Key Points
Increased mitochondrial ROS production, adaptation to intrinsic oxidative stress, and mitochondrial biogenesis are interconnected in CLL.
Targeting the respiratory chain and promoting mitochondrial ROS lead to selective cytotoxicity in CLL cells.
Abstract
Alterations of cellular metabolism represent a hallmark of cancer. Numerous metabolic changes are required for malignant transformation, and they render malignant cells more prone to disturbances in the metabolic framework. Despite the high incidence of chronic lymphocytic leukemia (CLL), metabolism of CLL cells remains a relatively unexplored area. The examined untreated CLL patients displayed a metabolic condition known as oxidative stress, which was linked to alterations in their lymphoid compartment. Our studies identified mitochondrial metabolism as the key source for abundant reactive oxygen species (ROS). Unlike in other malignant cells, we found increased oxidative phosphorylation in CLL cells but not increased aerobic glycolysis. Furthermore, CLL cells adapted to intrinsic oxidative stress by upregulating the stress-responsive heme-oxygenase-1 (HO-1). Our data implicate that HO-1 was, beyond its function as an antioxidant, involved in promoting mitochondrial biogenesis. Thus ROS, adaptation to ROS, and mitochondrial biogenesis appear to form a self-amplifying feedback loop in CLL cells. Taking advantage of the altered metabolic profile, we were able to selectively target CLL cells by PK11195. This benzodiazepine derivate blocks the mitochondrial F1F0-ATPase, leads to a surplus production of mitochondrial superoxide, and thereby induces cell death in CLL cells. Taken together, our findings depict how bioenergetics and redox characteristics could be therapeutically exploited in CLL.
Introduction
Metabolic reprogramming is a central feature of cancer cells and has emerged as an attractive target in novel therapeutic strategies.1 Chronic lymphocytic leukemia (CLL) is the leukemia with the highest incidence among adults in the Western countries. However, CLL cells’ metabolism remains a relatively unexplored area.
Results from few studies indicate that CLL cells might contribute to the so-called cancer-associated oxidative stress.2,3 This metabolic condition is regularly seen in cancer patients and results from accumulating reactive oxygen species (ROS).4 The underlying mechanisms of oxidative stress in cancer patients often remain elusive. Tumor cells5 as well as tumor-associated cells (eg, activated granulocytes6 ) can generate abundant ROS. Under physiological conditions, the mitochondrial respiratory chain is the main site of ROS production.7 Cancer-related mitochondrial alterations such as defective oxidative phosphorylation8 or an enhanced mitochondrial biogenesis9,10 promote (mitochondrial) ROS output.11 However, nonmitochondrial ROS production by the membrane-bound NADPH-oxidase (NOX) complex can also play a role in cancer. An enhanced NOX activity has actually been identified in several entities.12,13
Escalated ROS levels promote genetic instability and development of drug resistance, harness cell-signaling,4,14 and altogether account for cancer cells’ aggressive behavior.5 Oxidative stress additionally attenuates immune responses by leading to dysfunctions and even apoptosis of NK- and T-cells,15-17 suggesting a role in tumor immune-escape.18
Malignant cells on their part appear adapted to permanent (intrinsic) oxidative stress. They upregulate protective pathways,19 which in addition favor resistance toward anticancer agents.20,21
Increasing evidence suggests a connection between tumor-specific metabolism and a surplus of ROS production. Metabolic alterations can render malignant cells more susceptible to perturbations within the metabolic framework and therefore pose an opportunity for novel therapeutic interventions.1
We first sought to investigate the prevalence of oxidative stress in CLL patients and its systemic impact, namely the link to immune deregulations already found at early disease stages.22 Next, we focused on deciphering the metabolic processes responsible for the observed ROS overproduction. Using a comprehensive and multimodal approach, we identified an interrelationship between the altered mitochondrial metabolism in CLL cells, ROS generation, their adaptation to intrinsic oxidative stress, and mitochondrial biogenesis. Taking advantage of their peculiar bioenergetic profile, we were able to selectively target CLL cells by pharmacologically interfering with their mitochondrial metabolism.
A better understanding of the CLL-specific metabolic pathways is inevitable for developing novel targeted approaches. Agents directed against the unique metabolic features of CLL cells might represent an opportunity to improve current treatments and even to overcome drug resistance within cancer cells.
Material and methods
Human subjects
Samples were collected (during October 2011 through April 2013) on approval from the Ethics Committee of the University of Erlangen-Nuremberg (protocol number: 200_12) and after receiving patients’ written informed consent. This study was conducted in accordance with the Declaration of Helsinki. Peripheral blood mononuclear cells (PBMCs) and serum from a total of 75 untreated CLL patients and from 16 healthy donors (HDs) were collected prospectively (patients’ characteristics are detailed in supplemental Table 1).
Cell preparations
PBMCs were obtained using Ficoll-Paque (GE Healthcare, Piscataway Township, NJ). CLL cells and B-cells were purified by magnetic bead-based negative selection (B-cell Isolation Kit II, Miltenyi Biotec, Bergisch Gladbach, Germany).
Biomarker for oxidative stress
Products of DNA (8-hydroxydeoxyguanosine/8-OdHG, Cell Biolabs, San Diego, CA) and lipid (malondialdehyde/MDA, TBARS Assay Kit, Cayman Chemical, Ann Arbor, MI) oxidation were quantified in sera using a VersaMax ELISA (enzyme-linked immunosorbent assay) microplate reader (Molecular Devices, Sunnyvale, CA).
ATP levels
ATP content was assessed in B-cells and CLL cells using a colorimetric ATP Assay Kit (Abcam, Cambridge, United Kingdom).
NADPH measurement
NADPH levels were determined in 5 × 106 purified HD B-cells and CLL cells using a colorimetric NADP/NADPH assay kit (BioAssay Systems, Hayward, CA).
Antioxidant capacity
Antioxidant capacity was measured in cell lysates using a colorimetric Antioxidant Assay Kit (Cayman Chemical) following the manufacturer’s instructions.
Cytokines
The IFN-γ concentration was quantified using an IFN-γ Quantikine ELISA Kit (R&D, Minneapolis, MN).
NOX activity
Superoxide generation by membrane-bound NOX was measured in cytochrome c reduction assays as described previously23 using a Multiscan device (ThermoScientific, Waltham, MA).
Antibodies and flow cytometry (FACS)
Cells were stained according to the manufacturer's recommendations using fluorochrome-coupled antibodies (supplemental Table 2). Staining was performed in presence of Fc-receptor blocking solution (Biolegend, San Diego, CA). The LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Life Technologies, Carlsbad, CA) was used for dead-cell exclusion. Cell counts were determined using BD Trucount tubes (BD Biosciences, San Jose, CA). For intracellular staining, cells were treated with BD Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences). Cells were analyzed using an FACS Canto II cytometer (BD Biosciences) and the FlowJo Version 9.5 software (TreeStar, Ashland, OR).
Specific cell death
Viable cells are defined as the 7-AADneg/Annexin-Vneg cells in FACS. Percentage of specific cell death was calculated according to the following formula: specific cell death (%) = 100 × (% dead cells − % baseline dead cells) / (100% − % baseline dead cells).
Cellular ROS
Total and mitochondrial ROS were quantified by FACS using CellROX and MitoSOX dyes, respectively (Life Technologies). For evaluating the contribution of NOX and mitochondria to ROS production, cells were pretreated for 1 hour with 0.5 μM of the NOX inhibitor diphenyleneiodonium chloride (DPI, Sigma Aldrich) or 500 nM of the mitochondria-targeting antioxidant MitoQ (kindly provided by M. Murphy).
Glutathione levels
Glutathione was stained with Thioltracker Violet (Life Technologies) and was semiquantified using FACS.
Mitochondrial mass
Cells were stained with the mitochondrial specific dye MitoTracker Green (Life Technologies) according to the manufacturer’s instructions and were analyzed by FACS.
Mitochondrial membrane potential
Mitochondrial electrochemical membrane potential (ΔψM) was assessed using the potentiometric dye JC-1 (JC-1 Mitochondrial Assay Kit, Cayman Chemical). Cells were stained with JC-1 according to the manufacturer’s instructions for 15 minutes at 37°C followed by a FACS analysis.
DNA, RNA preparation, and quantitative polymerase chain reaction (qPCR)
RNA and DNA were extracted from cell lysates (AllPrep DNA/RNA/Protein Mini Kit, Qiagen, Hilden, Germany) and DNA was prepared (Superscript First Strand Synthesis System, Life Technologies) using a Mastercycler nexus (Eppendorf, Hamburg, Germany). The mRNA levels were quantified by qPCR (Quantitect SYBR Green PCR Kit; Qiagen) on a Rotor Gene Q (Qiagen). Relative gene expression was determined by normalizing the expression of each target gene to β2-microglobulin and/or β-actin using gene-specific primers (supplemental Table 3).
Extracellular flux assays
Bioenergetics of CLL cells and B-cells were determined using the XF96e Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA). Cells were seeded in specialized tissue culture plates in an optimized concentration of 240,000 cells per well and were subsequently immobilized using CELL-TAK (BD Biosciences). One hour before measurement, cells were incubated at 37°C in a CO2-free atmosphere. First, basal oxygen consumption rate (OCR) (an indicator for mitochondrial respiration) and extracellular acidification rate (ECAR) (an indicator for lactic acid production or glycolysis) were detected. Next, OCR and ECAR responses toward the application of oligomycin (1 µM), FCCP (2.5 µM), and the combination of antimycin (3 µM) and rotenone (3 µM) (XF Cell Mito Stress Test Kit, Seahorse Bioscience) as well as glucose and PK11195 (Sigma Aldrich) were evaluated. All experiments were performed in at least hexaplicates.
Fluorescence microscopy of mitochondria
Cells were stained with anti-CD19-APC (BD Biosciences). Mitochondria were visualized using MitoTracker Green FM (Life Technologies), and Hoechst 33258 (Sigma Aldrich) was added for nuclear staining. Cells were immediately analyzed using a fluorescence microscope (Axio-Imager M2, Zeiss, Jena, Germany) at a 630-fold magnification.
Transmission electron microscopy (TEM) of mitochondria
CLL cells and B-cells were pelleted and prepared for TEM as described previously24 with 1 modification: after fixation with osmium tetroxide, cells were washed and the pellets were embedded in agar (2% in H2O). Following hardening on ice, the agar block was cut in pieces and washed before performing dehydration. Samples were analyzed and photographed with a Zeiss EM10 electron microscope (Zeiss) and a Gatan SC1000 OriusTM CCD camera in combination with the Digital Micrograph TM software (Gatan, Pleasanton, CA).
Reagents
Dimethyl sulfoxide, phorbol-12-myristate-13-acetate (PMA), N-acetyl-cysteine (NAC), cytochrome c, and PK11195 were obtained from Sigma Aldrich, Ca2+ and Mg2+ from PAA (Piscataway, NJ), NADPH from Roth (Karlsruhe, Germany), and tin-protoporphyrin (SnPP) from Frontier Scientific (Logan, UT).
Statistical analyses
Differences in means were evaluated with parametric (2-tailed Student or paired t test) or nonparametric (Mann-Whitney U or Wilcoxon) tests based on the distribution levels. All statistical analyses were performed using GraphPad Prism Version 5 (GraphPad Prism Software Inc.) at a significance level of P < .05.
Results
Oxidative stress in CLL patients
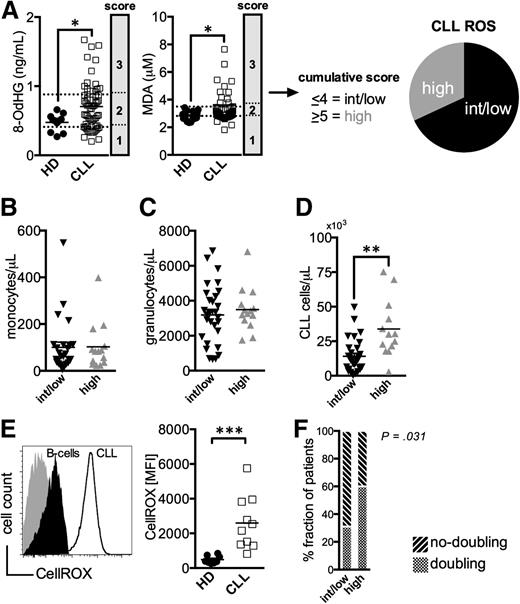
First, we evaluated whether CLL patients display signs of oxidative stress. Because chemotherapy induces ROS,25 only untreated patients were included in all our investigations. Established surrogate markers for ROS, namely products of DNA- (8-OHdG) and lipid-oxidation (MDA), were measured in patients’ and HDs’ sera. As anticipated, increased levels of oxidized biomolecules were prevalent in the CLL patients when compared with controls (Figure 1A). CLL samples were scored according to the 8-OdHG and MDA concentrations and were grouped into a cohort of intermediate/low (ROSint/low) and high ROS (ROShigh) levels (Figure 1A).
Increased markers for oxidative stress in untreated CLL patients. (A) Serum levels of 8-OdHG and MDA were quantified in HDs (n = 11-15) and CLL patients (n = 63) by ELISA. CLL samples were scored according to the percentile rank of the measured 8-OdHG and MDA concentrations (25th percentile: 1 point, 50th percentile: 2 points, 75th percentile: 3 points), respectively. Based on the cumulative score, patients were grouped into a cohort with intermediate/low (<4, ROSint/low) and high ROS (>5, ROShigh) levels. The absolute number of (B) CD14+ monocytes, of (C) granulocytes, and of (D) CD5+CD19+CD23+ CLL cells per μL peripheral blood of ROSint/low and ROShigh patients as measured by FACS. (E) A representative histogram of a FACS-analysis showing healthy B-cells (filled black) and CLL cells (black line) stained with a ROS-sensitive dye (CellROX) and an unstained control (filled gray). An overview of the CellROX-mean fluorescence intensity (MFI) in freshly isolated regular B-cells (n = 8) and CLL cells (n = 10) depicted as a scatter plot. (F) The proportion of patients with and without lymphocyte doubling among the ROSint/low and ROShigh groups is shown. The statistical analysis was performed using a χ2-test. Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001.
Increased markers for oxidative stress in untreated CLL patients. (A) Serum levels of 8-OdHG and MDA were quantified in HDs (n = 11-15) and CLL patients (n = 63) by ELISA. CLL samples were scored according to the percentile rank of the measured 8-OdHG and MDA concentrations (25th percentile: 1 point, 50th percentile: 2 points, 75th percentile: 3 points), respectively. Based on the cumulative score, patients were grouped into a cohort with intermediate/low (<4, ROSint/low) and high ROS (>5, ROShigh) levels. The absolute number of (B) CD14+ monocytes, of (C) granulocytes, and of (D) CD5+CD19+CD23+ CLL cells per μL peripheral blood of ROSint/low and ROShigh patients as measured by FACS. (E) A representative histogram of a FACS-analysis showing healthy B-cells (filled black) and CLL cells (black line) stained with a ROS-sensitive dye (CellROX) and an unstained control (filled gray). An overview of the CellROX-mean fluorescence intensity (MFI) in freshly isolated regular B-cells (n = 8) and CLL cells (n = 10) depicted as a scatter plot. (F) The proportion of patients with and without lymphocyte doubling among the ROSint/low and ROShigh groups is shown. The statistical analysis was performed using a χ2-test. Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001.
Production of ROS is a hallmark of myeloid cells, and their accumulation can lead to oxidative stress.6 Numbers of monocytes and neutrophils were comparable in ROShigh and ROSint/low patients (Figure 1B-C). Actually, higher ROS levels correlated with the number of circulating CLL cells (Figure 1D). This finding could reflect the higher ROS production in CLL cells compared with B-cells (Figure 1E). In addition, a larger fraction of ROShigh patients had experienced lymphocyte doubling (Figure 1F, supplemental Figure 1).
Lymphocyte alterations in patients with high ROS levels
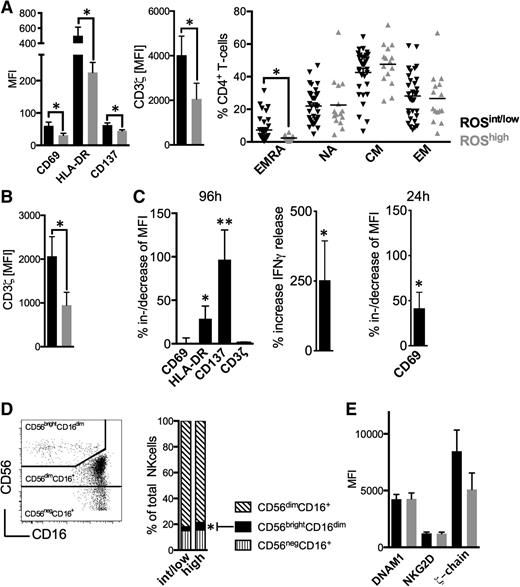
Next, we analyzed the lymphocyte compartment in conjunction with ROS levels. ROShigh patients had CD4+ T-cells of a less activated phenotype accompanied by a reduced CD3ζ-chain expression, and less circulating terminally differentiated effector memory cells (Figure 2A). Furthermore, CD8+ T-cells of ROShigh patients displayed significantly lower CD3ζ-chain levels (Figure 2B, supplemental Figures 3 and 4). Number of T-cells, CD4+/CD8+ T-cell ratio, and proliferative status were not different (supplemental Figures 4-6). These alterations are in line with reports on ROS effects on T-cells26 and observations that CD4+ T-cells are particularly deregulated in CLL.22 The addition of the antioxidant NAC when stimulating T-cells in the presence of CLL cells significantly bolstered T-cell activation as exemplified by an increased expression of HLA-DR, CD137 (both after 96 hours), CD69 (after 24 hours as an early activation marker), and IFN-γ production (Figure 2C). It is well established that NK-cells are sensitive toward ROS.16 We observed that increased ROS levels were associated with higher proportions of CD56brightCD16dim NK-cells (Figure 2D). Circulating NK-cells (supplemental Figure 7), expression of activating molecules such as DNAM1 and NKG2D, and CD3ζ-chain (Figure 2E) were similar in ROShigh and ROSint/low patients.
Oxidative stress-associated immune alterations in CLL patients. (A) The MFI of CD69, HLA-DR, CD137, CD3ζ-chain, and the maturation status of CD4+ T-cells were assessed in ROSint/low and ROShigh patients using FACS. (B) The mean MFI of the CD3ζ-chain on CD8+ T-cells as measured by FACS in ROSint/low and ROShigh patients. (C) The %al MFI-increase of CD69, HLA-DR, CD137, and CD3ζ-chain on (in presence of CLL cells) anti-CD2, anti-CD3, and anti-CD28 bead-activated CD4+ T-cells on addition of 2 mM NAC (n = 12) and the increase of IFNγ release (n = 7) as measured by FACS and ELISA, respectively, after 96 hours of culture. The %al MFI-increase of CD69 as assessed by FACS also after 24 hours (n = 5). (D) A representative FACS analysis of NK-cell subsets (left panel). The mean proportion of the NK-cell subpopulations in ROSint/low and ROShigh patients is shown as a column diagram (right panel). (E) The MFI of DNAM1, NKG2D, and CD3ζ-chain on NK-cells as measured by FACS in ROSint/low and ROShigh patients. Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005. CM, central memory; EM, effector memory; EMRA, terminally differentiated effector memory cells; NA, naïve.
Oxidative stress-associated immune alterations in CLL patients. (A) The MFI of CD69, HLA-DR, CD137, CD3ζ-chain, and the maturation status of CD4+ T-cells were assessed in ROSint/low and ROShigh patients using FACS. (B) The mean MFI of the CD3ζ-chain on CD8+ T-cells as measured by FACS in ROSint/low and ROShigh patients. (C) The %al MFI-increase of CD69, HLA-DR, CD137, and CD3ζ-chain on (in presence of CLL cells) anti-CD2, anti-CD3, and anti-CD28 bead-activated CD4+ T-cells on addition of 2 mM NAC (n = 12) and the increase of IFNγ release (n = 7) as measured by FACS and ELISA, respectively, after 96 hours of culture. The %al MFI-increase of CD69 as assessed by FACS also after 24 hours (n = 5). (D) A representative FACS analysis of NK-cell subsets (left panel). The mean proportion of the NK-cell subpopulations in ROSint/low and ROShigh patients is shown as a column diagram (right panel). (E) The MFI of DNAM1, NKG2D, and CD3ζ-chain on NK-cells as measured by FACS in ROSint/low and ROShigh patients. Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005. CM, central memory; EM, effector memory; EMRA, terminally differentiated effector memory cells; NA, naïve.
NOX activity and antioxidant capacity of CLL cells
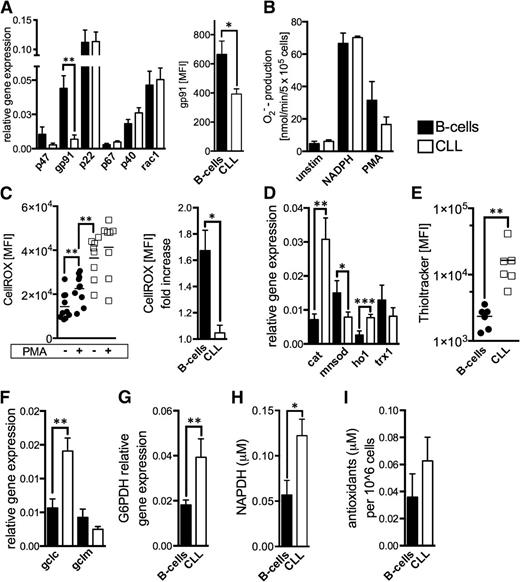
Malignant cells (eg, melanoma13 or chronic myelogenous leukemia27 ) can generate large amounts of superoxide through an increased activity of NOX. We evaluated NOX components in CLL cells and B-cells and found that the gp91 subunit is significantly lower expressed in CLL cells (Figure 3A). We measured superoxide production by NOX based on extracellular cytochrome c reduction.23 Superoxide levels were similar in B-cells and CLL cells before (baseline) and after the addition of a NOX substrate (ie, NADPH) as well as on respiratory burst induction by PMA (Figure 3B). The respiratory burst elicited only in B-cells a significant increase of cellular ROS (Figure 3C), which further corroborates the notion that NOX is not responsible for the ROS overproduction seen in CLL cells.
NADPH-oxidase activity and antioxidant machinery in CLL cells. (A) Relative gene expression of the NOX subunits is shown comparatively for purified HD B-cells (n = 15) and CLL cells (n = 22). Differences of the relative gene expression of gp91 were validated using FACS and determining the gp91 MFI of normal B-cells (n = 5) and CLL cells (n = 5). (B) Superoxide anions generated by the membrane-bound NOX were quantified based on cytochrome c reduction in purified HD B-cells (n = 4) and CLL cells (n = 5) at baseline conditions (unstim), on addition of the NOX-substrate NADPH, and after direct NOX activation using PMA. (C) Intracellular ROS levels are shown (left panel) as the CellROX-MFI in purified HD B-cells (n = 10) and CLL cells (n = 8) with and without the addition of NOX-activating PMA. The mean fold-increase of CellROX-MFI in B-cells and CLL cells on NOX activation is displayed (right panel). (D) The relative gene expression of key cellular antioxidants (catalase, cat; manganese superoxide dismutase, mnsod; heme-oxygenase-1, HO1; thioredoxin-1, trx1) is shown comparatively for purified HD B-cells (n = 10) and CLL cells (n = 22) as quantified by qPCR. (E) Cellular glutathione (GSH) levels were detected in HD B-cells (n = 6) and CLL cells (n = 6) using the Thioltracker probe and were semiquantified based on the Thioltracker-MFI by FACS. (F) The relative gene expression of key molecules involved in GSH-synthesis and (G) the regeneration of reduced GSH were evaluated in purified HD B-cells (n = 8-10) and CLL cells (n = 10-17). (H) Mean intracellular levels of the important cellular reducing molecule NADPH that is produced within the pentose phosphate pathway are shown for purified HD B-cells (n = 3) and CLL cells (n = 4) as measured by ELISA. (I) The overall antioxidant capacity was measured in lysates from normal B-cells (n = 7) and CLL cells (n = 9). Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001. GCLC, catalytic subunit of the glutamate-cysteine ligase; GCLM, modifier subunit of the glutamate-cysteine ligase; G6PDH, glucose-6-phosphate dehydrogenase.
NADPH-oxidase activity and antioxidant machinery in CLL cells. (A) Relative gene expression of the NOX subunits is shown comparatively for purified HD B-cells (n = 15) and CLL cells (n = 22). Differences of the relative gene expression of gp91 were validated using FACS and determining the gp91 MFI of normal B-cells (n = 5) and CLL cells (n = 5). (B) Superoxide anions generated by the membrane-bound NOX were quantified based on cytochrome c reduction in purified HD B-cells (n = 4) and CLL cells (n = 5) at baseline conditions (unstim), on addition of the NOX-substrate NADPH, and after direct NOX activation using PMA. (C) Intracellular ROS levels are shown (left panel) as the CellROX-MFI in purified HD B-cells (n = 10) and CLL cells (n = 8) with and without the addition of NOX-activating PMA. The mean fold-increase of CellROX-MFI in B-cells and CLL cells on NOX activation is displayed (right panel). (D) The relative gene expression of key cellular antioxidants (catalase, cat; manganese superoxide dismutase, mnsod; heme-oxygenase-1, HO1; thioredoxin-1, trx1) is shown comparatively for purified HD B-cells (n = 10) and CLL cells (n = 22) as quantified by qPCR. (E) Cellular glutathione (GSH) levels were detected in HD B-cells (n = 6) and CLL cells (n = 6) using the Thioltracker probe and were semiquantified based on the Thioltracker-MFI by FACS. (F) The relative gene expression of key molecules involved in GSH-synthesis and (G) the regeneration of reduced GSH were evaluated in purified HD B-cells (n = 8-10) and CLL cells (n = 10-17). (H) Mean intracellular levels of the important cellular reducing molecule NADPH that is produced within the pentose phosphate pathway are shown for purified HD B-cells (n = 3) and CLL cells (n = 4) as measured by ELISA. (I) The overall antioxidant capacity was measured in lysates from normal B-cells (n = 7) and CLL cells (n = 9). Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001. GCLC, catalytic subunit of the glutamate-cysteine ligase; GCLM, modifier subunit of the glutamate-cysteine ligase; G6PDH, glucose-6-phosphate dehydrogenase.
Suppression of antioxidant molecules28 can also lead to increased ROS levels in cancer cells. We assessed major antioxidants and found in CLL cells a significantly increased expression of catalase and heme oxygenase-1 (HO-1), whereas manganese superoxide dismutase (MnSOD) located in the mitochondria was found reduced (Figure 3D). Thiols represent the major nonenzymatic antioxidant scavenging ROS. CLL cells possessed significantly more thiols (Figure 3E) and expressed higher levels of the catalytic subunit of glutamate-cysteine ligase (GCLC) and glucose-6-phosphate dehydrogenase (G6PDH) (Figure 3F-G). GCLC is the rate-limiting enzyme for glutathione synthesis, the most abundant thiol. G6PDH is the key enzyme of the pentose phosphate pathway, which provides the principal cellular reductant NADPH (Figure 3H), which is necessary for glutathione recycling. Overall, redox-buffering capacity of CLL cells appears increased by trend (Figure 3I). In fact, these seemingly contradictory findings could be explained by a compensatory adaptation of CLL cells to persistent oxidative stress.4
Mitochondrial ROS levels and mitochondrial biogenesis in CLL cells
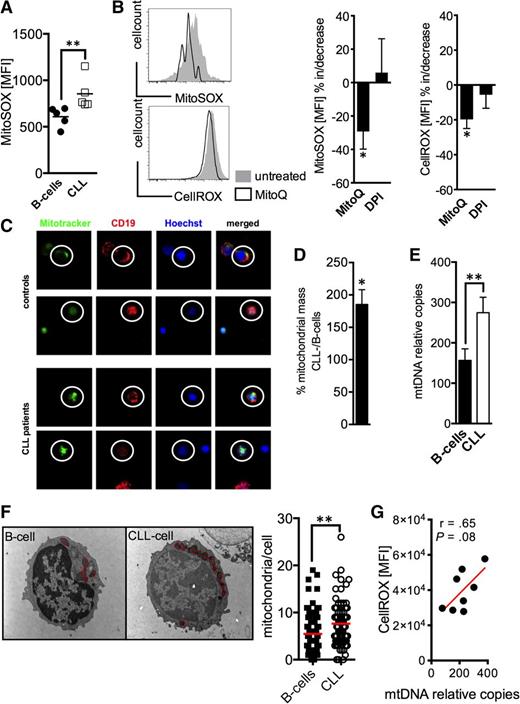
Complexes I and III of the respiratory chain are considered the main generators of mitochondrial ROS.11 Using a probe that selectively detects mitochondrial superoxide, we found significantly increased levels in CLL cells (Figure 4A). MnSOD plays a decisive role in metabolizing superoxide. Therefore, reduced MnSOD expression (Figure 3D) could contribute to increased mitochondrial ROS in CLL cells. To dissect better the respective role of mitochondria and NOX as ROS sources, we treated CLL cells with MitoQ and DPI, respectively. MitoQ is a mitochondria-targeted derivate of the antioxidant ubiquinone.29 DPI at the used dosage blocks NOX enzymes without affecting mitochondrial ROS production.30 Only MitoQ led to a significant reduction (of mitochondrial and at the same time) of total cellular ROS (Figure 4B). These data strongly point to mitochondria as the source of ROS overproduction in CLL cells (Figure 1E), supporting our previous results (Figure 3).
Mitochondrial ROS and mitochondrial biogenesis in CLL cells. (A) Mitochondrial-specific ROS production was detected by the MitoSOX probe and was semiquantified based on the MFI of MitoSOX by FACS in HD B-cells (n = 5) and CLL cells (n = 5). (B) (left panels) 2 representative histograms of a FACS analysis of CLL cells stained with MitoSOX (for mitochondrial ROS) and CellROX (for total cellular ROS) with (black line)/without (filled gray) MitoQ treatment. The mean increase/decrease of the MitoSOX- and CellROX-MFI is shown for CLL cells (n = 5) treated with MitoQ or diphenylen iodonium (DPI) blocking NOX activity. (C) Mitochondrial mass was visualized using Mitotracker (green) in CD19+ (red, encircled) B-cells and CLL cells by fluorescence microscopy as shown for 2 HDs and CLL patients, respectively. Samples were counterstained with the vital nuclear dye Hoechst (blue). (D-E) The 2 panels display the semiquantification of the mitochondrial mass in normal B-cells (n = 6-10) and CLL cells (n = 6-10) based on the CLL cell/B-cell quotient (in %) of the Mitotracker-MFI (left panel) and the relative (to nuclear DNA) mitochondrial DNA (mtDNA) copy number (right panel). (F) A representative electron-microscopic depiction of the increased number of mitochondria (red circles) in B-cells and CLL cells. The mean number of mitochondria per B-cell and CLL cell is shown as a dot plot. Thirty cells per donor (n = 3 for each cohort) were analyzed using electron microscopy. (G) Total cellular ROS as determined based on the CellROX MFI in CLL cells measured by FACS was correlated with the according mtDNA content as quantified by real time PCR. Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001.
Mitochondrial ROS and mitochondrial biogenesis in CLL cells. (A) Mitochondrial-specific ROS production was detected by the MitoSOX probe and was semiquantified based on the MFI of MitoSOX by FACS in HD B-cells (n = 5) and CLL cells (n = 5). (B) (left panels) 2 representative histograms of a FACS analysis of CLL cells stained with MitoSOX (for mitochondrial ROS) and CellROX (for total cellular ROS) with (black line)/without (filled gray) MitoQ treatment. The mean increase/decrease of the MitoSOX- and CellROX-MFI is shown for CLL cells (n = 5) treated with MitoQ or diphenylen iodonium (DPI) blocking NOX activity. (C) Mitochondrial mass was visualized using Mitotracker (green) in CD19+ (red, encircled) B-cells and CLL cells by fluorescence microscopy as shown for 2 HDs and CLL patients, respectively. Samples were counterstained with the vital nuclear dye Hoechst (blue). (D-E) The 2 panels display the semiquantification of the mitochondrial mass in normal B-cells (n = 6-10) and CLL cells (n = 6-10) based on the CLL cell/B-cell quotient (in %) of the Mitotracker-MFI (left panel) and the relative (to nuclear DNA) mitochondrial DNA (mtDNA) copy number (right panel). (F) A representative electron-microscopic depiction of the increased number of mitochondria (red circles) in B-cells and CLL cells. The mean number of mitochondria per B-cell and CLL cell is shown as a dot plot. Thirty cells per donor (n = 3 for each cohort) were analyzed using electron microscopy. (G) Total cellular ROS as determined based on the CellROX MFI in CLL cells measured by FACS was correlated with the according mtDNA content as quantified by real time PCR. Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001.
Next, we were interested in whether mitochondrial mass was altered in CLL cells. Using various modalities we found a substantial increase of mitochondrial mass, of relative mitochondrial DNA copies, and of mitochondrial number in CLL cells (Figure 4C-F), which is in line with previous reports.10 An increased mitochondrial biogenesis could by itself at least contribute to cellular oxidative stress (Figure 4G), because mitochondria represent the main source for (basal) ROS.
Increased respiratory rates and respiratory capacity in CLL cells
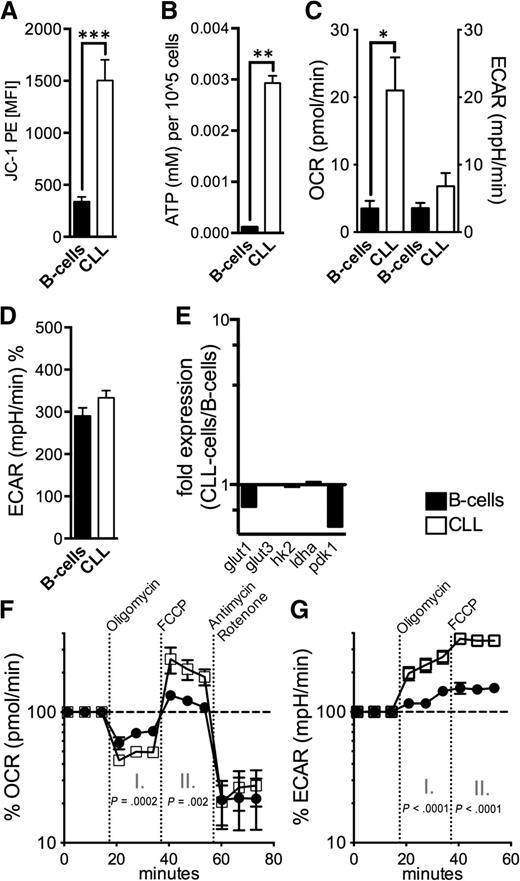
Defective mitochondria promote a compensatory increase of mitochondrial biogenesis.7 For assessing mitochondrial fitness we measured the mitochondrial membrane potential (∆ΨM) and cellular ATP production. Energy produced during mitochondrial respiration is maintained as a high ∆ΨM, whereas declining mitochondrial function is associated with ∆ΨM loss. We found a significantly higher ∆ΨM in CLL cells accompanied by a significantly elevated energy production (Figure 5A-B). Both are in opposition to a disrupted mitochondrial function. The functionality of the CLL cells’ mitochondria is further underlined by the increased respiratory rate (21.5+/−5 pmol/min OCR in CLL cells vs 3.5+/−1 pmol/L OCR in B-cells, P = .01) (Figure 5C). Aerobic glycolysis characterized by lactate production as measured by ECAR, unlike in many other cancer entities,31 does not appear overly activated in CLL cells (Figure 5C-D). Molecules of the glucose metabolism including glut1, glut3, hexokinase 2/hk2, lactate dehydrogenase a/ldha, and pyruvate dehydrogenase kinase 1/pdk1 were not increased in CLL cells (Figure 5E).
Mitochondrial metabolism in CLL cells. (A) The mitochondrial electrochemical membrane potential (∆ΨM) was semiquantified using the potentiometric dye JC-1 for HD B-cells (n = 10) and CLL cells (n = 10) by FACS. (B) ATP levels were measured in lysates from purified normal B-cells (n = 5) and CLL cells (n = 5) by ELISA. (C) Baseline OCR and ECAR were measured by an XFe96 flux analyzer in purified normal B-cells (n = 4) and CLL cells (n = 4). OCR is an indicator of mitochondrial respiration, and ECAR is predominantly the result of anaerobic glycolysis. (D) Mean ECAR levels on administration of glucose in purified normal B-cells (n = 4) and CLL cells (n = 4) as measured by an XFe96 flux analyzer. (E) The relative gene expression of key molecules of glycolysis (glut1; glut2; hexokinase-2, hk2; lactate dehydrogenase a, ldha; pyruvate dehydrogenase kinase 1, pdk1) is shown comparatively for purified HD B-cells (n = 10) and CLL cells (n = 10) as quantified by qPCR. (F) Respiration (OCR) is measured under basal conditions and in response to the indicated mitochondrial inhibitors in purified normal B-cells (n = 4) and CLL cells (n = 4). The resulting effects on OCR are shown as a percentage of the baseline measurement (set as 100%) for each treatment. Changes after oligomycin (I.) and FCCP (II.) application are indicative for respiration linked to ATP synthesis and the maximal respiratory capacity, respectively. (G.) ECAR is measured under basal conditions and in response to the indicated mitochondrial inhibitors in purified normal B-cells (n = 4) and CLL cells (n = 4). The resulting (compensatory) effects on ECAR are shown as a percentage of the baseline measurement (set as 100%). Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001.
Mitochondrial metabolism in CLL cells. (A) The mitochondrial electrochemical membrane potential (∆ΨM) was semiquantified using the potentiometric dye JC-1 for HD B-cells (n = 10) and CLL cells (n = 10) by FACS. (B) ATP levels were measured in lysates from purified normal B-cells (n = 5) and CLL cells (n = 5) by ELISA. (C) Baseline OCR and ECAR were measured by an XFe96 flux analyzer in purified normal B-cells (n = 4) and CLL cells (n = 4). OCR is an indicator of mitochondrial respiration, and ECAR is predominantly the result of anaerobic glycolysis. (D) Mean ECAR levels on administration of glucose in purified normal B-cells (n = 4) and CLL cells (n = 4) as measured by an XFe96 flux analyzer. (E) The relative gene expression of key molecules of glycolysis (glut1; glut2; hexokinase-2, hk2; lactate dehydrogenase a, ldha; pyruvate dehydrogenase kinase 1, pdk1) is shown comparatively for purified HD B-cells (n = 10) and CLL cells (n = 10) as quantified by qPCR. (F) Respiration (OCR) is measured under basal conditions and in response to the indicated mitochondrial inhibitors in purified normal B-cells (n = 4) and CLL cells (n = 4). The resulting effects on OCR are shown as a percentage of the baseline measurement (set as 100%) for each treatment. Changes after oligomycin (I.) and FCCP (II.) application are indicative for respiration linked to ATP synthesis and the maximal respiratory capacity, respectively. (G.) ECAR is measured under basal conditions and in response to the indicated mitochondrial inhibitors in purified normal B-cells (n = 4) and CLL cells (n = 4). The resulting (compensatory) effects on ECAR are shown as a percentage of the baseline measurement (set as 100%). Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001.
Treatment with the complex V inhibitor oligomycin reduced average OCR to 47+/−7% (of the baseline level) in CLL cells and to 66+/−12% in B-cells, respectively (Figure 5F). Because mitochondrial complex V (F0F1-ATPase) couples the electron chain to ATP synthesis, these data indicate that in CLL cells a significantly higher fraction of oxygen consumption was coupled to ATP-synthesis than in B-cells (53% vs 33%, P = .0002). We found no differences regarding the coupling efficacy and the expression of key uncoupling proteins (supplemental Figure 8). Next, we examined the effect of uncoupling respiration from energy generating oxidative phosphorylation on OCR so as to measure the cells’ maximal respiratory capacity. Addition of the uncoupling agent FCCP resulted in a significantly stronger OCR increase in CLL cells compared with B-cells (218+/−82% vs 121+/−14%, P = .002) (Figure 5F). This elevated spared or reserve respiratory capacity can be well explained by the observed increased mitochondrial biogenesis (Figure 4).32 Furthermore, CLL cells’ stronger dependency on maintaining the mitochondrial energetic flux was further highlighted by the effects elicited after the cells’ sequential treatment with oligomycin and FCCP, which block mitochondrial function (Figure 5G). CLL cells displayed a significantly higher compensatory increase of the aerobic glycolysis for meeting their energetic demands (230+/−62% in CLL cells vs 125+/−21% in B-cells after oligomycin and 354+/−57% in CLL cells vs 151+/−28% in B-cells after FCCP).
Targeting mitochondrial metabolism in CLL cells
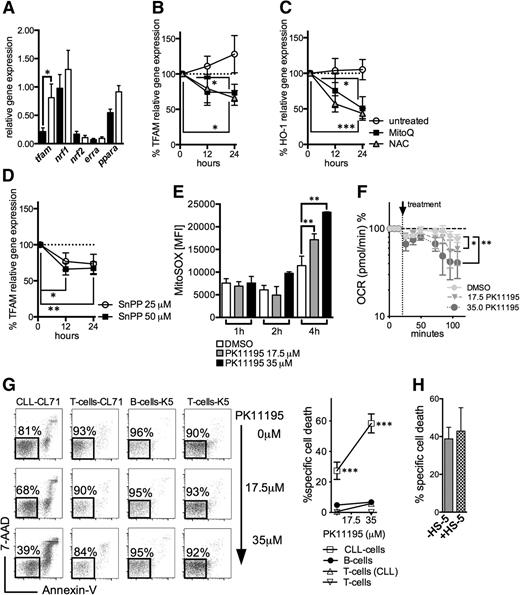
We analyzed the expression of molecules involved in mitochondrial biogenesis and/or mitochondrial translation including tfam, nuclear respiratory factor 1/nrf1, nuclear respiratory factor 2/nrf2, estrogen-related receptor α/erra, and the peroxisome proliferator-activated factor α/ppara.10 We found that tfam was significantly higher as expressed in CLL cells (Figure 6A). Previous reports have shown that ROS can act as an activator of tfam expression.33,34 Treating CLL cells with the antioxidants NAC and MitoQ resulted in a significant downregulation of tfam (Figure 6B). Moreover, redox modulation also led to a diminution of the highly expressed antioxidant HO-1 (Figures 3D and 6C).21 HO-1 is a stress-responsive molecule with strong antioxidant properties. Recent data suggest that its product CO is involved in mitochondrial biogenesis.35,36 In line with these observations, blocking HO-1-activity by SnPP resulted in a significant reduction of tfam expression (Figure 6D), further corroborating the (indirect) interconnection between ROS, cellular adaptation to ROS, and mitochondrial biogenesis.
Regulation of mitochondrial biogenesis and mitochondrial targeting in CLL cells. (A) The relative gene expression of key molecules governing mitochondrial biogenesis (mitochondrial transcription factor A, tfam; nuclear respiratory factor 1, nrf1; nuclear respiratory factor 2, nrf2; estrogen-related receptor a, erra; peroxisome proliferator-activated receptor a, ppara) as analyzed in purified HD B-cells (n = 8) and CLL cells (n = 10). (B) The %al changes of the TFAM relative gene expression in purified CLL cells (n = 6) on treatment with the mitochondrial-specific antioxidant MitoQ and the general antioxidant NAC in comparison with untreated cells over 12 and 24 hours. (C) The %al changes of the HO-1 relative gene expression in purified CLL cells (n = 6) on treatment with the mitochondrial-specific antioxidant MitoQ and the general antioxidant NAC in comparison with untreated cells over 12 and 24 hours. (D) The %al changes of the TFAM relative gene expression in purified CLL cells (n = 6) on treatment with 25 μM and 50 μM of the HO-1 inhibitor tin-protoporphyrin (SnPP) over 12 and 24 hours. Bars indicate the standard error mean. (E) Effects of 17.5 µM and 35 µM PK11195 on the generation of mitochondrial ROS in CLL cells (n = 4) 1, 2, and 4 hours after its application based on the MFI of MitoSOX as quantified by FACS. (F) Respiration (OCR) is measured under basal conditions and in response to the indicated F1F0-ATPase inhibitor PK11195 in CLL cells (n = 4). The resulting effects on OCR are shown as a percentage of the baseline measurement (set as 100%). (G) A representative FACS analysis of the effects of a 24-hour PK11195 treatment on the viability of a CLL patient’s (CL71) CLL cells and T-cells and HDs’ (K5) B- and T-cells, respectively, is shown (left panel). Viable cells are 7-AAD and Annexin-V negative (highlighted box). (Right panel) the mean values of the %al specific death elicited by 17.5 µM and 35 µM PK11195 (for 24 hours) in CLL patients’ (n = 5) CLL cells and T-cells and HDs’ (n = 5) B- and T-cells. (H) Bone marrow stromal cells (eg, the HS-5 cell line) can protect CLL cells from drug-induced apoptosis. The %al specific cell death as assessed by (FACS) is shown for purified CLL cells (n = 4) treated with 35 µM PK11195 for 24 hours in the presence or absence of HS-5 cells. Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001.
Regulation of mitochondrial biogenesis and mitochondrial targeting in CLL cells. (A) The relative gene expression of key molecules governing mitochondrial biogenesis (mitochondrial transcription factor A, tfam; nuclear respiratory factor 1, nrf1; nuclear respiratory factor 2, nrf2; estrogen-related receptor a, erra; peroxisome proliferator-activated receptor a, ppara) as analyzed in purified HD B-cells (n = 8) and CLL cells (n = 10). (B) The %al changes of the TFAM relative gene expression in purified CLL cells (n = 6) on treatment with the mitochondrial-specific antioxidant MitoQ and the general antioxidant NAC in comparison with untreated cells over 12 and 24 hours. (C) The %al changes of the HO-1 relative gene expression in purified CLL cells (n = 6) on treatment with the mitochondrial-specific antioxidant MitoQ and the general antioxidant NAC in comparison with untreated cells over 12 and 24 hours. (D) The %al changes of the TFAM relative gene expression in purified CLL cells (n = 6) on treatment with 25 μM and 50 μM of the HO-1 inhibitor tin-protoporphyrin (SnPP) over 12 and 24 hours. Bars indicate the standard error mean. (E) Effects of 17.5 µM and 35 µM PK11195 on the generation of mitochondrial ROS in CLL cells (n = 4) 1, 2, and 4 hours after its application based on the MFI of MitoSOX as quantified by FACS. (F) Respiration (OCR) is measured under basal conditions and in response to the indicated F1F0-ATPase inhibitor PK11195 in CLL cells (n = 4). The resulting effects on OCR are shown as a percentage of the baseline measurement (set as 100%). (G) A representative FACS analysis of the effects of a 24-hour PK11195 treatment on the viability of a CLL patient’s (CL71) CLL cells and T-cells and HDs’ (K5) B- and T-cells, respectively, is shown (left panel). Viable cells are 7-AAD and Annexin-V negative (highlighted box). (Right panel) the mean values of the %al specific death elicited by 17.5 µM and 35 µM PK11195 (for 24 hours) in CLL patients’ (n = 5) CLL cells and T-cells and HDs’ (n = 5) B- and T-cells. (H) Bone marrow stromal cells (eg, the HS-5 cell line) can protect CLL cells from drug-induced apoptosis. The %al specific cell death as assessed by (FACS) is shown for purified CLL cells (n = 4) treated with 35 µM PK11195 for 24 hours in the presence or absence of HS-5 cells. Bars indicate the standard error mean. Abbreviations: P, P-value; *P < .05; **P < .005; ***P < .001.
The idea of killing cancer cells by ROS-mediated mechanisms that take advantage of their intrinsic oxidative stress was proposed a decade ago.37 We hypothesized that the mitochondrial alterations (Figure 4) together with the CLL cells’ energetic reliance on oxidative phosphorylation (Figure 5F-G) would render them preferentially vulnerable to agents that induce further mitochondrial ROS by affecting the mitochondrial respiratory chain. The benzodiazepine derivate PK11195 generates mitochondrial superoxide in a concentration-dependent manner by inhibiting the mitochondrial F1F0-ATPase38 (Figure 6E-F). Treating CLL cells with PK11195 appears to shift the redox balance above their increased tolerability threshold and thereby induces cell death (Figure 6G). Furthermore, PK11195 did not elicit increased cell death in patients’ T-cells and healthy control-derived B- and T-cells (Figure 6G). CLL cells could not be rescued by human bone marrow stromal cells that modulate their redox status and harness their resilience toward ROS-inducing agents39 (Figure 6H).
Discussion
Malignant transformation is inevitably linked to metabolic reprogramming in cancer cells.1 As yet, and despite the high incidence of CLL, little is known about CLL cells’ metabolism. Oxidative stress represents a metabolic imbalance, which is regularly observed in cancer patients.25 In line with findings from a recent study evaluating ROS-related damages in lymphocytes from patients with monoclonal B-lymphocytosis and CLL,3 we found increased amounts of oxidatively modified DNA and lipids in the sera of untreated CLL patients. In other entities (eg, pancreatic cancer) it has been shown that the oxidative phenotype of cancer cells can correlate with poor prognosis.5,40 In a similar fashion, a significantly larger proportion of patients from the ROShigh group have experienced lymphocyte count doubling, which is an early marker for disease activity that has prognostic relevance.
Increased ROS levels can be detrimental for immune cells resulting in loss of function or even apoptosis of, for example, effector T-cells6,15 and NK-cells.16,27 On the basis of these observations, tumor-associated oxidative stress appears as a contributor to tumor immune escape.18 In fact, increased ROS levels in CLL patients were associated with a less activated phenotype of CD4+ T-cells and downregulation of the signal-transducing CD3ζ-chain in both the CD4+ and CD8+ T-cells. This is in line with data from other types of malignancies such as breast, colon, and pancreatic cancer.6 Furthermore, this link could at least partly explain the observed deregulations of CD4+ T-cells in patients with very early CLL.22 Interestingly, we found a higher proportion of circulating CD56bright NK-cells in patients with higher ROS levels. This circumstance additionally highlights how ROS fine-tune immune balances: this NK-cell subset has been shown to be more resilient toward ROS-induced cell death,16 which could be decisive for its preferential enrichment in CLL and other cancers.41 We could furthermore show that stimulating patients’ T-cells in the presence of the antioxidant NAC reverted some of the ROS-mediated effects. This provides on the one hand a strong indication that oxidative stress has a relevant impact on the CLL patients’ immune system and suggests on the other hand pharmacological redox-remodeling as a potential mean for harnessing immune responses in CLL. Such strategies are already exploited in various entities with the most promising results being achieved in AML by combining antioxidants with mild immune stimulation.42
Pathomechanisms responsible for ROS overproduction often remain vague. Our data suggest that CLL cells are strong contributors to patients’ oxidative stress. This is in line with studies showing that B-cell stimulation promotes ROS production23 and CLL cells partially resemble chronically activated B-cells.43 Several types of solid (eg, melanoma13 ) and hematological malignancies (eg, hairy cell leukemia12 ) display increased superoxide production by the membrane-bound NOX. Expression of NOX subunits and NOX activity were not increased in CLL cells. In fact, we found lower levels of the gp91 subunit. Interestingly, it has been recently shown that gp91-deficiency in B-cells is associated with a stronger proliferative capacity,44 which is a prerequisite for CLL development.
Although lower antioxidant levels have been described, for example, in lung cancer cells,28 in most malignancies the opposite is the case. Because cancer cells are continuously exposed to high amounts of endogenous ROS, cells capable of an according stress adaptation are preferentially selected. Consequently, an enhanced antioxidant capacity is regularly seen in tumors,19 which also has implications for the development of drug resistance.20 Several antioxidants were increased in CLL cells, which is partly (eg, for HO-1) in line with previous reports.21 Expression of G6PDH catalyzing the entry of glucose-6-phosphate into the pentose phosphate pathway to generate nucleotides, NAPDH, and thiols was also increased. Thiols themselves hold crucial antioxidant functions45 and were elevated in CLL cells, potentially contributing to the (by trend) enhanced antioxidant capacity.
Our data altogether point to mitochondria as the main cause of oxidative stress in CLL cells. This is in accord with recent studies showing that B-cell stimulation leads to an increase in mitochondrial ROS, which are essential for an adequate B-cell response.23 We could show that CLL cells similar to AML-blasts9 exhibit increased mitochondrial biogenesis that translates into increased cellular respiratory rate and respiratory reserve capacity. In contrast to many other types of cancer, CLL cells do not appear to be overly utilizing aerobic glycolysis, also known as the Warburg effect.31 Our results suggest that CLL cells use primarily oxidative phosphorylation for generating energy. The increased mitochondrial biogenesis could theoretically reflect feedback mechanisms compensating for mitochondrial defects.33 However, CLL cells’ mitochondria appeared functional. In fact, increased respiratory rates (eg, by leading to more leaking protons in the electron transport chain) and high ∆ΨM (eg, by increasing stability of ROS) can positively correlate with mitochondrial ROS levels as established in previous studies11 and could thereby explain the observed oxidative stress.
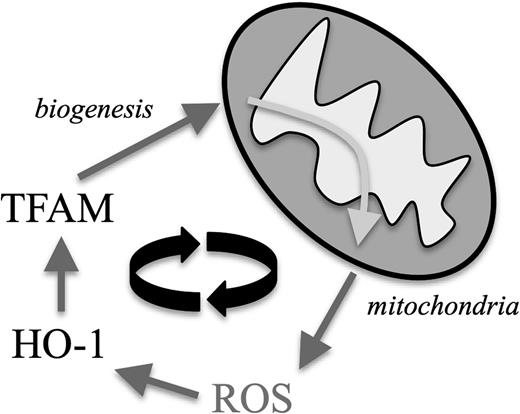
Furthermore, increasing evidence suggests a close interrelationship between oxidative stress and mitochondrial biogenesis. Cellular adaptation mechanisms—in our case the upregulation of the stress-responsive HO-135 —toward abundant ROS can act as positive regulators of mitochondrial transcription factors such as TFAM,34 which was upregulated in CLL cells (Figure 7). This process is thought to represent a protective measure: increased mitochondrial ROS lead to mitochondrial damages and subsequent decrease of energy production. The according activation of the redox adaptation cascade would then additionally promote biogenesis of novel and functional mitochondria.46 Because more mitochondria can result in more mitochondrial ROS, this process turns into a vicious cycle in CLL cells (Figure 7). As a proof of principle, we were able to target this self-amplifying loop in vitro by treating CLL cells with antioxidants that led to downregulation of both HO-1 and TFAM. This interventional approach could represent an interesting strategy aiming (a) directly to antagonize ROS-mediated effects and (b) indirectly to prevent mitochondrial biogenesis.
The proposed mechanism by which mitochondrial ROS and mitochondrial biogenesis form a self-amplifying feedback loop in CLL cells. Increased mitochondrial ROS production leads to an intrinsic compensatory upregulation of cellular antioxidants, including HO-1. HO-1 represents a positive regulatory signal for TFAM, which drives mitochondrial biogenesis leading subsequently to more ROS-generating mitochondria.
The proposed mechanism by which mitochondrial ROS and mitochondrial biogenesis form a self-amplifying feedback loop in CLL cells. Increased mitochondrial ROS production leads to an intrinsic compensatory upregulation of cellular antioxidants, including HO-1. HO-1 represents a positive regulatory signal for TFAM, which drives mitochondrial biogenesis leading subsequently to more ROS-generating mitochondria.
Furthermore, this oxidative phenotype of CLL cells provides the mechanistic basis for the efficacy and specificity of PK11195. This benzodiazepine derivate inhibits the F1F0-ATPase,38 thereby blocking oxidative phosphorylation. The resulting surplus production of mitochondrial superoxide leads to a rather selective apoptosis in CLL cells. The targeted action of PK11195 is most likely attributed to the metabolic differences between malignant and nonmalignant cells. Our data are consistent with recent studies evaluating F1F0-ATPase inhibitors for eliminating alloreactive T-cells in graft-versus-host disease, which share metabolic similarities with CLL cells.47
In conclusion, this study shows that the CLL cells’ mitochondria are a key source for the patients’ oxidative stress linked to immune alterations. Our observations provide a rationale for 2 opposite therapeutic redox-modulating strategies in CLL. First, cellular adaptation to oxidative stress appears as an indirect promoter of ROS production; a vicious cycle that could be breached by redox-manipulation using antioxidants that could also prove beneficial for enhancing intrinsic immune responses. Second, using agents that further aggravate mitochondrial ROS production and thereby overwhelm the cancer cells’ protective systems could selectively impact CLL cells. Targeting metabolic pathways could help to increase the efficacy of already-existing therapeutic approaches and to overcome drug resistances in CLL.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are very grateful to M. Murphy from Cambridge University for providing MitoQ and to N. Schröder-Kreß for assisting with electron microscopy.
This work was supported by the IZKF Erlangen, the Max-Eder program of the Deutsche Krebshilfe, the European Hematology Association, and the Ria-Freifrau von Fritsch Foundation.
Authorship
Contribution: R.J. and A.D.H. performed research, analyzed data, and helped to write the manuscript; H.B., A.G., J. Bricks, J. Berger, and D.S. performed research; M.J.E. collected patient material and provided patient data; A.M. helped to design the study and to write the manuscript; and D.M. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dimitrios Mougiakakos, Department of Internal Medicine 5, Hematology and Oncology, University of Erlangen-Nuremberg, Ulmenweg 18, 90154 Erlangen, Germany; e-mail: dimitrios.mougiakakos@uk-erlangen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal