Key Points
HDAC inhibitors downregulate expression of the activating NKp30 ligand B7-H6, reducing NKp30-dependent tumor cell recognition by NK cells.
Our results have implications for the design of potential cancer treatments combining immunotherapy with HDAC inhibitors.
Abstract
Natural killer (NK) cells are central effector cells during innate immune responses against cancer. Natural cytotoxicity receptors expressed by NK cells such as NKp30 are involved in the recognition of transformed cells. Recently, the novel B7 family member B7-H6, which is expressed on the cell surface of various tumor cells including hematological malignancies, was identified as an activating ligand for NKp30. To investigate expression and regulation of B7-H6, we generated monoclonal antibodies. Our study reveals that B7-H6 surface protein and messenger RNA (mRNA) expression in various tumor cell lines was downregulated upon treatment with pan- or class I histone deacetylase inhibitors (HDACi) as well as after small interfering RNA–mediated knockdown of the class I histone deacetylases (HDAC) 2 or 3. B7-H6 downregulation was associated with decreased B7-H6 reporter activity and reduced histone acetylation at the B7-H6 promoter. In certain primary lymphoma and hepatocellular carcinoma samples, B7-H6 mRNA levels were elevated and correlated with HDAC3 expression. Finally, downregulation of B7-H6 on tumor cells by HDACi reduced NKp30-dependent effector functions of NK cells. Thus, we identified a novel mechanism that governs B7-H6 expression in tumor cells that has implications for potential cancer treatments combining immunotherapy with HDACi.
Introduction
Natural killer (NK) cells are innate immune cells that are able to recognize and kill target cells by an array of specialized cell surface receptors that mediate activating or inhibitory signals.1-3 Stressed, infected, or transformed cells often downregulate inhibitory NK-cell ligands such as major histocompatibility complex class I and upregulate activating ligands on the cell surface; this results in the killing of target cells.3 The activating NK-cell receptor NKG2D binds to the stress-induced ligands major histocompatibility complex class I polypeptide-related sequence A/B (MICA/B) and UL16 binding proteins (ULBPs), and this receptor–ligand interaction was shown to play an important role in tumor cell recognition and killing.4,5 The natural cytotoxicity receptor NKp30 (natural cytotoxicity triggering receptor 3, NCR3) is involved in NK-cell–mediated killing of various tumor cell lines6 and virus-infected cells7,8 and also in the interaction with dendritic cells.9 In patients with acute myeloid leukemia (AML), decreased NKp30 expression was shown to correlate with poor prognosis and survival.10 Alternatively-spliced NKp30 isoforms exist and were shown to be of prognostic value for patients with gastrointestinal stromal tumors.11 Several ligands have been proposed to bind to NKp30, including the intracellular proteins BCL2-associated anthanogene 612 and pp65 of human cytomegalovirus.7 The NKp30 ligand on the surface of tumor cells has recently been identified to be B7-H6 (natural cytotoxicity triggering receptor 3 ligand 1, NCR3LG1), a novel B7 family member.13-15 The interaction of B7-H6 on target cells with NKp30 on NK cells leads to interferon-gamma production and target cell killing.13 Expression studies using a polyclonal anti–B7-H6 antibody revealed that B7-H6 was detectable on the surface of tumor cell lines of various origins as well as in a small percentage of patients with AML, marginal zone lymphoma, and acute T-cell leukemia.13 Healthy tissues were reported to lack B7-H6 transcripts,13 suggesting that B7-H6 could be a potential tumor marker. To date, the mechanisms that regulate B7-H6 expression in tumor cells are poorly understood. A comprehensive understanding of these mechanisms is crucial for the evaluation of B7-H6 as a potential target for tumor therapies.
Histone deacetylases (HDACs) are enzymes that remove acetyl groups of histone and nonhistone proteins.16-18 They regulate key cellular processes, including proliferation and survival, by modulating gene expression and protein activity. In general, HDACs act as transcriptional repressors, since histone hypoacetylation in promoter regions is associated with reduced transcription and epigenetic silencing.19 This process is counteracted by histone acetyltransferases (HATs), for example, cyclic adenosine monophosphate response element-binding protein (CREBBP, also known as CBP) and p300 (EP300). However, several genes, including certain genes of the host immune response, require HDAC activity for their induction, indicating that HDACs can also function as transcriptional activators.20,21 Additionally, genome-wide studies have shown that HDACs and HATs colocalize at the promoters of active genes, correlating with histone hyperacetylation and transcription.22 The HDACs primarily involved in regulation of transcription belong to the class I HDACs, which consists of HDACs 1–3 and HDAC8. HDACs 1-3 are ubiquitously expressed and frequently overexpressed or dysregulated in human malignancies.18,23
Small molecular compounds (HDAC inhibitors [HDACi]) have been developed for tumor therapy that induce cell-cycle arrest, differentiation, or apoptosis in tumor cells.17,24,25 HDACi induce the expression of tumor-suppressing genes, including the cyclin-dependent kinase inhibitor 1A,26 as well as NKG2D ligands, resulting in enhanced NKG2D-mediated killing of tumor cells.27-31 Additionally, HDACi can downregulate certain genes with tumor-promoting activity.24,32,33 This downregulation was shown to correlate with reduced transcription due to local histone hypoacetylation after HDAC and HAT dissociation from the respective promoter regions.32,34 The HDACi suberoylanilide hydroxamic acid (SAHA), also known as vorinostat, has been approved for the treatment of cutaneous T-cell lymphoma.35 Clinical trials are currently being performed to evaluate the application of HDACi for the treatment of other hematological malignancies as well as solid tumors.
The purpose of our study was to identify mechanisms that regulate the expression of the activating NKp30 ligand B7-H6 in tumor cells. Our results demonstrate that class I–specific HDACi downregulate B7-H6 cell surface expression on tumor cells derived from hematological diseases and solid tumors and thereby affect tumor cell recognition of primary NK cells. Since elevated levels of B7-H6 were also found in a subset of primary tumor samples that correlated with HDAC3 expression, our findings might have implications for the rational design of novel strategies of cancer immunotherapy.
Materials and methods
Cells, reagents, and treatments
X63-Ag8.653, Ba/F3, HeLa, K562, Ramos, HepG2 (all from American Type Culture Collection), and SK-Mel-37 (provided by H. Bernhard, Darmstadt Clinic, Germany) were grown in RPMI 1640 containing 2 mM l-glutamine and 10% fetal calf serum (FCS; all Invitrogen). NK-92CI36 cells were cultured in α-minimal essential medium containing 12.5% FCS, 12.5% horse serum (Invitrogen), 2 mM l-glutamine, and 50 µM β-mercaptoethanol (both Sigma-Aldrich). Ba/F3 that overexpress B7-H6 were generated by retroviral transduction (provided by C. Watzl, Technical University Dortmund, Germany). Human foreskin keratinocyte (Invitrogen) culture and NK-cell isolation from healthy donors were performed as described.37 NK cells were cultured overnight in CellGro stem cell growth medium (CellGenix) with 10% human AB serum (PAA Laboratories) and 200 U/mL recombinant human interleukin-2 (IL-2; National Institutes of Health). All media contained 100 U/mL penicillin and 100 µg/mL streptomycin (Sigma-Aldrich), and cell lines were routinely tested by the Multiplex Cell Contamination Testing Service at the German Cancer Research Center. Cells were treated for 16 hours with the HDACi SAHA (1 µM; Cayman Chemical), trichostatin A ([TSA] 100 µM; Sigma-Aldrich), apicidin (1 µM; AppliChem), compound 2 (40 µM, Cpd238 ), which were dissolved in dimethylsulfoxide (DMSO; Sigma-Aldrich), or DMSO solvent control. Valproic acid ([VPA] 10 mM; Sigma-Aldrich) was dissolved in water. Cells were treated for 16 hours with the HAT inhibitors (HATi), curcumin39 or C64640 (both 10 µM; Sigma-Aldrich), or DMSO solvent control. For inhibition of transcription, actinomycin D (10 µg/mL; Sigma-Aldrich) was added to cultures 30 minutes prior to treatment with HDACi. All antibodies and fusion proteins (FPs) used are described in the supplemental Methods (available on the Blood Web site). Animal experiments were approved by the Regierungspräsidium Karlsruhe. The study involving samples of human livers (n = 3) and hepatocellular carcinoma (HCC; n = 27) was approved by the institutional ethics committee of the Medical Faculty of Heidelberg University (application no. 206/05). This study was conducted in accordance with the Declaration of Helsinki.
siRNA transfection
Cells were transfected with small interfering RNAs (siRNAs) as described.41 The percentage of B7-H6 messenger RNA (mRNA) and surface expression was calculated as follows: 100 × (B7-H6 expression specific siRNA)/(B7-H6 expression negative control siRNA). All siRNAs used are described in the supplemental Methods. Knockdown efficiency was determined by western blot as described.42
Luciferase reporter assay
A B7-H6 reporter construct was generated by cloning a fragment of the B7-H6 5′-untranslated region ([UTR] −1270 to +208 according to National Center for Biotechnology Information Reference Sequence NM_001202439.1) that was approximately 1.5 kb into the pGLuc-basic vector (New England Biolabs). Cells were cotransfected in 96-well plates with the B7-H6 reporter construct or an empty vector control (VC; 50 ng each) and the pSV40-Cluc vector (10 ng; New England Biolabs) using Lipofectamine 2000 (Invitrogen). Thirty-two hours posttransfection, the cells were treated with SAHA or DMSO for 16 hours. Luciferase activity was measured in cell culture supernatants using an Orion L luminometer (Titertek Berthold). Normalization was performed as follows: (relative light units pGLuc-basic)/(relative light units pSV40-Cluc). The fold-change luciferase activity was calculated as follows: (normalized activity B7-H6 5′-UTR)/(normalized activity VC).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed using the SimpleChIP enzymatic chromatin IP kit (Cell Signaling Technology) according to the manufacturer’s instructions. Chromatin was isolated from HeLa cells treated with SAHA or DMSO, and 10 µg each was used for IP. The precipitated chromatin was analyzed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) using primers that amplify a 155-bp region containing the transcription start site in the B7-H6 5′-UTR (−24 to +131).
NK-cell degranulation assay
Degranulation assays were performed with target cells pretreated with SAHA or DMSO and washed twice to remove the drugs. NK-92CI or healthy donor NK cells activated overnight with IL-2 were used as effectors after blocking of NK-cell receptors with specific monoclonal antibodies (mAbs) or respective isotype controls. Targets and effectors were cocultured at an 1:1 effector to target ratio as described.42 The effectors were analyzed for expression of CD107a/b using flow cytometry by gating on CD56+ CD3− 7-AAD− cells. The percentage of specific degranulation was calculated as follows: (% experimental degranulation) − (% spontaneous degranulation).
Statistical analysis
Statistical significances were determined using a 2-tailed Student t test. For analysis of B7-H6 mRNA expression in primary tumor samples, the mean ± the standard error of the mean (SEM) was calculated, and statistical significance was determined using a 2-tailed Mann-Whitney test. Correlation analysis was performed using a 2-tailed Pearson correlation of log10-transformed values.
Results
Generation and validation of the anti-B7-H6 mAbs 1.18 and 5.51.18
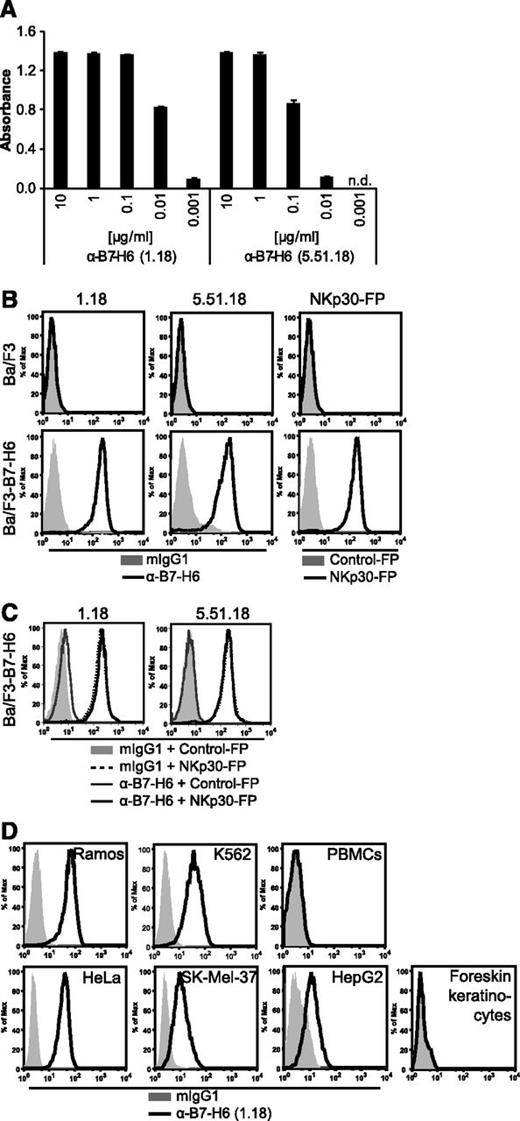
To investigate the expression and regulation of B7-H6 in tumor cell lines, B7-H6–specific mAbs were generated in mice. Both mAbs 1.18 and 5.51.18 bound to immobilized B7-H6-FP in a dose-dependent manner in enzyme-linked immunosorbent assay (Figure 1A) and stained Ba/F3 transductants expressing B7-H6, but not the parental cell line in flow cytometry (Figure 1B). Similar staining patterns were obtained using an NKp30-FP (Figure 1B). Mutational analyses revealed that the mAbs bound to different epitopes of the B7-H6 extracellular domain (data not shown). Additionally, we evaluated the mAbs for their ability to block NKp30-FP binding. Preincubation of Ba/F3-B7-H6 cells with neither 1.18 nor 5.51.18 anti-B7-H6 mAb reduced NKp30-FP staining of B7-H6–expressing Ba/F3 cells (Figure 1C). Tumor cell lines of various origins, including B-cell lymphoma (Ramos), erythroleukemia (K562), cervical carcinoma (HeLa), melanoma (SK-Mel-37), and HCC (HepG2) were stained by mAbs 1.18 (Figure 1D) and 5.51.18 (data not shown) in flow cytometry. However, no cell surface expression of B7-H6 was detectable on peripheral blood mononuclear cells (PBMCs) and foreskin keratinocytes from healthy donors (Figure 1D; data not shown). The closest relatives of B7-H6 are B7-H1, B7-H3, and B7-DC (PD1L2)13 ; however, in vitro differentiated macrophages that express these molecules were not bound by the 1.18 mAb (data not shown), indicating that the generated mAbs did not cross-react with related B7 family members.
Generation and validation of the anti-B7-H6 mAbs 1.18 and 5.51.18. (A) Binding of mAbs 1.18 and 5.51.18 at the indicated concentrations to immobilized B7-H6-FP was measured by enzyme-linked immunosorbent assay. The mean ± standard deviation absorbance at 492 nm of duplicates is shown. n.d., not detectable. (B) Binding of 1.18, 5.51.18, mIgG1 isotype control, NKp30-FP, and a control-FP to Ba/F3 and Ba/F3-B7-H6 transductants in flow cytometry. (C) Binding of NKp30-FP and a control-FP after preincubation with 1.18, 5.51.18 anti–B7-H6 mAbs, or mIgG1 isotype control to Ba/F3-B7-H6 cells was assessed by flow cytometry. (D) B7-H6 surface expression was analyzed on Ramos, K562, HeLa, SK-Mel-37, and HepG2 tumor cell lines; PBMCs and foreskin keratinocytes from healthy donors by the mAb 1.18 in flow cytometry. Representative histograms from 1 out of at least 3 experiments are shown.
Generation and validation of the anti-B7-H6 mAbs 1.18 and 5.51.18. (A) Binding of mAbs 1.18 and 5.51.18 at the indicated concentrations to immobilized B7-H6-FP was measured by enzyme-linked immunosorbent assay. The mean ± standard deviation absorbance at 492 nm of duplicates is shown. n.d., not detectable. (B) Binding of 1.18, 5.51.18, mIgG1 isotype control, NKp30-FP, and a control-FP to Ba/F3 and Ba/F3-B7-H6 transductants in flow cytometry. (C) Binding of NKp30-FP and a control-FP after preincubation with 1.18, 5.51.18 anti–B7-H6 mAbs, or mIgG1 isotype control to Ba/F3-B7-H6 cells was assessed by flow cytometry. (D) B7-H6 surface expression was analyzed on Ramos, K562, HeLa, SK-Mel-37, and HepG2 tumor cell lines; PBMCs and foreskin keratinocytes from healthy donors by the mAb 1.18 in flow cytometry. Representative histograms from 1 out of at least 3 experiments are shown.
B7-H6 expression in tumor cell lines is downregulated by treatment with HDACi
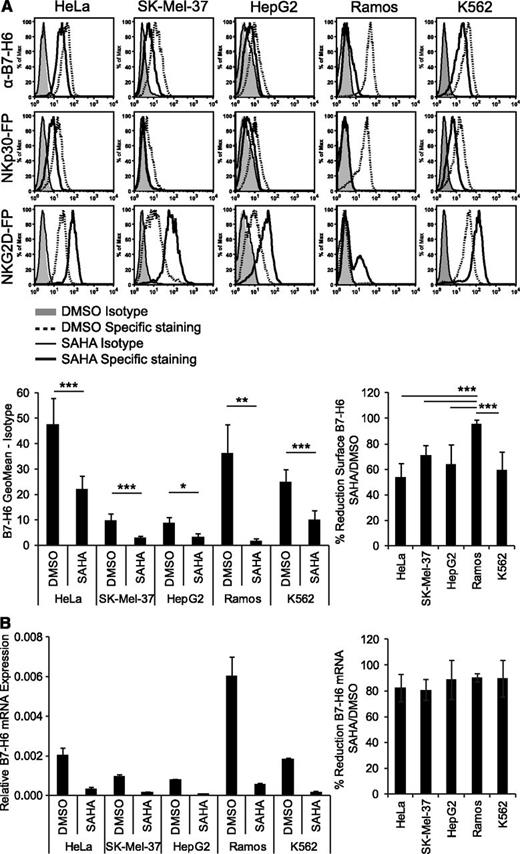
To identify mechanisms of B7-H6 regulation, different tumor cell lines were treated with several cytostatic drugs, proteasome inhibitors, or proinflammatory cytokines and analyzed by flow cytometry. However, no effect on the surface expression of B7-H6 was observed (data not shown). In contrast, incubation with the pan-HDACi SAHA led to a significant reduction in B7-H6 surface expression compared with the solvent control DMSO in all tumor cell lines of hematological and solid tumor origin tested (Figure 2A). B7-H6 surface expression was reduced from 53.7 ± 10.6% (in HeLa) up to 95.6 ± 3.2% (in Ramos cells; Figure 2A). Staining with an NKp30-FP also showed reduced expression of NKp30 ligands, while the expression of NKG2D ligands was induced after SAHA treatment (Figure 2A) as previously published.27-30 Moreover, B7-H6 mRNA expression was reduced by at least 82.5% after SAHA treatment compared with DMSO control in the tested cell lines (Figure 2B). The percentages of dead cells were not significantly altered by SAHA treatment compared with the DMSO control (supplemental Figure 1A). Together, our data demonstrate that cell surface and mRNA expression of B7-H6 are downregulated by treatment with the HDACi SAHA.
B7-H6 expression in tumor cell lines is downregulated by treatment with HDACi. HeLa, SK-Mel-37, HepG2, Ramos, and K562 cells were treated for 16 hours with the pan-HDACi SAHA or solvent control (DMSO). (A) Treated cells were stained with anti–B7-H6 mAb 1.18, mIgG1 isotype control, NKp30-FP, NKG2D-FP, and a control-FP for flow cytometry. Dead cells were excluded by gating on 7-AAD− cells. Representative histograms of at least 2 independent experiments are shown. The mean geometrical mean and % reduction in B7-H6 surface expression after SAHA treatment ± standard deviation (SD) from at least four 4 experiments is shown. **P ≤ .01; ***P ≤ .001. (B) B7-H6 mRNA expression in treated cell lines was determined by qRT-PCR. Mean (expression of B7-H6 relative to GAPDH) and % reduction in B7-H6 mRNA expression after SAHA treatment ± SD of duplicates is shown for 1 representative experiment out of 2. The values from DMSO treatment were set as 100%.
B7-H6 expression in tumor cell lines is downregulated by treatment with HDACi. HeLa, SK-Mel-37, HepG2, Ramos, and K562 cells were treated for 16 hours with the pan-HDACi SAHA or solvent control (DMSO). (A) Treated cells were stained with anti–B7-H6 mAb 1.18, mIgG1 isotype control, NKp30-FP, NKG2D-FP, and a control-FP for flow cytometry. Dead cells were excluded by gating on 7-AAD− cells. Representative histograms of at least 2 independent experiments are shown. The mean geometrical mean and % reduction in B7-H6 surface expression after SAHA treatment ± standard deviation (SD) from at least four 4 experiments is shown. **P ≤ .01; ***P ≤ .001. (B) B7-H6 mRNA expression in treated cell lines was determined by qRT-PCR. Mean (expression of B7-H6 relative to GAPDH) and % reduction in B7-H6 mRNA expression after SAHA treatment ± SD of duplicates is shown for 1 representative experiment out of 2. The values from DMSO treatment were set as 100%.
B7-H6 expression is reduced by inhibitors of class I HDACs and by knockdown of HDAC2 or HDAC3
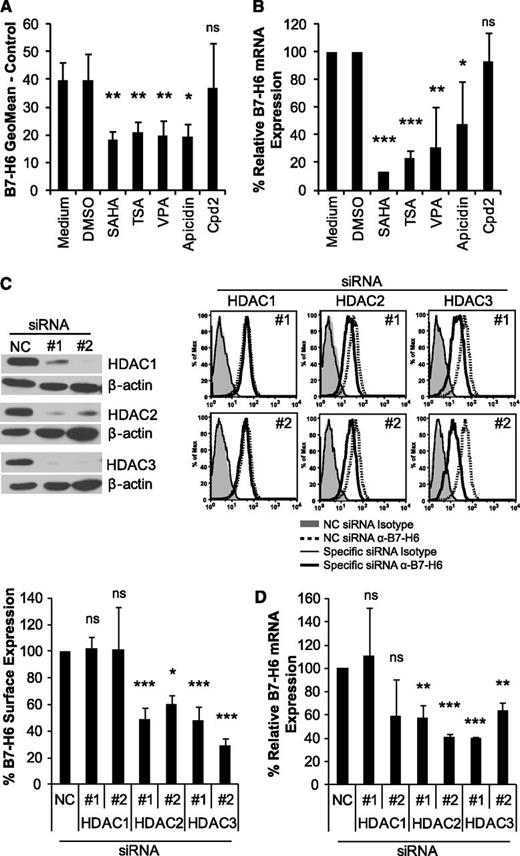
Next, we investigated whether other HDACi downregulate B7-H6 expression. Indeed, the pan-HDACi TSA led to a similar reduction of B7-H6 cell surface and mRNA expression in HeLa (Figure 3A-B) and Ramos cells (supplemental Figure 2) as SAHA. To further characterize the HDACs involved, HeLa and Ramos cells were treated with the class I–specific HDACi VPA or apicidin, which resulted in downregulation of B7-H6 to levels similar to those with the pan-HDACi (Figure 3A-B; supplemental Figure 2). In contrast, incubation with Cpd2, an inhibitor that targets the class I HDAC8,38 did not lead to significant downregulation, indicating that this HDAC was not involved in B7-H6 modulation. None of the inhibitors significantly affected cell viability (supplemental Figure 1B). To identify the class I HDAC that regulates B7-H6 expression, 2 siRNAs (1 and 2) each targeting HDAC1, HDAC2, or HDAC3, were transfected into HeLa cells. These siRNAs led to an almost complete knockdown of the respective HDACs at the mRNA (data not shown) and protein level compared with a negative control siRNA (Figure 3C). The expression of HDACs that were not targeted by the specific siRNAs was not significantly affected (data not shown). Importantly, knockdown of HDAC2 or HDAC3 reduced B7-H6 surface and mRNA expression to an extent similar to that found with the class I–specific HDACi, whereas knockdown of HDAC1 did not affect B7-H6 expression (Figure 3C-D). Similar results were obtained using the SK-Mel-37 cell line (data not shown). These data demonstrate that not only inhibition of class I HDACs by small molecular compounds but also siRNA-mediated knockdown of HDAC2 or HDAC3 lead to downregulation of B7-H6 in tumor cell lines.
B7-H6 expression is reduced by inhibitors of class I HDACs and by HDAC2 or HDAC3 knockdown. HeLa cells were treated for 16 hours with the following HDACi: pan-HDACi SAHA and TSA, class I–specific VPA, and apicidin as well as HDAC8-specific Cpd2. DMSO was used as a solvent control for all inhibitors except for VPA, which was dissolved in water. (A) HeLa cells were stained with anti-B7-H6 mAb 1.18 or isotype control for flow cytometry. Dead cells were excluded by gating on 7-AAD− cells. The mean geometric mean ± standard deviation (SD) of at least 3 independent experiments is shown. (B) B7-H6 mRNA expression in treated HeLa cells was determined by qRT-PCR. The mean (% expression of B7-H6 relative to GAPDH) ± SD is shown for at least 2 independent experiments. The values from medium (control for VPA) and DMSO (control for the other inhibitors) were set as 100%, respectively. (C) HeLa cells were transfected with 2 different siRNAs (1 and 2) targeting HDAC1, HDAC2, or HDAC3 and a negative control (NC) siRNA. Knockdown was confirmed by western blot with β-actin staining as a loading control. Cropped blot images are shown. For flow cytometry, cells were stained with anti-B7-H6 mAb 1.18 or isotype control as in A. Representative histograms and the mean % B7-H6 surface expression compared with NC siRNA (set as 100%) ± SD of at least 2 experiments are shown. (D) B7-H6 mRNA expression in siRNA-transfected HeLa cells was determined by qRT-PCR. Mean (% expression of B7-H6 relative to GAPDH) compared with the NC siRNA (set as 100%) ± SD is shown from at least 2 experiments. ns, not significant; *P ≤ .05; **P ≤ .01; and ***P ≤ .001.
B7-H6 expression is reduced by inhibitors of class I HDACs and by HDAC2 or HDAC3 knockdown. HeLa cells were treated for 16 hours with the following HDACi: pan-HDACi SAHA and TSA, class I–specific VPA, and apicidin as well as HDAC8-specific Cpd2. DMSO was used as a solvent control for all inhibitors except for VPA, which was dissolved in water. (A) HeLa cells were stained with anti-B7-H6 mAb 1.18 or isotype control for flow cytometry. Dead cells were excluded by gating on 7-AAD− cells. The mean geometric mean ± standard deviation (SD) of at least 3 independent experiments is shown. (B) B7-H6 mRNA expression in treated HeLa cells was determined by qRT-PCR. The mean (% expression of B7-H6 relative to GAPDH) ± SD is shown for at least 2 independent experiments. The values from medium (control for VPA) and DMSO (control for the other inhibitors) were set as 100%, respectively. (C) HeLa cells were transfected with 2 different siRNAs (1 and 2) targeting HDAC1, HDAC2, or HDAC3 and a negative control (NC) siRNA. Knockdown was confirmed by western blot with β-actin staining as a loading control. Cropped blot images are shown. For flow cytometry, cells were stained with anti-B7-H6 mAb 1.18 or isotype control as in A. Representative histograms and the mean % B7-H6 surface expression compared with NC siRNA (set as 100%) ± SD of at least 2 experiments are shown. (D) B7-H6 mRNA expression in siRNA-transfected HeLa cells was determined by qRT-PCR. Mean (% expression of B7-H6 relative to GAPDH) compared with the NC siRNA (set as 100%) ± SD is shown from at least 2 experiments. ns, not significant; *P ≤ .05; **P ≤ .01; and ***P ≤ .001.
Downregulation of B7-H6 by HDACi is associated with reduced B7-H6 reporter activity and decreased histone acetylation
HDAC1, HDAC2, and HDAC3 regulate histone acetylation and transcription by binding to promoters. To determine whether HDACi treatment affects B7-H6 promoter activity, luciferase reporter assays were performed by transfecting HeLa cells with a reporter construct containing the B7-H6 5′-UTR (−1270 to +208). Upon SAHA treatment, a significant decrease of luciferase activity compared with the DMSO solvent control was detected (Figure 4A). To determine whether HDAC1, HDAC2, and HDAC3 bind to the region in the B7-H6 5′-UTR containing the transcription start site, ChIP assays were performed. Binding of all 3 HDACs was detected in this region (Figure 4B). ChIP assays for acetylated histones H3 and H4 (acetyl-H3 and -H4) revealed that these histones were acetylated at the amplified region of the B7-H6 5′-UTR (Figure 4C). After 16 hours of treatment with SAHA, acetylation of histones H3 and H4 was strongly reduced (Figure 4C). In parallel, we detected hyperacetylation of these histones at the ULBP2 promoter after SAHA treatment (data not shown), as previously reported.28 The reduced histone acetylation upon SAHA treatment at the B7-H6 5′-UTR was paralleled with reduced binding of HDAC3 and the HAT CBP, whereas binding of HDAC1 and HDAC2 was not significantly changed (Figure 4D). Accordingly, treatment with the HATi curcumin and C646 also led to reduced B7-H6 cell surface expression in HeLa and SK-Mel-37 cells (supplemental Figure 3). In addition to transcriptional regulation, HDACi have been reported to affect mRNA stability.43,44 However, SAHA treatment did not further reduce B7-H6 mRNA levels when de novo transcription was blocked by actinomycin D, indicating that SAHA does not affect B7-H6 mRNA stability (Figure 4E). Together, these data show that HDACi treatment reduces B7-H6 reporter activity as well as the binding of HDAC3 and the HAT CBP to the B7-H6 5′-UTR and leads to local histone hypoacetylation.
Downregulation of B7-H6 by HDACi is associated with reduced B7-H6 reporter activity and decreased histone acetylation. (A) HeLa cells were transfected with a luciferase (GLuc) reporter construct containing the B7-H6 5′-UTR (−1270 to +208, see insert) or a VC followed by DMSO/SAHA treatment. The mean (fold change luciferase activity relative to VC) ± standard deviation (SD) is shown for 1 representative experiment out of 2. The value obtained in the VC sample was set as 1. (B) ChIP assays were performed using chromatin from HeLa cells for IP with the indicated mAbs or isotype control. Binding of HDAC1-3 to a region including the transcription start site in the B7-H6 5′-UTR (see insert in B) was assessed. The mean (signal relative to the input) ± SD of duplicates from 1 experiment out of 4 is shown. (C) Acetylated histone H3 and H4 were analyzed by ChIP 16 hours after DMSO or SAHA treatment. The mean (signal relative to the input) ± SD of duplicates from 1 experiment out of 3 is shown. (D) The binding of HDAC1-3 and the HAT CBP was analyzed by ChIP after DMSO/SAHA treatment. The mean (signal relative to the input) was calculated as: (signal with indicated mAb) – (signal with isotype control); ± SD from at least 3 experiments is shown. (E) B7-H6 mRNA levels were determined after DMSO/SAHA treatment in the presence of actinomycin D (ActD). The mean (% expression of B7-H6 relative to GAPDH) ± SD is shown for each time point after ActD addition of 2 experiments. The value before ActD treatment was set as 100%. ns, not significant; *P ≤ .05; ***P ≤ .001.
Downregulation of B7-H6 by HDACi is associated with reduced B7-H6 reporter activity and decreased histone acetylation. (A) HeLa cells were transfected with a luciferase (GLuc) reporter construct containing the B7-H6 5′-UTR (−1270 to +208, see insert) or a VC followed by DMSO/SAHA treatment. The mean (fold change luciferase activity relative to VC) ± standard deviation (SD) is shown for 1 representative experiment out of 2. The value obtained in the VC sample was set as 1. (B) ChIP assays were performed using chromatin from HeLa cells for IP with the indicated mAbs or isotype control. Binding of HDAC1-3 to a region including the transcription start site in the B7-H6 5′-UTR (see insert in B) was assessed. The mean (signal relative to the input) ± SD of duplicates from 1 experiment out of 4 is shown. (C) Acetylated histone H3 and H4 were analyzed by ChIP 16 hours after DMSO or SAHA treatment. The mean (signal relative to the input) ± SD of duplicates from 1 experiment out of 3 is shown. (D) The binding of HDAC1-3 and the HAT CBP was analyzed by ChIP after DMSO/SAHA treatment. The mean (signal relative to the input) was calculated as: (signal with indicated mAb) – (signal with isotype control); ± SD from at least 3 experiments is shown. (E) B7-H6 mRNA levels were determined after DMSO/SAHA treatment in the presence of actinomycin D (ActD). The mean (% expression of B7-H6 relative to GAPDH) ± SD is shown for each time point after ActD addition of 2 experiments. The value before ActD treatment was set as 100%. ns, not significant; *P ≤ .05; ***P ≤ .001.
B7-H6 mRNA levels positively correlate with HDAC3 levels in follicular lymphoma and HCC
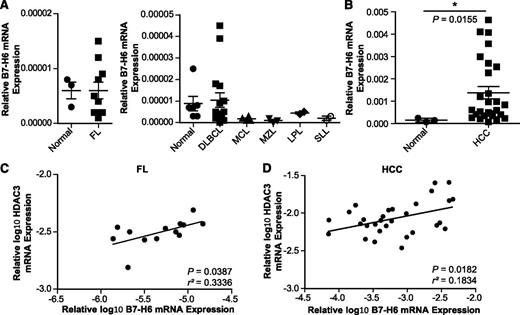
B7-H6 expression in primary tumors remains poorly explored. In our study, we analyzed B7-H6 mRNA expression in different lymphomas, including follicular lymphoma (FL, n = 10), diffuse large B-cell lymphoma (DLBCL, n = 16), mantle cell lymphoma (MCL, n = 4), marginal zone lymphoma (MZL, n = 3), lymphoplasmacytic and small lymphocytic lymphoma (LPL and SLL, each n = 2), as well as HCC45 (n = 27). Our data reveal that B7-H6 mRNA was detectable at low levels in healthy tissues consisting of normal lymph node or spleen (n = 6; Figure 5A) and liver (n = 3; Figure 5B), respectively. However, B7-H6 mRNA was elevated in a subset of the FL and DLBCL specimens analyzed (Figure 5A). In the other lymphoma samples, low levels of B7-H6 mRNA comparable to those found in the healthy controls were observed (Figure 5A). In HCC, B7-H6 mRNA levels were significantly elevated compared with normal liver controls (Figure 5B). Because HDACs are often overexpressed in tumors23 and because we found that HDAC3 affected B7-H6 expression, HDAC3 transcript levels in primary tumor samples were determined. Indeed, we observed a moderate but significant positive correlation of B7-H6 and HDAC3 mRNA expression levels in FL and HCC (Figure 5C-D), whereas no significant correlation was observed in DLBCL (data not shown). Also, HDAC1 and HDAC2 mRNA levels did not correlate with B7-H6 levels (data not shown). These data suggest that B7-H6 mRNA levels correlate with HDAC3 mRNA levels in certain lymphomas and liver cancer samples.
B7-H6 mRNA levels positively correlate with HDAC3 levels in follicular lymphoma and HCC. B7-H6 mRNA expression was quantified by qRT-PCR in samples of (A) FL (n = 10), DLBCL (n = 16), MCL (n = 4), MZL (n = 3), LPL and SLL (each n = 2), normal lymph node or spleen tissue (n = 6) as well as (B) HCCs (n = 27) and normal liver tissues (n = 3). The mean (expression of B7-H6 relative to GAPDH and HDAC3 relative to β-actin) ± standard error of the mean is shown. The statistical significance determined by the Mann-Whitney test with *P ≤ .05 is indicated. (C-D) Correlation between B7-H6 and HDAC3 transcript levels of samples analyzed in A and B. Mean (log10-transformed expression relative to GAPDH or β-actin) is shown for each sample. The statistical significance and r squared as determined by Pearson correlation are indicated.
B7-H6 mRNA levels positively correlate with HDAC3 levels in follicular lymphoma and HCC. B7-H6 mRNA expression was quantified by qRT-PCR in samples of (A) FL (n = 10), DLBCL (n = 16), MCL (n = 4), MZL (n = 3), LPL and SLL (each n = 2), normal lymph node or spleen tissue (n = 6) as well as (B) HCCs (n = 27) and normal liver tissues (n = 3). The mean (expression of B7-H6 relative to GAPDH and HDAC3 relative to β-actin) ± standard error of the mean is shown. The statistical significance determined by the Mann-Whitney test with *P ≤ .05 is indicated. (C-D) Correlation between B7-H6 and HDAC3 transcript levels of samples analyzed in A and B. Mean (log10-transformed expression relative to GAPDH or β-actin) is shown for each sample. The statistical significance and r squared as determined by Pearson correlation are indicated.
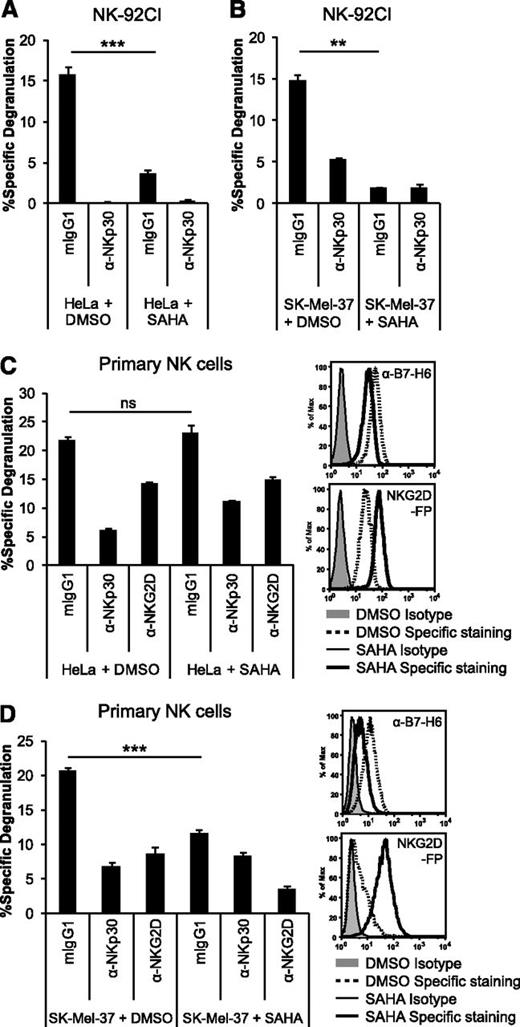
Downregulation of B7-H6 by HDACi impairs NKp30-dependent degranulation by NK cells
Finally, we investigated the effect of HDACi-mediated downregulation of B7-H6 on tumor cell recognition by NK cells. After pretreatment of HeLa cells with SAHA or DMSO solvent control, cells were washed and cocultured with NK-92CI effector cells that express high levels of NKp30 (supplemental Figure 4A). The degranulation (Figure 6A) and cytotoxicity (supplemental Figure 4B) of NK-92CI cells were strongly reduced in cocultures with HeLa cells pretreated with SAHA. Blocking with an anti-NKp30 mAb abrogated degranulation, indicating that HeLa cells were primarily recognized via NKp30 (Figure 6A). Similar results were obtained with the melanoma cell line SK-Mel-37 (Figure 6B) and the leukemia cell line K562 (supplemental Figure 4C).
Downregulation of B7-H6 by HDACi leads to reduced NKp30-dependent degranulation by NK cells. Degranulation of NK-92CI cells in (A-B) or primary NK cells activated with IL-2 in (C-D), in a 4-hour coculture at an 1:1 ratio with HeLa (A,C), or SK-Mel-37 cells (B,D) pretreated with the HDACi SAHA or DMSO solvent control is depicted. NK-cell receptors were blocked with the indicated mAbs. The mean (% specific degranulation) of triplicates ± standard deviation from 1 representative experiment with NK-92CI or of 1 NK-cell donor out of 3 is shown. Representative histograms show the surface staining using anti-B7-H6 mAb 1.18, mIgG1 isotype control, NKG2D-FP, or control-FP of the targets after pretreatment. ns, not significant; **P ≤ .01; ***P ≤ .001.
Downregulation of B7-H6 by HDACi leads to reduced NKp30-dependent degranulation by NK cells. Degranulation of NK-92CI cells in (A-B) or primary NK cells activated with IL-2 in (C-D), in a 4-hour coculture at an 1:1 ratio with HeLa (A,C), or SK-Mel-37 cells (B,D) pretreated with the HDACi SAHA or DMSO solvent control is depicted. NK-cell receptors were blocked with the indicated mAbs. The mean (% specific degranulation) of triplicates ± standard deviation from 1 representative experiment with NK-92CI or of 1 NK-cell donor out of 3 is shown. Representative histograms show the surface staining using anti-B7-H6 mAb 1.18, mIgG1 isotype control, NKG2D-FP, or control-FP of the targets after pretreatment. ns, not significant; **P ≤ .01; ***P ≤ .001.
Next, we assessed the impact of SAHA-mediated B7-H6 downregulation on the degranulation of primary IL-2–activated NK cells from healthy donors. Previous publications have shown that HDACi treatment induces NKG2D ligands on tumor cells, resulting in enhanced NKG2D-dependent effector functions.27-31 Indeed, upon SAHA treatment, HeLa cells upregulated NKG2D ligands while B7-H6 was downregulated (Figure 6C). Coculture of primary NK cells with HeLa cells resulted in degranulation that was largely inhibited by NKp30 blocking (72.1% ± 3.6% inhibition) and to a lesser extent by NKG2D blocking (Figure 6C). After SAHA treatment, similar overall degranulation was observed, but NKp30-dependent degranulation was significantly reduced, as indicated by reduced NKp30 blockade (51.3% ± 0.7% inhibition). In SK-Mel-37 cells, SAHA treatment also resulted in upregulation of NKG2D ligands and downregulation of B7-H6. Importantly, pretreatment of this cell line with SAHA led to reduced overall degranulation of the NK cells compared with the DMSO-treated cells (Figure 6D). Together, these data demonstrate that the downregulation of B7-H6 by SAHA inhibits NKp30-dependent activation of the NK cell line NK-92CI as well as primary NK cells.
Discussion
The novel B7 family member B7-H6 has recently been identified as an activating ligand on tumor cells for the NK-cell receptor NKp30. The aim of our study was to investigate mechanisms of B7-H6 regulation. We show that B7-H6 expression in tumor cell lines of different origin was downregulated by HDACi treatment, resulting in impaired NKp30-mediated NK-cell effector function.
Using 2 B7-H6–specific mAbs generated in our laboratory, we found a similar expression pattern on tumor cells and absence on healthy PBMCs as previously described using a polyclonal anti–B7-H6 antibody.13 We observed elevated B7-H6 mRNA levels in a subset of samples of primary human FL, DLBCL, and HCC. Similarly, Brandt et al13 determined that only a small percentage of patients with certain hematological malignancies expressed B7-H6. Further studies with larger sample sizes are required for a comprehensive evaluation of B7-H6 expression in hematological malignancies. Currently, we are establishing protocols for the detection of B7-H6 by immunohistochemistry on tissue sections of various solid tumors as well as in healthy controls. It will be important to determine whether cell surface expression of B7-H6 is also elevated in lymphomas and HCC compared with healthy tissues and if B7-H6 could serve as a potential drug target in these malignancies.
Our data reveal that treatment with pan- and class I HDACi, as well as siRNA-mediated knockdown of class I HDAC2 or HDAC3, leads to downregulation of B7-H6 on the cell surface and at the mRNA level in different tumor cell lines. These results suggest that HDAC2 and HDAC3 are required for B7-H6 expression in these tumor cells. Although downregulation of B7-H6 mRNA upon SAHA treatment was similar in all cell lines tested, downregulation of cell surface B7-H6 was most pronounced in Ramos cells. Therefore, it is likely that in Ramos cells additional posttranscriptional mechanisms, for example, lower B7-H6 protein stability, exist. Importantly, our data reveal a positive correlation of HDAC3 and B7-H6 mRNA expression in primary FL and HCC samples. Thus, HDAC3 might affect the expression of B7-H6 in certain primary tumors and be involved in its induction or regulation during tumorigenesis. HDAC2 levels did not correlate with B7-H6, suggesting that HDAC3 is more important in regulating B7-H6 expression in primary tumor cells. In DLBCL no significant correlation between HDAC3 and B7-H6 expression was observed (data not shown), indicating that additional mechanisms of B7-H6 regulation in these tumor cells exist. In contrast, HDACi were shown to induce ligands for the NKG2D receptor such as ULBPs and MICA.27-30 HDAC3 was shown to be involved in the repression of ULBP expression, whereas its knockdown led to induction of ULBPs in HeLa cells.28 Thus, HDAC3 differentially affects the expression of NKG2D and NKp30 ligands on tumor cells.
We show that HDAC1, HDAC2, and HDAC3 as well as the HAT CBP were bound to the region containing the B7-H6 transcription start site in HeLa cells, which is in line with published data on HDAC binding to promoters of actively transcribed genes.22 Treatment with SAHA led to dissociation of HDAC3 and CBP at the investigated region. In addition, histone acetylation was reduced in the B7-H6 5′-UTR, even though SAHA, like other HDACi, strongly enhances global histone acetylation levels.26,46 The observed histone hypoacetylation could be the result of reduced cooperative HDAC/HAT binding and activity at the B7-H6 5′-UTR. Indeed, our data revealed that the CBP/p300 inhibitors curcumin and C646 downregulated B7-H6 on the cell surface, demonstrating that HATs are involved in the regulation of B7-H6. The contribution of individual HATs to B7-H6 modulation requires further investigation. A similar mechanism has been proposed for other genes that are negatively regulated by HDACi.32,34 Local histone hypoacetylation induced by HDACi affects transcription by preventing recruitment of RNA polymerase II34,47 as well as by reducing the binding of transcription factors like Sp1.32,34 In our study, SAHA treatment did not affect B7-H6 mRNA stability; however, it did lead to the reduction of B7-H6 reporter activity in a luciferase reporter assay. This could be due to the ability of HDACi to enhance the acetylation of transcription factors themselves, which can modulate their DNA binding and activity.17 In future studies, it will be important to perform a detailed characterization of the B7-H6 promoter to identify the transcription factors that regulate B7-H6 expression and are involved in the regulation by HDACi.
Importantly, our results show that the downregulation of B7-H6 by the HDACi SAHA leads to reduced NKp30-dependent effector functions of the NK-92CI cell line as well as IL-2–activated NK cells from healthy donors. In these experiments, the outcome of NK-cell activation depended on the relative contribution of B7-H6 downregulation and NKG2D ligand induction in the tumor cells. In previous studies, an increase of overall NK-cell cytotoxicity in response to certain HDACi-treated target cells was reported.27-31,48 In these studies, effector populations that primarily exert NKG2D-dependent functions28,29 or target cells that express little or no B7-H6 on the cell surface27,30 were used, which contributed to the observed overall increase in NK-cell–mediated killing of the HDACi-treated targets. In other studies, treatment of leukemic blasts with the HDACi VPA increased their susceptibility to killing by KIR-mismatched NK cell lines in many but not all AML samples.31 Samples from only a small percentage of AML patients were reported to express B7-H6,13 suggesting that the NKp30/B7-H6 axis might play an important role in the killing of HDACi-treated blasts from only a subset of AML patients. Of note, IL-2 activation of NK cells was shown to be negatively affected by the presence of HDACi.49,50 In order to exclude this negative effect on NK cells in our study, target cells were washed extensively to remove the HDACi before the coculture. Additionally, we observed enhanced degranulation of NK cells in coculture with other HDACi-treated tumor cells including the leukemia cell line HL-60, as previously described.50 The functional outcome of B7-H6 modulation by HDACi on NK-cell activation could also be potentially affected by the differential expression of NKp30 isoforms in cancer patients, since 1 isoform (NKp30c) was described to be immunosuppressive.11
HDACi are currently in phase I and II clinical trials for the treatment of patients with different tumor entities such as lymphomas and HCC. It was anticipated that NK-cell responses would be enhanced by HDACi treatment by upregulation of NKG2D ligands.31,48 Our data, however, reveal that HDACi downregulate the activating ligand B7-H6 and thereby inhibit NKp30-mediated recognition of tumor cells. Thus, the NK ligands expressed by the tumor cells might determine the clinical outcome of potential HDACi treatment protocols combined with NK-cell–based therapy.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Carsten Watzl, Prof Helga Bernhard, Dr Thomas Hofmann, and Prof Christoph Plass for providing cell lines or reagents; Prof Annette Kopp-Schneider for advice on statistical analysis; and Prof Olaf Witt for valuable discussion. The authors also thank collaboration partners at Bayer Health Care and Dr Margareta Correia for critical reading of the manuscript.
This work was supported by the Deutsche José Carreras Leukämie-Stiftung e.V. (to A.C. and M.W.H.; November 2006), by the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum, and the Israel Ministry of Science and Technology (to A.C.). This study was also supported by the Helmholtz Alliance Immunotherapy of Cancer (to K.B.) and by the tissue bank of the National Center for Tumor Disease–Heidelberg (to K.B.).
Authorship
Contribution: N.F. designed and performed experiments, analyzed data, and wrote the manuscript; S.T. designed luciferase reporter assays and performed qPCR of primary HCC and liver cDNA; A.R. performed cytotoxicity assays; S.T. and A.A. generated B7-H6-FP; G.M., N.F., and A.A. generated monoclonal antibodies; G.M. performed enzyme-linked immunosorbent assays; I.O. provided reagents, protocols, and advice; K.B. provided primary HCC and liver cDNA; M.W.H. provided primary tumor material and valuable advice; and A.C. oversaw the project, designed experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adelheid Cerwenka, Innate Immunity Group, German Cancer Research Center, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; e-mail: a.cerwenka@dkfz.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal