In this issue of Blood, Cheah et al provide long-term data on the use of imatinib to treat patients with eosinophilia and myeloid neoplasms carrying the PDGFRB fusion gene, which is a very rare condition in medicine.1
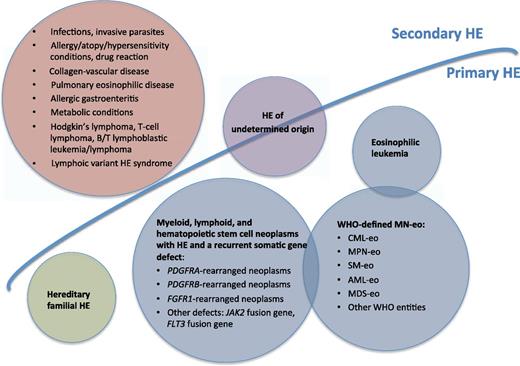
Classification of HE. AML-eo, CBFbeta-fusion gene-related acute myeloid leukemia; CML-eo, Ph-positive/BCR/ABL1-positive chronic myeloid leukemia; MDS-eo, myelodysplastic syndromes with HE; MN-eo, myeloid neoplasm with HE A; MPN-oe, JAK2 V617F-positive myeloproliferative neoplasms; SM-eo, KIT D816V1-positive systemic mastocytosis.
Classification of HE. AML-eo, CBFbeta-fusion gene-related acute myeloid leukemia; CML-eo, Ph-positive/BCR/ABL1-positive chronic myeloid leukemia; MDS-eo, myelodysplastic syndromes with HE; MN-eo, myeloid neoplasm with HE A; MPN-oe, JAK2 V617F-positive myeloproliferative neoplasms; SM-eo, KIT D816V1-positive systemic mastocytosis.
Eosinophils constitute 3% to 5% of the differential white blood cell count, with a corresponding absolute eosinophil number of 0.3 to 0.5 × 109/L. Eosinophil production from hematopoietic precursor cells is under the control of transcription factors such as GATA-1, PU.1, CCAAT-enhancer-binding proteins, and Signal Transducer and Activator of Transcription 5, and in cooperation with this, cytokines such as interleukin 5, granulocyte macrophage-colony-stimulating factor, and interleukin 3 promote proliferation, differentiation, and survival of normal and neoplastic eosinophils.2 Hypereosinophilia (HE) is conventionally defined as an eosinophil count persistently exceeding 1.5 × 109/L.2
The classification of HE is based on the World Health Organization criteria3 subsequently updated by the panelists of the Working Conference on Eosinophil Disorder and Syndromes,4 with the addition of cytohistomorphological criteria for HE. The rarity of the condition described by Cheah et al1 requires us to know exactly how to diagnose this disease. As illustrated in the figure, 4 main categories have been recognized: secondary HE (the most frequent, with a nonclonal eosinophil production), primary HE (clonal, with an underlying stem cell disorder), hereditary (familial) HE, and HE of undetermined significance (without an underlying cause of HE). The first step in the diagnostic work-up of HE is to exclude secondary causes of eosinophilia, looking also for nonmyeloid malignancies (mainly lymphomas) associated with secondary eosinophilia. Diagnosis of clonal HE unequivocally requires blood and marrow evaluation with morphologic, cytogenetic, and immunophenotypic analysis. To be practical, well-defined World Health Organization myeloid neoplasms such chronic myeloid leukemia, myeloproliferative neoplasms,5 systemic mastocytosis, acute myelogenous leukemia, myelodysplastic syndromes, or myelodysplastic syndrome/myeloproliferative neoplasm overlap disorder should be carried out. The succeeding step is to screen peripheral blood for the distinct driving molecular alterations of HE; namely, PDGFRA, PDGFRB, and FGFR1.
The first is the FIP1L1-PDGFRA fusion gene, which is the most frequent recurrent aberration and is detectable in 5% to 15% of all cases with clonal HE. The fusion transcript results from an 800-kilobase internal deletion on band 4q12 containing the gene CHIC2.6 Reverse transcription-polymerase chain reaction or interphase/metaphase fluorescence in situ hybridization probes that hybridize to the region between the FIP1L1 and PDGFRA genes are commonly used to detect the presence of this cytogenetically occult deletion. Alternatively, high levels of serum tryptase may be of diagnostic value. In addition to FIP1L1, other partner genes or activating point mutations in PDGFRA have been described.7 Low-dose imatinib, by inhibiting the phosphorylation of PDGFRA, is effective in HE with FIP1L1-PDGFRA, achieving clinical and hematological responses in few weeks.8 Very few patients experience acquired imatinib resistance resulting from point mutations in the PDGFRA kinase domain (T674I is the most frequent).
The absence of FIP1L1-PDGFRA indicates the need to promptly evaluate other recurrent abnormalities as PDGFRB or FGFR1 fusion genes. Molecular evidence for a PDGFRB or FGFR1 fusion gene is accompanied by its abnormal karyotype equivalent: rearrangement of 5q31-33 for PDGFRB and rearrangement of 8p11-13 for FGFR1. Cheah et al1 report that all PDGFRB patients had 5q31-33 abnormalities underlying the relevance to test cytogenetics in suspected clonal HE. In detail, 54% of their patients had t(5;12)(q33;p13), and overall, 78% had a t(5;12) translocation.1 The t(5;12) (q33;p13) involves the ETV6, the pointed domain of which replaces the extracellular ligand-binding domain of PDGFRB, which is required for oligomerization and activation of the kinase domain. Experiments demonstrated that ETV6-PDGFRB stimulates the proliferation of Ba/F3 cell and in vivo promotes hematopoietic cell proliferation in mouse transplantation models, and that in transducing human CD34-positive cord blood hematopoietic stem cells with ETV6-PDGFRB, an eosinophil differentiation is induced. Hence, this is an oncogenetic alteration, probably acting through the nuclear factor κB pathway. More than 20 fusion partner genes have been describes so far,9 as confirmed by Cheah et al.1 From a diagnostic point of view, these PDGFRB diseases present with a clinical picture of chronic myelomonocytic leukemia with eosinophilia, atypical chronic myelogenous leukemia, or myelodysplastic syndrome/myeloproliferative neoplasm overlap disorders. Imatinib is considered a definitive treatment of PDGFR-activated diseases.8,10 Cheah et al1 reported a very long-term study (median follow-up, 10 years) on efficacy/safety of imatinib in 26 patients with HE.1 Response was achieved in 96% of patients, without relapse or clonal evolution for those achieving cytogenetic or molecular response. The 10-year overall survival is 90%, with a 6-year progression-free survival rate of 88%. Safety profile is good, without HE-specific toxicity.
FGFR1 fusion genes is the last in terms of frequency among clonal HE. The most frequent translocation is t(8;13)(p11;q12), involving ZNF198 at 13q12, resulting in a chimeric protein with constitutive activation of FGFR1. This disease is also known as 8p11 myeloproliferative syndrome or stem cell leukemia/lymphoma, as lesion biopsy shows T-cell lymphoblastic leukemia/lymphoma or mixed myeloid/T-cell lineage, and the disease has an aggressive clinical course.
In conclusion, imatinib in PDGFRB disease really changed the natural history of HE: Patients passed from a 2-year survival of 55% without imatinib to a 10-year survival of 90% with imatinib. Given these therapeutic implications of this long-term observation, physicians must aggressively investigate the molecular basis of these eosinophilic diseases, with emphasis on the PDGFR family, and when identified, imatinib must be started promptly.
Conflict-of-interest disclosure: The author declares no competing financial interests.