6, 176.2-176.8, Chr6 161.9-162.9, Chr7 p-arm | Yes | Yes (JAK1, POLE, PARK2-PACRG) |
| AMKL16rem | Remission | Trisomy | wt | Yes | Yes | No | ||
| AMKL20 | AML-M7 | Trisomy | p.P50Fs | Yes | Chr1 174-247, Chr15 62-99.5 | Chr7 0-10.1 | (SCNAs seen in leukemias) | |
| AMKL21 | AML-M7 | Trisomy | p.F33Fs | Yes | Chr1 200-247 | Chr17 0-7.7, 75.9-77.3 | (SCNAs seen in leukemias) | |
| AMKL24 | AML-M7 | Trisomy | p.P50Fs | Yes | Chr1 222-247 | (SCNAs seen in leukemias) | ||
| AMKL25 | AML-M7 | Trisomy | p.T52Fs (dupl) | Yes | t(X;16)(q26;q23) (associated with multiple myeloma) |
dupl, duploid; wt, wild type.
DS AMKL is characterized by additional driver mutations and SCNAs common with other types of leukemia
In all DS AMKL samples in which both exome sequencing and high-resolution SCNA analysis were performed, we could identify mutations/deletions in genes proven as functional drivers of tumorigenesis in non-DS leukemia (Figure 1; Table 2).
Model of progression of 7 cases of DS-related TMD/AMKL. Boxes indicate the genes with putative driver mutations. Genes in bold type are common with other leukemias; those in italics are not reported in leukemia, but in other types of malignancy. Red bars indicate the time point at which the tumor sample was taken. Numbers indicate percentages of leukemic cells based on the quantifications of reads with the somatic mutations from exome sequencing. Tumors presented in blue are self-regressed TMDs, and those in green are AMKLs. T21-HP, hematopoietic precursors with trisomy 21; ?, for AMKLs 15 and 16, a TMD sample was not available and it is unknown whether there were mutations acquired in this phase.
Model of progression of 7 cases of DS-related TMD/AMKL. Boxes indicate the genes with putative driver mutations. Genes in bold type are common with other leukemias; those in italics are not reported in leukemia, but in other types of malignancy. Red bars indicate the time point at which the tumor sample was taken. Numbers indicate percentages of leukemic cells based on the quantifications of reads with the somatic mutations from exome sequencing. Tumors presented in blue are self-regressed TMDs, and those in green are AMKLs. T21-HP, hematopoietic precursors with trisomy 21; ?, for AMKLs 15 and 16, a TMD sample was not available and it is unknown whether there were mutations acquired in this phase.
Putative third-hit driver mutations/SCNAs in the studied DS TMDs/AMKLs
| Tumor . | Gene/locus . | Somatic mutation . | SCNA . | Pathway . | Catalog of Somatic Mutations in Cancer ID . | Other cancers* . | Reference PMID* . |
|---|---|---|---|---|---|---|---|
| AMKL15 | DLEC1 | p.D1772del_3nt | COSM308378 | AML, ccRCC, lymphoma | 23050586, 22461374, 22976956 | ||
| AMKL15 | EXT1 | Hemizygous loss, 6.2 Mb | WNT | AML, ALL, skin cancers | 15385438, 15797385 | ||
| AMKL15 | Chromosome 14 | Tetrasomy 14 | AML, CML, ALL | 21331167, 15942938, 19429805, 19148138, 15262443 | |||
| AMKL15 | Chromosome 21 | Tetrasomy 21 | AML,CML, ALL | 15339695, 15390279, 17431878 | |||
| AMKL16 | SMC3 | p.G1118R | Chromatin remodeling, TP53 signaling | AML | 22237025, 22237025 | ||
| AMKL16 | DHX29 | p.E178K | GBM, melanoma, ovarian cancer | 20018725 | |||
| AMKL16 | POLE | p.R976L | MMR | COSM937303 (p.R976S) | Endometrial, colorectal cancers | 23636398, 22810696 | |
| AMKL16 | JAK1 | p.S703T | JAK-STAT | COSM96541 | ALL | 21390130, 22237106 | |
| AMKL16 | PARK2/PAKRG | Hemizygous loss, 1 Mb | TP53 signaling | ALL/CLL | 16287063 | ||
| AMKL16 | Chromosome 7 | i(7q) | AML,ALL | 12959859, 3873587 | |||
| AMKL4 | EZH2 | p.D674_Y675ins_1nt | Chromatin remodeling | COSM52999 | AML | 21921040, 23099237, 20601953 | |
| AMKL4 | APC | Hemizygous loss, 35 Mb | WNT | AML | 21765021, 21339756, 20733155 | ||
| TMD4 | PIK3C2A | p.R1461× | PI3K | — | 22983395 | ||
| TMD12 | FLT3 | p.K663N | MAPK | COSM24667 (p.K663Q) | AML | 17359372 |
| Tumor . | Gene/locus . | Somatic mutation . | SCNA . | Pathway . | Catalog of Somatic Mutations in Cancer ID . | Other cancers* . | Reference PMID* . |
|---|---|---|---|---|---|---|---|
| AMKL15 | DLEC1 | p.D1772del_3nt | COSM308378 | AML, ccRCC, lymphoma | 23050586, 22461374, 22976956 | ||
| AMKL15 | EXT1 | Hemizygous loss, 6.2 Mb | WNT | AML, ALL, skin cancers | 15385438, 15797385 | ||
| AMKL15 | Chromosome 14 | Tetrasomy 14 | AML, CML, ALL | 21331167, 15942938, 19429805, 19148138, 15262443 | |||
| AMKL15 | Chromosome 21 | Tetrasomy 21 | AML,CML, ALL | 15339695, 15390279, 17431878 | |||
| AMKL16 | SMC3 | p.G1118R | Chromatin remodeling, TP53 signaling | AML | 22237025, 22237025 | ||
| AMKL16 | DHX29 | p.E178K | GBM, melanoma, ovarian cancer | 20018725 | |||
| AMKL16 | POLE | p.R976L | MMR | COSM937303 (p.R976S) | Endometrial, colorectal cancers | 23636398, 22810696 | |
| AMKL16 | JAK1 | p.S703T | JAK-STAT | COSM96541 | ALL | 21390130, 22237106 | |
| AMKL16 | PARK2/PAKRG | Hemizygous loss, 1 Mb | TP53 signaling | ALL/CLL | 16287063 | ||
| AMKL16 | Chromosome 7 | i(7q) | AML,ALL | 12959859, 3873587 | |||
| AMKL4 | EZH2 | p.D674_Y675ins_1nt | Chromatin remodeling | COSM52999 | AML | 21921040, 23099237, 20601953 | |
| AMKL4 | APC | Hemizygous loss, 35 Mb | WNT | AML | 21765021, 21339756, 20733155 | ||
| TMD4 | PIK3C2A | p.R1461× | PI3K | — | 22983395 | ||
| TMD12 | FLT3 | p.K663N | MAPK | COSM24667 (p.K663Q) | AML | 17359372 |
ccRCC, clear cell renal cell carcinoma; GBM, glioblastoma multiforme; MMR, mismatch repair pathway.
Mutations affecting the same amino acid are denoted in bold type.
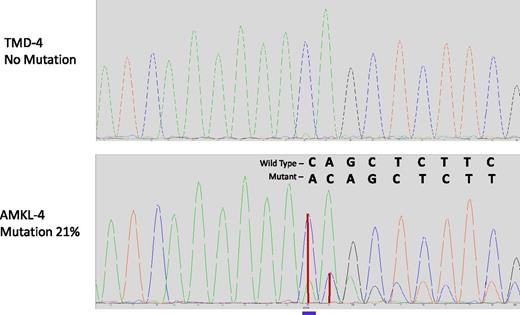
In AMKL4 (Table 3), we identified several putative driver mutations in addition to GATA1 mutations, all of which were absent at the TMD stage (TMD4). Among these mutations, there was a frameshift insertion in EZH2 (p.D674_Y675delinsX) (Figure 2). This mutation is a known recurrent mutation (Catalog of Somatic Mutations in Cancer: 52999) in patients with myeloid disorders18,19 ; moreover, mono- or biallelic EZH2 mutations were detected in 12% of myeloproliferative disorders.18,19 We also detected a p.G1118R mutation in SMC3, which is found to be recurrently mutated in AMLs.20 Both EZH2 and SMC3 encode the proteins involved in chromatin remodeling.20,21 Another candidate driver mutation (p.E178K) was observed in DHX29, a gene previously reported to be associated with cancerogenic pathways in nonleukemic tumors.22 In addition, this AMKL sample acquired 2 deletions of Chr5 (a 2.4-Mbp and a 37-Mbp segment, see Figure 3); intriguingly, neither of these deleted segments overlapped the two critical deleted regions on Chr5q in non-DS myeloid leukemias23 (Table 1). The larger of the two deleted regions in AMKL4 did, however, encompass the tumor suppressor APC gene, deleted in ≤15% of non-DS AMLs.24
Mutations (percentage of mutation) in TMD and AMKL of patient 4
| Gene (mutation) . | TMD . | AMKL . |
|---|---|---|
| GATA1 (p.67ins_20nt) | + (12) | — |
| PIK3C2A (p.R1461×) | + (12) | — |
| GATA1 (p.71_74del_12nt) | + (8) | + (20) |
| EZH2 (p.D730_Y731ins_1nt) | — | + (18) |
| DHX29 (p.E178K) | — | + (25) |
| SMC3 (p.G1118R) | — | + (23) |
| SLC6A5 (p.L488L) | — | + (26) |
| PIEZO1 (p.T2127M) | — | + (20) |
| FCER1G (exon2:c.141+5C>A) | — | + (21) |
| CAPN11 (p.R687K) | — | + (28) |
| Gene (mutation) . | TMD . | AMKL . |
|---|---|---|
| GATA1 (p.67ins_20nt) | + (12) | — |
| PIK3C2A (p.R1461×) | + (12) | — |
| GATA1 (p.71_74del_12nt) | + (8) | + (20) |
| EZH2 (p.D730_Y731ins_1nt) | — | + (18) |
| DHX29 (p.E178K) | — | + (25) |
| SMC3 (p.G1118R) | — | + (23) |
| SLC6A5 (p.L488L) | — | + (26) |
| PIEZO1 (p.T2127M) | — | + (20) |
| FCER1G (exon2:c.141+5C>A) | — | + (21) |
| CAPN11 (p.R687K) | — | + (28) |
Percentage of mutation calculated based on the Sanger sequencing data.
Chromatograms of DNA region of EZH2 harboring the mutation in AMKL4. Frameshift insertion p.D674_Y675 in EZH2 is absent in TMD (upper panel) and is present in AMKL (lower panel). Blue line, position of the insertion; vertical red lines, height of peaks of wild-type and mutant sequences.
Chromatograms of DNA region of EZH2 harboring the mutation in AMKL4. Frameshift insertion p.D674_Y675 in EZH2 is absent in TMD (upper panel) and is present in AMKL (lower panel). Blue line, position of the insertion; vertical red lines, height of peaks of wild-type and mutant sequences.
SNP microarray analysis of samples from a single DS patient at two stages of disease, the TMD and the AMKL stage. SNP microarray analysis of AMKL4 (A) in comparison with TMD4 (B) for chromosome 5, with relative chromosomal position indicated by the ideogram (C). For each DNA, an allele difference plot (similar to B-allele frequency) is plotted in log2 scale for all SNP genotypes (top row), and a log2-ratio for the probe intensity of all probes (lower row). Red boxes indicate regions of SCNAs that are detected by differences in allele distribution and simultaneous loss of probe intensity, ie, lower log2-ratio. Here 2 deletions are detected on chromosome 5 (chr5:1.4-3.8Mbp and chr5:85.4-122.1Mbp).
SNP microarray analysis of samples from a single DS patient at two stages of disease, the TMD and the AMKL stage. SNP microarray analysis of AMKL4 (A) in comparison with TMD4 (B) for chromosome 5, with relative chromosomal position indicated by the ideogram (C). For each DNA, an allele difference plot (similar to B-allele frequency) is plotted in log2 scale for all SNP genotypes (top row), and a log2-ratio for the probe intensity of all probes (lower row). Red boxes indicate regions of SCNAs that are detected by differences in allele distribution and simultaneous loss of probe intensity, ie, lower log2-ratio. Here 2 deletions are detected on chromosome 5 (chr5:1.4-3.8Mbp and chr5:85.4-122.1Mbp).
Patient 15 was diagnosed with AMKL at the age of 17 months. All the somatic events detected in AMKL15 were absent in the remission sample. This leukemia harbors 1 additional copy of chromosome 21 (tetrasomy 21) and 2 extra copies of chromosome 14 (tetrasomy 14). Chromosome 21 has 3 copies of 1 allele and 1 copy of the second allele; chromosome 14 has 2 copies of each allele (Tables 1 and 2). Additionally, we detected a submicroscopic 6.2-Mbp deletion on Chr8q that harbors only 4 genes: CSMD3, TRP51, EXT1, and SAMD12 (supplemental Figure 1A). Interestingly, EXT1 could be considered a putative driver gene, since it was found that EXT1 CpG island hypermethylation is common in leukemia, especially in acute promyelocytic leukemia and acute lymphoblastic leukemia, and nonmelanoma skin cancer25 (Figure 1). We also found a putative driver mutation in DLEC1 (p.D1772delD) which has already been reported for an AML case26 and in kidney tumor (Catalog of Somatic Mutations in Cancer: COSM308378); moreover, epigenetic silencing of this gene was detected in the majority of Hodgkin and non-Hodgkin lymphomas.27
Patient 16 was diagnosed with AMKL at age 21 months. We were able to identify several putative driver mutations in addition to GATA1, all of which were absent in the remission sample (Table 2): one was a mutation in JAK1 p.S703T (Catalog of Somatic Mutations in Cancer: COSM96541), which has already been observed in lymphoblastic leukemias28,29 (Figure 1; Table 2). Another was a putative driver p.R976L mutation in the POLE gene encoding the polymerase ε, which is involved in defective DNA mismatch repair pathway. AMKL16 also acquired several chromosomal abnormalities affecting chromosomes other than Chr21: balanced loss of heterozygosity (LOH) in 3q (starting at Mbp 109) including 2 small deletions; furthermore, a small likely homozygous deletion was identified on chromosome 3p including coding exons of the gene FHIT. Chromosomal rearrangements in chromosome 7 resulted in 1 copy of 7p arm and 3 copies of 7q arm, most likely due to isochromosome 7q, as well as submicroscopic deletions on Chr6. Chromosomal aberrations in chromosomes 3 and 7 are frequently detected in leukemias. A deletion on Chr6 involved 3 genes, 2 of which were PARK2 and PACRG (supplemental Figure 1B), whose common promoter was found to be frequently abnormally methylated, resulting in downregulation of gene expression in non-DS acute lymphoblastic leukemia and chronic myeloid leukemia (CML).30
DS TMD: low mutation rates and intact genome integrity
In 5 TMD samples analyzed by exome sequencing, between 1 and 3 mutations were detected, including different GATA1 mutations in the same patient (supplemental Table 3). In 2 of 5 TMD cases (TMD3 and TMD4), we detected mutations other than GATA1 (Tables 1 and 2).These data indicate very low mutation rates in DS TMDs. None of the 10 DS TMD DNA samples showed any SCNAs by high-resolution SNP and CNV array analysis (Table 1). In AMKL4, 2 SCNAs observed at the AMKL stage of the disease (deletions of 2 regions on 5q) were not present at the TMD stage (TMD4) (Figure 3). This suggests that segmental chromosomal instability is not a major contributing factor to the pathogenesis of DS TMD. A recent study31 mentions that 4 of 7 DS TMD cases did show some SCNAs (lower than DS AMKLs), but no patient details or data were given for a detailed comparison with our study.
DS AMKL: mutation rates typical of leukemia and high chromosomal instability
DS AMKLs studied here harbor a few somatic coding mutations in the whole exome (between 8 and 14) (supplemental Table 3), indicating that mutation rates are comparable with the other types of leukemia.20,32,33 On the other hand, high-resolution SCNA analysis using genome-wide combined SNP/CNV array has revealed multiple acquired segmental gains, losses, or LOH events in 6 of 7 DS AMKL cases (Table 1), which is concordant with the previous observations in DS AMKLs and is higher than in other pediatric AML subgroups, with the exception of non-DS AMKL.31,34 In the single case where no SCNAs were found (AMKL25), karyotyping revealed a balanced translocation, t(X;16)(q26;q23).
GATA1 mutations are often sole acquired events in DS TMD
We performed conventional sequencing of the second exon (and neighboring introns) of the GATA1 gene, and found, as expected, typical GATA1 mutations in all DS TMD and all DS AMKL cases (Table 1; supplemental Table 3). Exome sequencing of 8 TMD/AMKL samples verified the presence of somatic mutations in GATA1 and identified additional somatic coding mutations (supplemental Table 3). In 1 sample for which both TMD and AMKL stages of the disease were analyzed (patient TMD4→AMKL4), we were able to establish the clonal competition of megakaryoblastic clones identified by their different GATA1 mutations at the TMD stage and record the fate of clones that accumulated different additional mutations (Figures 1 and 2).
Patient 4 was diagnosed with TMD at 8 days of age, and after spontaneous regression, 24 months later the disease had progressed to AMKL (sample AMKL4). In TMD4, we detected 2 distinct mutations in the second exon of GATA1 (20-nt insertion and 12-nt deletion) as well as a nonsense mutation in PIK3C2A (p.R1461X) (Table 3). In AMKL4, only one of these mutations, namely the 12-nt deletion in GATA1, was present, suggesting that: (i) the two GATA1 mutations in TMD4 represent different megakaryoblastic clonal populations; and (ii) the 20-nt duplication in GATA1 and the mutation in PIK3C2A coexist in the same clone of megakaryoblasts in TMD, which eventually self regressed (here: clone A); however, the clone with 12-nt deletion in GATA1 progressed to AMKL (here: clone B). Additional evidence favoring colocalization of GATA1 20-nt duplication and PIK3C2A mutation comes from the fact that both mutations were present in TMD in 12% of DNA, as verified by Sanger sequencing. Clone B in TMD4 was characterized by only 1 GATA1 mutation (12-nt deletion) present in 8% of DNA. However, in AMKL4, this clone acquired 7 additional somatic mutations. All these mutations, as well as the GATA1 12-nt deletion, were present in 21% to 26% of DNA of the AMKL sample, as validated with Sanger sequencing, which suggests their origin within the same clone of megakaryoblasts.
The facts that truncating GATA1 mutations are invariably associated with DS TMD and DS AMKL and that clone B in TMD4 contains only 1 GATA1 mutation strongly suggest that the GATA1 mutation in the context of T21 is both necessary and sufficient for the clonal expansions presenting as DS TMD (Figure 1).
Given that GATA1 was the first mutation in clone A of TMD4, and that all cells of this clone contain a PIK3C2A nonsense mutation, we suggest that this mutation provides a selective advantage to these megakaryoblasts over megakaryoblasts with only the GATA1 20-nt duplication. Indeed PIK3C2A is known to be essential for endothelial cell migration, proliferation, and survival, indicating its putative tumorigenic properties. However, it is not clear from our data if the PIK3C2A p.R1461X mutation is activating or loss of function due to nonsense-mediated decay. Additionally we identified an activating mutation in receptor tyrosine kinase FLT3 (K663N) in TMD12. The presence of additional driver mutations in TMDs reveals the mutational process and competition of clones even at preleukemic stages in DS megakaryoblastic leukemias. Most importantly, this indicates that additional driver mutations may accumulate in TMDs without immediate progression to AMKL but instead can regress thereafter. The fate of TMDs may depend on the tumorigenic potential of these secondary mutations. However, for patient 12, only a TMD sample is available (no germline DNA), and we cannot fully exclude the germline origin of K663N FLT3 variant. In any case, this variant is of potential interest in the context of tumorigenesis, since in the literature a somatic mutation in the same codon of FLT3 (K663Q) was described in AML and was confirmed to result in a constitutively activated protein.35
Altogether these data suggest that GATA1 mutation is the primary event in DS TMDs and is mostly seen in neonatal TMD samples in the absence of any other mutation. However, in some cases, progression of the disease is characterized by accumulation of additional driver mutations occurring in different subpopulations of megakaryoblasts, resulting in competition between clones. The analysis of patient TMD4→AMKL4 demonstrates that despite accumulation at the DS TMD stage of an additional mutation in PIK3C2A (affecting the phosphatidylinositol-3 kinase [PIK3]/Akt pathway, cooperating with ERG and GATA1s in leukemogenesis36 ), this clone was not the one transformed to AMKL later at infant stage (Figure 1), but rather the other TMD clone, containing just a GATA1 mutation, accumulated multiple other mutations leading to DS AMKL. This indicates that factors that might have provided selective advantage to the double (GATA1 + PIK3C2A) over single (GATA1) mutant cells at TMD stage (most probably in utero, in the fetal liver) no longer existed in the bone marrow in infancy. Evidence for differences in these two environments has been previously suggested from mouse-model studies.6,37
Discussion
Multiple putative third-hit driver mutations for TMD-to-AMKL transformation observed in all sequenced DS AMKL samples
Additional putative driver mutations in genes such as EZH2, JAK1, EXT1, DLEC1, or APC revealed in this work may cause the transitions of TMD to AMKL. The mutation in EZH2, as well as hemizygous loss of APC in AMKL4, was absent in its TMD stage (TMD4) (Tables 1 and 2; Figures 1 and 2). Our hypothesis that these mutations are capable of triggering progression of TMD to AMKL is reinforced by the observation that 2 of 21 cell lines (∼10%) derived from individuals with MDSs, myelodysplastic-myeloproliferative neoplasms, or myeloproliferative neoplasms that later progressed to AML harbored EZH2 mutations.19 Additionally, it was observed that mutations in EZH2 almost double the risk of progression of primary myelofibrosis to acute leukemia in humans (from 17.6% to 31.8%),18 and they are sufficient to cause T-cell acute lymphoblastic leukemia in mice.38 Loss of APC is also associated with poor prognosis in non-DS AMLs. Therefore, in this study, we established that EZH2 mutation and APC loss are secondary hits compared with the initial GATA1 driver mutation. Because EZH2 and APC are strongly implicated in leukemogenesis, we hypothesized that they triggered the transition to AMKL of clone B in patient 4.39 However, we cannot assess the relative contribution of each event to the malignant transformation. Similar observations were made in AMKL15 with the deletion of the likely driver gene EXT1, small deletion in DLEC1, and tetrasomies 14 and 21; as well as in AMKL16 where more than one putative additional driver mutation was detected: oncogenic mutation in JAK1 p.S703T,40 mutation in POLE (R976L), and deletion of PARKIN2/PARCG (Figure 2; supplemental Figure 1B). Mutation in POLE (R976L) could potentially be considered a putative driver, as a mutation in the same codon in POLE (R976S) was detected in endometrial tumor (COSM937303); however, the implication of POLE in DS AMKL needs additional confirmation. For patients 15 and 16, no TMD samples were available, so we cannot rule out the possibility that some of the likely driver mutations detected in AMKL were present at the TMD stage. Thus the question of the minimal set of secondary driver events sufficient for the transformation of TMD to AMKL remains open.
The combined SNP/SCNA analysis revealed multiple acquired segmental gains, losses, or LOH events in 6 of 7 DS AMKL cases (Table 1). In the single case where no SCNAs were found (AMKL25), karyotyping revealed a balanced translocation, t(X;16)(q26;q23). Translocations involving 16q23 have been described in multiple myeloma.41 The implication of detected SCNAs in AMKLs, all of which are common in other types of leukemia, also remains to be investigated (Table 1). However, the fact that all detected SCNAs rendered poor prognoses to the patients with non–DS-associated leukemia suggests their causative role in TMD-to-AMKL progression.23,30,42
Predicted effects of observed third-hit driver events for DS AMKL converge on upregulation of MYC
One of the tumorigenic consequences of mutated GATA1 is overexpression of MYC.13-15,43 We have reasoned that the secondary mutations detected in this study targeting JAK1, APC, PIK3C2A, FLT3, and EXT1, as well as mutations in DS AMKL observed by others (JAK2, JAK3, MPL), may activate mitogen-activated kinase (MAPK)/PI3K, Janus kinase/signal transducer and activator of transcription (JAK-STAT), and wingless-related integration site (WNT) pathways, which also result in overexpression of MYC. The fact that putative driver mutations were observed in all 3 DS AMKL samples analyzed by exome sequencing allows us to speculate that these secondary mutations may have an additive effect on GATA1 mutation in overexpression of MYC. In line with this hypothesis, there is evidence of overexpression of NMYC in RNA from DS TMD, as well as MYC RNA from AMKLs.44
Additional driver mutations may be functioning in leukemogenesis through strengthening of the effect of GATA1 mutations. Activating mutations in JAK1 result in aberrant phosphorylation of STAT1 and STAT3, which are involved in megakaryoblastic differentiation and control of cell cycle through transcriptional repression of MYC (supplemental Figure 2).45 Mutations in PIK3C2A and FLT3 revealed in this study point to the MAPK/PI3K pathway in DS leukemia. In T-cell ALL, aberrant activation of the PI3K pathway has been shown to function through MYC activation.46 Activating mutation of FLT3 have also been shown to increase expression of MYC in AMLs.47 Hemizygous loss of APC detected in this work is known to cause aberrant activation of the WNT pathway in leukemia,48,49 which may also lead to overexpression of MYC. EXT1 gene was found to be essential for the coordinated activation of WNT signaling during Drosophila development.50 Interestingly EZH2, a component of Polycomb repressive complex, found to be mutated in 1 patient in this study, is known to be directly controlled by MYC in AMLs.51
Our study cannot of course rule out other driver hits conferred by epigenetic events. A previous transcriptome comparison found the expression of testis-cancer antigen PRAME as the discriminative marker of DS AMKL compared with DS TMD,44 and the overexpression of this gene is known to be due to demethylation of the regulatory regions and is established as a potential driver event in tumorigenesis52 and inhibitor of differentiation in myeloid leukemic precursor cells.53 Interestingly, overexpression of PRAME was found to be part of the progression program for CML, in synergy with deregulation of the WNT/β-catenin pathway.54
In conclusion, our data demonstrate that DS AMKLs form by accumulation of multiple somatic SCNAs and point mutations in genes known as drivers of leukemogenesis. These putative driver events can be detected in every DS AMKL case, affecting one or more typical leukemogenic signaling pathways, such as JAK/STAT, MAPK/PI3K, and WNT. GATA1 mutations could be the first, and often sole, acquired events in DS TMD. The DS TMD samples are devoid of acquired SCNAs but may contain additional mutations that provide selective advantage to the TMD clone at fetal/neonatal age. These additional driver mutations at DS TMD stage do not necessarily predict transformation to AMKL, indicating that factors determining the selective advantage for proliferation/elimination could be very different in fetal compared with infant hematopoiesis. The final validation of these observations will require analysis of a larger set of AMKL and TMD samples.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Dubois-Ferrière Dinu Lipatti grant to S.I.N.; SNF 144082, ERC 249968, and Foundation “ChildCare” grants to S.E.A.; and KKL632 grant from the Kay Kendall Leukaemia Fund to D.N.
Authorship
Contribution: S.I.N. designed and organized the project, processed and analyzed the data, and wrote the manuscript; S.E.A. organized and supervised the project and wrote the manuscript; D.N. designed and organized the project, assembled the sample collection, and wrote the manuscript; F.S. processed and analyzed the data; A.V. and E.F. ran the experiments; J.G. performed GATA1 sequencing and analyzed the data; E.G. and G.B. collected patient samples and isolated DNA; and A.H. and J.A.V. ran aCGH/SNP experiments and analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Correspondence: Stylianos Antonarakis, Department of Genetic Medicine and Development, University of Geneva Medical School, 1 Rue Michel-Servet, 1211 Geneva, Switzerland; e-mail: Stylianos.Antonarakis@unige.ch; and Dean Nizetic, Centre for Paediatrics, Blizard Institute, Barts and The London School of Medicine and Dentistry, 4 Newark St, London, E1 2AT, United Kingdom; e-mail: d.nizetic@qmul.ac.uk.