Key Points
MRD assessment by sequencing is prognostic of TTP and OS in multiple myeloma patients.
Among patients in complete response, MRD assessment by sequencing enables identification of 2 distinct subgroups with different TTP.
Abstract
We assessed the prognostic value of minimal residual disease (MRD) detection in multiple myeloma (MM) patients using a sequencing-based platform in bone marrow samples from 133 MM patients in at least very good partial response (VGPR) after front-line therapy. Deep sequencing was carried out in patients in whom a high-frequency myeloma clone was identified and MRD was assessed using the IGH-VDJH, IGH-DJH, and IGK assays. The results were contrasted with those of multiparametric flow cytometry (MFC) and allele-specific oligonucleotide polymerase chain reaction (ASO-PCR). The applicability of deep sequencing was 91%. Concordance between sequencing and MFC and ASO-PCR was 83% and 85%, respectively. Patients who were MRD– by sequencing had a significantly longer time to tumor progression (TTP) (median 80 vs 31 months; P < .0001) and overall survival (median not reached vs 81 months; P = .02), compared with patients who were MRD+. When stratifying patients by different levels of MRD, the respective TTP medians were: MRD ≥10−3 27 months, MRD 10−3 to 10−5 48 months, and MRD <10−5 80 months (P = .003 to .0001). Ninety-two percent of VGPR patients were MRD+. In complete response patients, the TTP remained significantly longer for MRD– compared with MRD+ patients (131 vs 35 months; P = .0009).
Introduction
Historically, the goal of multiple myeloma (MM) therapy has been to achieve a partial response or disease stabilization.1,2 The introduction of novel agents, including thalidomide, lenalidomide, and bortezomib, in combination with autologous stem cell transplantation, has dramatically altered the treatment paradigm and significantly improved overall survival (OS) in MM patients.3 As a result, increasing emphasis has been placed on the achievement of complete response (CR), which is defined as the absence of M-protein by immunofixation and <5% plasma cells (PCs) in bone marrow (BM).4,5 Unfortunately, most patients with MM ultimately relapse and die of the disease despite achieving CR.6 Although some reports have attributed the sustainability of MM to a minor population of clonogenic CD138− cells, the dominant population of CD138+ PCs contain clonogenic cells.7 Therefore, most but not all of MM relapses can be attributed to the persistence of undetectable minimal residual disease (MRD). Thus a CR definition that is based solely on protein analysis and conventional cytological cell morphology techniques is insufficient.
The identification of MRD is an emerging component of CR assessment in MM patients. Similar to some lymphoid8 or myeloid9 malignancies, traditional methods for measuring MRD in MM include allele-specific oligonucleotide polymerase chain reaction (ASO-PCR) of rearranged B-cell receptor genes and multiparametric flow cytometry (MFC). Such methods are based on the identification of clonotypic sequences or aberrant immunophenotypes, respectively. Previous studies have shown that molecular and MFC remissions are associated with significantly longer progression-free survival (PFS) and OS in MM patients.10,11 Moreover, such studies have demonstrated that in patients who achieved conventional CR, those who demonstrated a complete clearance of bone marrow tumor cells assessed by PCR or MFC have a significantly longer PFS and OS compared with those with persistent tumor cells. This suggests that molecular or immunophenotypic remissions are more sensitive than conventional CR by immunofixation.
However, both techniques have some disadvantages. ASO-PCR in MM is associated with high technical complexity and low applicability.12 Although some alternative PCR strategies for assessing MRD in MM could improve the applicability,13-16 they typically result in decreased sensitivity. MFC has a higher applicability, virtually covering all patients,17 and the current sensitivity ranges between 10−4 and 10−5; however, further standardization efforts are required. Moreover, no tumor cells are detectable by MFC or PCR in a fraction of patients who ultimately relapse, which indicates that there is room for improvement in the MRD detection limit and suggests that alternative techniques should be investigated.
Recent reports have demonstrated the utility of high-throughput sequencing (HTS)–based MRD assessment in lymphoid malignancies.18-22 This quantitative method, termed the LymphoSIGHT™ platform, relies on amplification and sequencing of immunoglobulin gene segments using consensus primers, with a demonstrated applicability higher than 90%. Standardized algorithms quantify the number of cancer molecules with an assay sensitivity ≤10−6, which represents at least 1 log greater sensitivity than the ASO-PCR and MFC methods, respectively.19 Here we assessed the prognostic value of MRD detection by sequencing in a cohort of 133 MM patients enrolled in GEM (Grupo Español de Mieloma) Myeloma Trials, as well as the concordance between MRD levels measured by MFC, ASO-PCR, and high-throughput sequencing.
Methods
Patients and clinical samples
Bone marrow samples from 133 patients included in GEM clinical trials were selected for inclusion in this study based on sample availability. Patients <65 years were treated within the GEM2000 or GEM05 <65 protocols, whereas elderly patients were treated within the GEM05 ≥65 or GEM10 ≥65 trials. Detailed descriptions of these trials have been published.23-26 A previous analysis27 demonstrated that the achievement of negative MRD by MFC had prognostic value in young and elderly MM patients, and the prognostic influence of MRD assessment is independent of the type of treatment. All patients included in this study achieved at least very good partial response (VGPR) after front-line therapy. The samples for MRD investigation were obtained either after induction in the elderly population or after induction plus autologous stem cell transplant in patients <65 years.
The studies were conducted in accordance with Declaration of Helsinki principles and were approved by the relevant institutional review boards. Written informed consent was obtained from all subjects.
MRD measurements by the HTS method
HTS was carried out according to the LymphoSIGHTTM method (Sequenta, Inc., San Francisco, CA), as previously described.19,20 For MM diagnostic samples, genomic DNA was amplified using locus-specific primer sets for the immunoglobulin heavy-chain locus (IGH) complete (IGH-VDJH), IGH incomplete (IGH-DJH), and immunoglobulin κ locus (IGK). The amplified product was subjected to sequencing, and the sequences and frequencies of the different clonotypes in the sample were obtained. Myeloma gene rearrangements were identified using a method as previously described.19,22 Patients in whom a high-frequency myeloma clone (>5%) was not identified were excluded from the MRD analysis. MRD was assessed in patients with a high-frequency myeloma clone using the IGH-VDJH and IGK or IGH-VDJH, IGH-DJH, and IGK assays. Once the absolute amount of total cancer-derived molecules present in a sample was determined, a final MRD measurement was calculated, providing the number of cancer-derived molecules per 1 million cell equivalents.19 In cases in which 2 or more tumor clones existed, the clone with the highest MRD value was reported. Molecular CR was defined according to the International Myeloma Working Group (IMWG) consensus recommendations.28
MRD studies by flow cytometry and ASO-PCR methods
Bone marrow samples were immunophenotyped using a 4-color direct immunofluorescence technique, as previously reported.10,17,29 The phenotypic aberrancies detected at diagnosis were used as patient-specific probes for MRD assessment after induction therapy. During follow-up, immunophenotypic response was defined as the absence of detectable MM plasma cells by MFC at a sensitivity level of 10−4 to 10−5, following the IMWG recommendations.28 In addition, BM MRDs were evaluated in a subset of patients by ASO real-time PCR using TaqMan technology as previously described.15,30
Statistical analysis
Statistical analyses were performed with the SPSS program version 21.0 (SPSS, Inc., IBM, Armonk, NY). Contingency tables were used to analyze associations between categorical variables, considering the χ2 test for statistical significance. The Student t test was used to compare averages of continuous variables between groups. A P value < .05 was considered significant. The concordance among sequencing, MFC, and ASO-PCR was analyzed in log space using the Pearson coefficient test.
For survival analyses, the end points examined were time to tumor progression (TTP) and OS, both assessed according to the IMWG criteria from the start of treatment.28 For the univariate analysis, survival curves were calculated according to the Kaplan-Meier method and differences between curves were compared with the Wilcoxon test. A multivariate analysis was also performed using the Cox proportional regression hazard model to identify the factors that might have a significant independent influence on OS and TTP. The variables studied in the multivariate model are included in Table 1, together with molecular response determined by sequencing (negative/positive).
Main patient characteristics at diagnosis grouped according to molecular response by deep sequencing
| Characteristic . | Non-MR (n = 80) . | MR (n = 30) . |
|---|---|---|
| Male/female, % | 51/49 | 52/48 |
| Mean age, y (range) | 62 (39-84) | 61 (42-75) |
| Mean β2-microglobulin, mg/L | 4.2 | 4.3 |
| Mean hemoglobin, mg/dL | 11.2 | 11 |
| Mean creatinine, mg/dL | 1 | 1.1 |
| Mean serum calcium, mg/dL | 9.5 | 9.8 |
| Mean albumin, g/dL | 3.7 | 3.4 |
| LDH high, % | 5 | 3 |
| Durie-Samon Stage (I/II/III), % | 10/48/42 | 4/36/60 |
| Durie-Salmon Stage A/B, % | 98/2 | 96/4 |
| IIS I/II/III, % | 49/39/12 | 77/23/0 |
| Bone lesions (nonlesion, minor lesions, major lesions) (%) | 26/42/32 | 33/30/37 |
| Maximum response after front-line therapy*, n | ||
| Complete remission (CR) | 36* | 26* |
| Very good partial remission (VGPR) | 42* | 4* |
| Characteristic . | Non-MR (n = 80) . | MR (n = 30) . |
|---|---|---|
| Male/female, % | 51/49 | 52/48 |
| Mean age, y (range) | 62 (39-84) | 61 (42-75) |
| Mean β2-microglobulin, mg/L | 4.2 | 4.3 |
| Mean hemoglobin, mg/dL | 11.2 | 11 |
| Mean creatinine, mg/dL | 1 | 1.1 |
| Mean serum calcium, mg/dL | 9.5 | 9.8 |
| Mean albumin, g/dL | 3.7 | 3.4 |
| LDH high, % | 5 | 3 |
| Durie-Samon Stage (I/II/III), % | 10/48/42 | 4/36/60 |
| Durie-Salmon Stage A/B, % | 98/2 | 96/4 |
| IIS I/II/III, % | 49/39/12 | 77/23/0 |
| Bone lesions (nonlesion, minor lesions, major lesions) (%) | 26/42/32 | 33/30/37 |
| Maximum response after front-line therapy*, n | ||
| Complete remission (CR) | 36* | 26* |
| Very good partial remission (VGPR) | 42* | 4* |
LDH, lactate dehydrogenase. MR, molecular response.
P(χ2) < .001.
Results
Identification of clonal rearrangements by deep sequencing
A clonal rearrangement was identified at a frequency >5% in the diagnostic BM aspirate in 121 of 133 patients. The sequencing method successfully identified a myeloma clone that was suitable for follow-up MRD assessment in 91% of MM patients. Thus, the MRD applicability of deep sequencing was 91%.
The IGH-VDJH assay was the most frequent gene rearrangement identified: at least one IGH-VDJH clonal rearrangement was detected in 69% (84/121) of MM diagnostic samples, and at least one IGK clonal rearrangement was detected in 55% (66/121) of the samples. In contrast, the incomplete IGH-DJH clonal rearrangement was identified in 48% (58/121).
Follow-up samples were available for analysis in 110 of 121 patients with a clonal rearrangement at diagnosis. During follow-up, most patients (n = 80, 73%) remained positive by sequencing at MRD levels of 10−5 or higher. Baseline clinical characteristics of patients grouped according to molecular remission status achieved by sequencing are summarized in Table 1.
Prognostic values of MRD assessment by sequencing
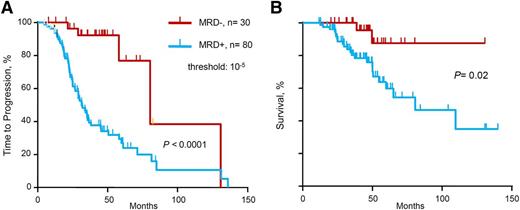
We evaluated the prognostic influence of MRD status by deep sequencing on TTP and OS (Table 2 and Figure 1). Molecular response by deep sequencing, which corresponds to MRD negativity defined as MRD level <10−5, was associated with significantly longer TTP (median 80 vs 31 months, P < .0001) and OS (median not reached vs 81 months, P = .02). In the multivariate analysis for TTP, MRD– status by deep sequencing was the single variable with statistical significance (hazard ratio [HR] 8.6, P = .012). Only 8 patients with high-risk cytogenetics were included in this cohort, and therefore cytogenetics risk-based subanalyses were not performed because of insufficient statistical power.
Survival values according to conventional responses and for different levels of minimal residual disease determined by deep sequencing, multiparametric flow cytometry, or ASO-PCR
| . | . | Time to progression . | Overall survival . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All series . | N . | 5 y, % . | Median mo (±95% CI) . | HR . | P value . | 5 y, % . | Median mo (±95% CI) . | HR . | P value . |
| SEQ <10−5 | 30 | 84 | 80 (48-112) | 5.39 | .0001 | 89 | NR | 4.88 | .019 |
| SEQ >10−5 | 80 | 29 | 31 (26-37) | 65 | 81 (40-122) | ||||
| MFC <10−5 | 34 | 66 | NR | 3.97 | .0001 | 82 | NR | 2.17 | .046 |
| MFC >10−5 | 65 | 32 | 32 (24-40) | 66 | 110 (50-170) | ||||
| ASO-PCR 10−5 | 22 | 65 | NR | 4.59 | .03 | 81 | NR | — | .340 |
| ASO-PCR >10−5 | 19 | 0 | 26 (20-32) | 81 | NR | ||||
| Conventional response and MRD | |||||||||
| VGPR | 48 | 31 | 29 (18-40) | 2.00 | .001 | 61 | 110 (35-184) | 2.17 | .022 |
| CR | 62 | 49 | 60 (39-80) | 78 | NR | ||||
| CR & SEQ <10−5 | 26 | 82 | 131 (51-154) | 2.87 | .001 | 88 | NR | — | .172 |
| CR & SEQ >10−5 | 36 | 31 | 35 (30-41) | 71 | NR | ||||
| CR & MFC <10−5 | 28 | 65 | NR | 2.87 | .06 | 78 | NR | — | .596 |
| CR & MFC >10−5 | 33 | 37 | 35 (30-41) | 77 | NR | ||||
| . | . | Time to progression . | Overall survival . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All series . | N . | 5 y, % . | Median mo (±95% CI) . | HR . | P value . | 5 y, % . | Median mo (±95% CI) . | HR . | P value . |
| SEQ <10−5 | 30 | 84 | 80 (48-112) | 5.39 | .0001 | 89 | NR | 4.88 | .019 |
| SEQ >10−5 | 80 | 29 | 31 (26-37) | 65 | 81 (40-122) | ||||
| MFC <10−5 | 34 | 66 | NR | 3.97 | .0001 | 82 | NR | 2.17 | .046 |
| MFC >10−5 | 65 | 32 | 32 (24-40) | 66 | 110 (50-170) | ||||
| ASO-PCR 10−5 | 22 | 65 | NR | 4.59 | .03 | 81 | NR | — | .340 |
| ASO-PCR >10−5 | 19 | 0 | 26 (20-32) | 81 | NR | ||||
| Conventional response and MRD | |||||||||
| VGPR | 48 | 31 | 29 (18-40) | 2.00 | .001 | 61 | 110 (35-184) | 2.17 | .022 |
| CR | 62 | 49 | 60 (39-80) | 78 | NR | ||||
| CR & SEQ <10−5 | 26 | 82 | 131 (51-154) | 2.87 | .001 | 88 | NR | — | .172 |
| CR & SEQ >10−5 | 36 | 31 | 35 (30-41) | 71 | NR | ||||
| CR & MFC <10−5 | 28 | 65 | NR | 2.87 | .06 | 78 | NR | — | .596 |
| CR & MFC >10−5 | 33 | 37 | 35 (30-41) | 77 | NR | ||||
SEQ, minimal residual disease by deep sequencing.
TTP and OS of series according to minimal MRD levels. (A) TTP and (B) OS for MRD levels ≤10−5 vs >10−5, as determined by deep sequencing.
TTP and OS of series according to minimal MRD levels. (A) TTP and (B) OS for MRD levels ≤10−5 vs >10−5, as determined by deep sequencing.
Subsequently, we evaluated the prognostic impact of different levels of MRD sensitivity defined by deep sequencing. Patients were grouped into 3 categories according to their MRD levels: (1) ≥10−3 (n = 43), (2) 10−3 to 10−5 (n = 37), and (3) <10−5 (n = 30). The median TTP were 27 months, 48 months, and 80 months, respectively (P from .003 to .0001) (Figure 2A). No significant differences were observed in the TTP of patients with MRD levels between <10−3 and >10−4 vs ≤10−4 and ≥10−5. This sensitivity analysis was extended to assess OS across the 3 categories of MRD levels. Similar to the TTP analysis, MRD levels of <10−5 were associated with significantly longer OS compared with patients with a high MRD level (defined as >10−3) (median not reached vs 55 months, P = .002). Analogous results were found when comparing patients with MRD levels of 10−3 to 10−5 with patients with a high MRD level (>10−3) (median not reached vs 55 months, P = .02) (Figure 2B).
TTP and OS of series stratified according to different MRD levels >10−3 vs 10−3 to 10−5 vs <10−5. (A) TTP and (B) OS, as determined by deep sequencing.
TTP and OS of series stratified according to different MRD levels >10−3 vs 10−3 to 10−5 vs <10−5. (A) TTP and (B) OS, as determined by deep sequencing.
We performed a subanalysis to assess the importance of MRD by deep sequencing across different clinical trials. In this analysis, we studied young patients (n = 76) and elderly patients (n = 34) separately, and the prognostic significance of achieving MRD negativity by deep sequencing persisted (data not shown).
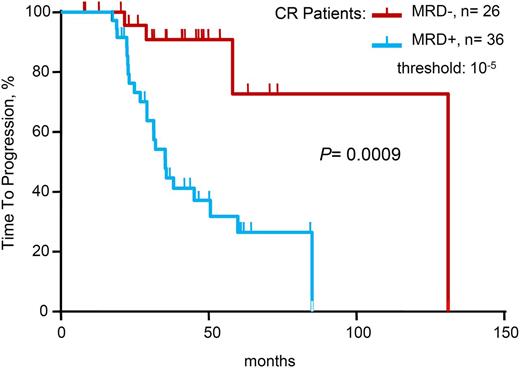
When limiting the study to the 62 patients in conventional CR at the time of analysis, 36 (58%) of 62 patients in CR were positive by sequencing at MRD levels at 10−5 and higher. With a median follow-up of 42 months, patients in CR who were MRD– by deep sequencing had significantly longer TTP (median 131 vs 35 months; P = .0009) compared with patients in CR who were MRD+ by sequencing. Regarding OS, the medians were not reached in either of the two groups (Table 2 and Figure 3).
Time to progression for patients achieving conventional complete remission (CR), according to minimal residual disease (MRD) status as determined by deep sequencing.
Time to progression for patients achieving conventional complete remission (CR), according to minimal residual disease (MRD) status as determined by deep sequencing.
All patients in VGPR were MRD+ by sequencing (92%), with 4 exceptions, patients who were MRD– by sequencing and positive by immunofixation. However, 3 of the 4 patients became immunofixation-negative shortly thereafter, based on follow-up evaluations.
Correlation among MFC, ASO-PCR, and sequencing results
MRD information by MFC and ASO-PCR analysis was available in 99 and 41 patients, respectively. The prognostic significance of MRD status determined by these techniques is summarized in Table 2.
Upon comparing the MRD results obtained by MFC and ASO-PCR with sequencing results, a good degree of concordance was observed across the 3 platforms. When comparing MFC vs sequencing, 83% of the samples yielded concordant results: 60 (61%) were MRD+ and 22 (22%) were MRD– by both methods, with an r2 of 0.58. For the comparison between ASO-PCR and sequencing, the concordance was 85%: 20 (49%) were MRD+ and 15 (36%) were MRD– by both methods, with an r2 of 0.54 for the comparison of quantitative MRD levels.
To assess the clinical significance of discrepancies among platforms, we focused on the 99 patients for whom we had MRD results using both sequencing and MFC. As mentioned before, 82 cases had concordant results (60 double-positives and 22 double-negatives), whereas 12 patients were negative by MFC but still remained positive by sequencing (MFC–/sequencing+); the 5 remaining patients had the opposite pattern (MFC+/sequencing–). The prognosis of sequencing– cases (median TTP not reached) was significantly better than MFC+/sequencing+ samples (median TTP 29 months, P = .0001). Importantly, patients who were MFC–/sequencing+ had an intermediate TTP (median 50 months, P = .05 for the comparison with sequencing– cases). Of the 5 cases with MFC+/sequencing–, only one patient has progressed so far.
Discussion
Over the last decade, there has been enormous progress in the treatment of MM. With the introduction of novel agents such as immunomodulatory drugs (thalidomide, lenalidomide, pomalidomide) and proteasome inhibitors (bortezomib, carfilzomib),31,32 CR can now be achieved in a significant fraction of newly diagnosed MM patients, which was unimaginable 10 years ago.33-37 Consequently, the use of more sensitive assays for detecting and monitoring clinically meaningful residual disease has become a relevant tool for assessing treatment efficacy and prognostication. MRD measurements can be used to predict OS and PFS, to inform consolidation and maintenance strategies, and to evaluate the comparative efficacy of novel therapies.38
Recent studies have shown that MFC, a sensitive method for detection of residual myeloma cells, is predictive of PFS and OS in MM patients.10 Moreover, MFC remission was shown to have prognostic value in patients in CR assessed by traditional immunofixation response criteria, which indicates that the deeper the response, the longer the survival. These studies have been replicated in larger cohorts11,29 and have contributed to the recent proposal by the IMWG to create a new response category, “immunophenotypic CR,” which is defined as the absence of phenotypically aberrant plasma cells in BM analyzed by MFC.28 These reports underscore the emerging importance of MRD assessment in MM patients and suggest that novel methods for MRD detection can play a role in the evolving MM treatment paradigm.
Similarly, ASO-PCR provides an accurate quantification of residual disease. Several recent reports using quantitative PCR have shown the ability to stratify MM patient cohorts in the transplant setting with different prognoses.30,39,40 Thus, the term molecular response has also been included in the IMWG criteria and is considered the highest degree of response.28 However, most patients who achieve an MRD– status eventually relapse, which indicates that the sensitivity and specificity of traditional techniques for MRD assessment can be improved.
In this study, we tested whether measurement of residual myeloma cells using the LymphoSIGHT™ sequencing platform19 provides a sensitive alternative to other MRD methods. The sequencing-based method uses consensus primers to universally amplify and sequence all rearranged immunoglobulin gene segments present in a myeloma clone. This method assesses stable and specific DNA tumor markers, providing a direct, sensitive, and objective quantification of myeloma disease burden. In this study, the HTS approach identified a tumor marker in 91% of MM patients, and thus was “applicable” to >90% of MM patients. Applicability is a critical factor in the evaluation of any MRD assessment technique, because a higher rate of applicability directly translates into more patients who can benefit from MRD investigations. In this study, applicability was based on the identification of a high-frequency (>5%) tumor sequence in the baseline BM aspirate that was suitable for analysis in follow-up samples. The sequencing method provides a significant improvement in applicability compared with ASO-PCR, which has an applicability rate typically <70%30,41,42 and avoids the necessity of designing individualized PCR probes. Only MFC has demonstrated a higher applicability (virtually 100% of patients with new MFC methods)17 ; however, the sensitivity of MFC (10−4 to 10−5) is 1 to 2 logs lower than that achieved by the HTS method (10−6).19 The sequencing method can detect a single cell and is limited only by input cell amount. Most of the samples analyzed by sequencing in this study had <300 000 input cells. Therefore, although the sequencing platform has the sensitivity to detect 1 in 1 million cells, the sequencing MRD positivity threshold for this study was set at 1 in 100 000 cells. Although sensitivities higher than 10−4 to 10−5 can be reached with the new MFC instruments,43,44 they will require analysis of up to 2 × 106 cells through an accurate measurement that has not been systematically evaluated to date.
We analyzed the prognostic value of different levels of MRD assessment by deep sequencing. The sequencing platform identified 3 groups of patients with different TTP: patients with high (>10−3), intermediate (10−3 to 10−5), and low (<10−5) MRD levels showed significantly different TTP: 27, 48, and 80 months, respectively, which indicates that deeper responses were associated with significantly longer survival. The possibility of stratifying patients into 3 risk categories by deep sequencing (low, intermediate, high) could be a significant advantage over less sensitive techniques such as fluorescence-PCR (sensitivity ∼10−3) that can only identify a subgroup of high-risk patients.13
When analyzing the correlation between MRD and conventional response status, the vast majority of patients in VGPR (92%) were MRD+, and although this would seem obvious, these are the first empirical data to establish this correlation. When limiting the analysis to patients in CR, MRD levels ≤10−5 by sequencing stratified the patients into 2 groups with significantly different TTPs. Specifically, there was significantly improved TTP in the MRD– group vs the MRD+ group among the CR patients (median 131 vs 35 months, P = .0009).
A high level of concordance was observed between MRD levels by deep sequencing and results obtained by MFC or ASO-PCR. All 3 platforms discriminated between MRD+ high-risk patients and MRD– cases that exhibited a favorable prognosis (Table 2). However, patients who were MRD– by sequencing showed increased TTP compared with those who were sequencing+/MFC– (P = .05). This analysis was performed using a threshold of 10−5 for defining detectable disease by MFC. These results support the interpretation that the low-level MRD detected by the higher sensitivity of the sequencing platform compared with MFC is clinically significant.
It is important to note that the sensitivity of all 3 approaches is limited by the amount and purity of DNA or the number of cells analyzed, critical factors that depend on the specimen characteristics. As a result, the sensitivity will improve if more BM material is obtained for analysis. Indeed, many of the cases in this study were evaluated by MFC by analyzing up to 2 × 106 cells, which translated into an improvement in MFC results (data not shown). Finally, molecular approaches use a stable DNA marker and a reproducible universal assay with a digital readout, which avoids the problems of MFC laboratory-to-laboratory variability.45,46 Therefore, the sequencing method has the potential to be distributed across multiple laboratories, because it relies on automated data analysis and does not involve expert interpretation by an operator.
Other challenges associated with the MFC platform are related to sample requirements. As opposed to the MFC platform, which requires live cells, the sequencing-based approach uses a stable DNA marker and can be performed on fresh samples, as well as on stored cells and DNA, facilitating retrospective analysis and flexible sample collection requirements and transfer in clinical settings. In turn, immunophenotyping has the advantage of speed with results available in only a few hours.
However, the use of BM samples for MRD assessment presents a potential challenge for all MRD methods. The pattern of BM infiltration in MM is not uniform, which is an important distinction from other hematologic disorders such as the acute leukemias. This, together with extramedullary disease, represents a potential pitfall common to all MRD techniques that use BM samples, as nonrepresentative samples of disease infiltration are sometimes obtained. For this reason, although MRD+ results are consistently informative, MRD– results may correspond to a false-negative case. The use of alternative methods for disease assessment such as imaging techniques (positron emission tomography, computed tomography),47 monitoring of clonogenic MM progenitors,48,49 or MM circulating tumor cells,22,49,50 could provide complementary information to MRD and improve the estimation of the risk of progression.
This study suggests that MRD assessment by sequencing is a useful method for patient risk stratification, and the definition of molecular CR in clinical trials can be extended to include the sequencing method. Indeed, achievement of MRD negativity may ultimately serve as a primary end point in clinical trials for MM. In addition, sensitive methods of MRD detection may contribute to the design of patient-specific treatment approaches, such as de-escalation of therapy for MRD– patients or continuous or escalation of treatment for MRD+ patients, which is consistent with the current standard of care in other hematologic malignancies. In summary, these data underscore the promise of sequencing-based MRD assessment in MM patients and provide a strong rationale for the use of improved MRD assessment in the evolving MM clinical paradigm.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research project was supported by grants PS09/01370, PS09/1450, PI12/01761, and PI12/023121 from the Fondo de Investigaciones Sanitarias; RD12/10 Red de Cáncer (Cancer Network of Excellence) from the Instituto de Salud Carlos III, Spain, and from the Fondo de Investigaciones Sanitarias, Asociación Española Contra el Cáncer (AECC, GCB120981SAN) and the CRIS foundation. This study was supported in part by research funding from Sequenta, Inc. (F.P., L.W., M.M., M.F.).
Authorship
Contribution: J.M.-L., J.J.L., J.S.M., M.F., and R.G.-S. designed research, analyzed data, and wrote the paper; F.P. and M.M. analyzed the data; and M.G., S.B., R.A., N.P., M.A.M., B.P., L.W., C.J., M.S., T.C., I.R., M.V.M., L.R., A.O., M.J.B., R.M., and J.B. performed the research.
Conflict-of-interest disclosure: F.P., L.W., M.M., and M.F. are employees of and holders of equity in Sequenta, Inc. The remaining authors declare no competing financial interests.
Correspondence: Joaquin Martinez-Lopez, Hematología Hospital 12 de Octubre, Universidad Complutense, Edificio Atención Ambulatoria. Planta Sexta Bloque D, Avenida de Córdoba s/n, Madrid. 28041, Spain; e-mail: jmartinezlo@hematologia12octubre.com.