Key Points
Liver-specific hepcidin KO mice fully recapitulate the severe iron overload phenotype observed in the total KO mice.
The hepcidin produced by hepatocytes is the main regulator of body iron homeostasis.
Abstract
Hepcidin is a 25-amino-acid peptide demonstrated to be the iron regulatory hormone capable of blocking iron absorption from the duodenum and iron release from macrophages. Mutations affecting hepcidin regulators or the hepcidin gene itself cause hemochromatosis, a common genetic disorder. Hepcidin is produced mainly by the liver, but many cells and tissues express low levels of the hormone. To determine the contribution of these hepcidin-producing tissues in body iron homeostasis, we have developed a new mouse model in which the hepcidin gene can be conditionally inactivated. Here we compare a liver-specific knockout (KO) mouse model with total KO mice. We show that the liver-specific KO mice fully recapitulate the severe iron overload phenotype observed in the total KO mice, with increased plasma iron and massive parenchymal iron accumulation. This result demonstrates that the hepatocyte constitutes the predominant reservoir for systemic hepcidin and that the other tissues are unable to compensate.
Introduction
Hepcidin expression is induced by iron accumulation and diminished in situations of iron needs (increased erythropoiesis and hypoxia).1 Hepcidin controls serum iron levels by binding to ferroportin (FPN), the only known iron exporter, and inducing its degradation.2 Even if the liver is the main site of hepcidin production, many tissues or cells (macrophages,3,4 brain,5 heart,6 retina,7 kidney,8,9 adipocytes,10 pancreas,11 etc) have been shown to express hepcidin at low levels. However, the contribution of these tissues to circulating hepcidin levels and therefore to body iron homeostasis is unknown. To address this question, we generated mice with floxed hamp1 alleles (with locus of crossover in P1 [loxP] sequences flanking exons 2 and 3 of the gene) allowing tissue-specific gene deletion with the classical Cre-loxP strategy. Using the ubiquitous Cre recombinase–deleter mice (E2a-Cre mice), we have validated our targeting strategy and reproduced the iron overload observed in the classical hepcidin knockout (KO) mice.12 Whether nonhepatocyte sources of hepcidin could affect iron homeostasis has not been documented yet. We therefore studied the effect of hepcidin deficiency in the liver by crossing the floxed hepcidin mice (hamp1lox/lox mice) with transgenic mice expressing the hepatocyte-specific albumin Cre recombinase (Alb-Cre).13 Liver-specific KO mice were compared with wild-type (WT) and total KO mice (E2a-Cre). The iron status of the animals was monitored.
Study design
Gene targeting and generation of conditional hamp1 KO mice
The hamp1 targeting construct was generated from polymerase chain reaction (PCR) products amplified from the DNA of 129/SV ES cells. The 5′ (4 kb) and 3′ (1.8 kb) homology arms (bold line) were inserted on either side of a phosphoglycerate kinase promoter-driven hygromycin selection cassette flanked by FLP recombinase target (FRT) sites, in the pL3-FRT-Hygro vector (Figure 1Aii). The generation of the hamp1-targeted clones is described in the supplemental Methods (see the Blood Web site). The hygromycin resistance cassette flanked by FRT sites was excised by crossing hamp1lox/+ mice with flippase-expressing mice (Figure 1Aiii). The resulting heterozygous offspring were bred with deleter E2a-Cre transgenic mice14 or hepatocyte-specific Alb-Cre mice13 (Figure 1Aiv). These mice were crossed to generate WT, hamp1−/− (Hepc∆total), or liver-specific KO (Hepc∆liver) mice. Only males were used in the study. The animal studies described here were reviewed and approved (Agreement no. CEEA34.CP.003.13) by the “Président du Comité d'Ethique pour l'Expérimentation Animale Paris Descartes” and are in accordance with the principles and guidelines established by the European Convention for the Protection of Laboratory Animals.
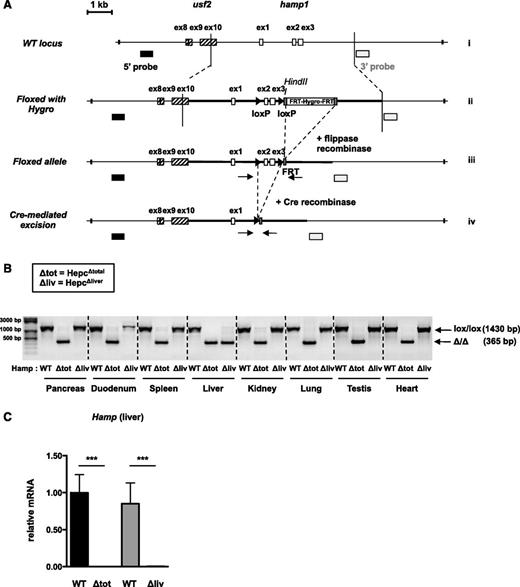
Generation of conditional KO mice for hepcidin. (A) Scheme of the mouse WT hamp1 genomic sequence (i), preceded by the usf2 gene loxP-flanked allele with FRT-flanked Hygro sequence (ii), floxed allele after flippase recombination (iii), and conditional KO allele after Cre-mediated excision (iv). Arrows: P1 and P2 primers used for deletion verification. (B) Recombination of the floxed hamp1 allele by genomic PCR in the pancreas, duodenum, spleen, liver, kidney, lung, testis, and heart of the Hepc∆total, Hepc∆liver and WT mice. (C) Hepcidin messenger RNA (mRNA) expression in the liver of Hepc∆total and Hepc∆liver mice and control littermates. n > 5. Values of WT mice were set up at 1. Samples were normalized to the threshold cycle value for cyclophilin. ***P < .001.
Generation of conditional KO mice for hepcidin. (A) Scheme of the mouse WT hamp1 genomic sequence (i), preceded by the usf2 gene loxP-flanked allele with FRT-flanked Hygro sequence (ii), floxed allele after flippase recombination (iii), and conditional KO allele after Cre-mediated excision (iv). Arrows: P1 and P2 primers used for deletion verification. (B) Recombination of the floxed hamp1 allele by genomic PCR in the pancreas, duodenum, spleen, liver, kidney, lung, testis, and heart of the Hepc∆total, Hepc∆liver and WT mice. (C) Hepcidin messenger RNA (mRNA) expression in the liver of Hepc∆total and Hepc∆liver mice and control littermates. n > 5. Values of WT mice were set up at 1. Samples were normalized to the threshold cycle value for cyclophilin. ***P < .001.
Reverse transcription and real-time quantitative PCR
RNA extraction, quantitative PCR, and sequences of the primers used have been previously described.15
Western blot
Extraction of membrane proteins was performed as previously described.15 The following antibodies were used: divalent metal transporter 1 (DMT1)16 (courtesy of François Cannone-Hergaux), duodenal cytochrome B (DCYTB) (DCYTB11-A; Alpha Diagnostic), FPN (MTP11-A; Alpha Diagnostic), and ferritin (SAB2500431; Sigma-Aldrich).
Iron measurements and immunostaining
Plasma and tissue iron levels were quantified colorimetrically by a previously described method.15 For histology, tissues were fixed in 4% formaldehyde and embedded in paraffin. Five micrometer slides were either stained with Perls’s Prussian blue and nuclear fast red counterstain or stained by immunohistochemistry using FPN antibody (MTP11-A; Alpha Diagnostic).
Hepcidin enzyme-linked immunosorbent assay
Plasma hepcidin was measured following the manufacturer’s instructions (Bachem).
Results and discussion
We introduced, in the mouse genome, loxP sites flanking exons 2 and 3 of hamp1 gene, which encodes the hepcidin protein (“Study design” and Figure 1A). Mice for floxed hamp1 alleles (hamp1lox/lox) were bred to a transgenic line (E2a-Cre) expressing a Cre recombinase under the control of the ubiquitous E2a promoter17 or to transgenic mice expressing the hepatocyte-specific Alb-Cre recombinase,13 to generate total (Hepc∆total) and liver-specific (Hepc∆liver) homozygote KO mice, respectively. We observed an efficient truncation of the floxed hamp1 allele in all the organs of the Hepc∆total mice we tested. As expected, deletion of the floxed hamp1 allele was observed only in the liver of the Hepc∆liver mice and not in the other organs (Figure 1B). Hepcidin expression, measured by quantitative PCR, was completely absent in the liver of the Hepc∆total and Hepc∆liver mice compared with their WT littermates (Figure 1C).
Plasma hepcidin inhibits iron efflux into plasma by directly binding to and inducing the degradation of FPN, which is present on macrophages and enterocytes. We therefore measured FPN expression in spleen and duodenum extracts of Hepc∆total and Hepc∆liver mice. FPN was similarly increased in the spleen of 6-month-old Hepc∆total and Hepc∆liver mice compared with control littermates, as assessed by western-blot analysis (with no alteration of FPN mRNA level; not shown). As a consequence, ferritin levels were decreased in both Hepc∆total and Hepc∆liver mice (Figure 2A), corroborated by the absence of iron staining in the spleen compared with WT animals (Figure 2D). In the duodenum, we showed by immunohistochemistry (Figure 2B) that FPN expression was also similarly increased in both mutant mice (with a modest twofold increase of FPN mRNA level; not shown). The increased rate of the duodenal iron absorption proteins (DCYTB and DMT1) was identical in the Hepc∆total and Hepc∆liver mice compared with their control littermates (Figure 2C).
The liver-specific KO mice recapitulate the phenotype of the total KO mice. (A) FPN and ferritin expression in the spleen of 6-month-old Hepc∆total and Hepc∆liver mice and their control littermates by western blotting. (B) Immunohistochemistry for FPN in the duodenum of Hepc∆total and Hepc∆liver mice and WT mice. Bars represent 50 μm. (40/0.60, Leica DMI3000B microscope, Leica DFC310FX camera [Leica LAS Core software]). (C) Western blotting in duodenum scrapings of 6-month-old Hepc∆total, Hepc∆liver, and control mice. (D) Liver, pancreas, and plasma iron in 2-, 3-, and 6-month-old Hepc∆total and Hepc∆liver mice and their control littermates. n > 5. *P < .05, ***P < .001. ns indicates not significant. Perls’s blue staining of the spleen of 6-month-old Hepc∆total and Hepc∆liver mice and WT mice. Bars represent 100 μm. (20/0.4, Leica DMI3000B microscope, Leica DFC310FX camera [Leica LAS Core software]). (E) Plasma hepcidin in 6-month-old control, Hepc∆total, and Hepc∆liver mice. n = 4.
The liver-specific KO mice recapitulate the phenotype of the total KO mice. (A) FPN and ferritin expression in the spleen of 6-month-old Hepc∆total and Hepc∆liver mice and their control littermates by western blotting. (B) Immunohistochemistry for FPN in the duodenum of Hepc∆total and Hepc∆liver mice and WT mice. Bars represent 50 μm. (40/0.60, Leica DMI3000B microscope, Leica DFC310FX camera [Leica LAS Core software]). (C) Western blotting in duodenum scrapings of 6-month-old Hepc∆total, Hepc∆liver, and control mice. (D) Liver, pancreas, and plasma iron in 2-, 3-, and 6-month-old Hepc∆total and Hepc∆liver mice and their control littermates. n > 5. *P < .05, ***P < .001. ns indicates not significant. Perls’s blue staining of the spleen of 6-month-old Hepc∆total and Hepc∆liver mice and WT mice. Bars represent 100 μm. (20/0.4, Leica DMI3000B microscope, Leica DFC310FX camera [Leica LAS Core software]). (E) Plasma hepcidin in 6-month-old control, Hepc∆total, and Hepc∆liver mice. n = 4.
Finally, we examined the age-dependent variation (2, 3, and 6 months) of the iron parameters in the Hepc∆total, Hepc∆liver, and control mice (Figure 2D). Even though liver iron content was higher in the liver-specific KO compared with the total KO mice at 2 months (with, however, a similar Perls’s blue staining; supplemental Figure 1), both mutant mice had a massive iron accumulation at 3 and 6 months. Iron gradually accumulated in the pancreas, in an age-dependent manner in both Hepc∆total and Hepc∆liver mice. Plasma iron increased at 2 months and remained constant during the following months. Hepcidin was undetectable in the plasma of Hepc∆total but also in Hepc∆liver mice (Figure 2E), confirming the lack of contribution of extrahepatic organs in the overall circulating hepcidin.
In conclusion, hepatic hepcidin is fundamental for the maintenance of systemic iron homeostasis in basal conditions. The liver-specific KO mice fully recapitulate the severe iron overload phenotype observed in the total KO mice demonstrating that the hepatocyte constitutes the predominant reservoir for systemic hepcidin, the other tissues being unable to compensate for the severe iron overload. Our results correlate with the liver-specific KOs of hepcidin regulators HFE,18 hemojuvelin,19 or transferrin receptor 2,20 which all display a severe iron overload, similar to that of the respective total KO mice.
This new mouse model, in which the hepcidin gene can be spatiotemporally inactivated, will be important for future research to determine the impact of hepcidin deficiency in different tissues in pathophysiological conditions. For example, hypoxia induces a strong increase of hepcidin expression in the heart,6 suggesting its implication in cardiac diseases. Inflammatory stimuli have been shown to strongly upregulate hepcidin in myeloid cells, such as macrophages and neutrophils,3,4 or in the brain.21 Therefore, nonhepatic hepcidin may be essential at the site of infections and/or in poorly perfused tissues, inaccessible by systemic hepcidin from the circulation.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Marc Foretz for helpful advice on the generation of the floxed construct, François Cannone-Hergaux for the DMT1 antibody, and Terrie and Philip Sangiorgio for the English proofreading.
This study was supported by a grant from the European Research Council under the European Community’s Seventh Framework Program (FP7/2011-2015 grant agreement no. 261296).
Authorship
Contribution: S.Z., J.R.R.M., S.D., M.H., L.V., and C.P. performed experiments; S.Z., J.R.R.M., S.D., M.H., L.V., S.V., and C.P. conceived, analyzed, and interpreted the experiments; and C.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carole Peyssonnaux, Institut Cochin, Département Endocrinologie Métabolisme et Diabète, INSERM U1016, CNRS UMR8104, 24 rue du Faubourg Saint Jacques, 75014 Paris, France; e-mail: carole.peyssonnaux@inserm.fr.

![Figure 2. The liver-specific KO mice recapitulate the phenotype of the total KO mice. (A) FPN and ferritin expression in the spleen of 6-month-old Hepc∆total and Hepc∆liver mice and their control littermates by western blotting. (B) Immunohistochemistry for FPN in the duodenum of Hepc∆total and Hepc∆liver mice and WT mice. Bars represent 50 μm. (40/0.60, Leica DMI3000B microscope, Leica DFC310FX camera [Leica LAS Core software]). (C) Western blotting in duodenum scrapings of 6-month-old Hepc∆total, Hepc∆liver, and control mice. (D) Liver, pancreas, and plasma iron in 2-, 3-, and 6-month-old Hepc∆total and Hepc∆liver mice and their control littermates. n > 5. *P < .05, ***P < .001. ns indicates not significant. Perls’s blue staining of the spleen of 6-month-old Hepc∆total and Hepc∆liver mice and WT mice. Bars represent 100 μm. (20/0.4, Leica DMI3000B microscope, Leica DFC310FX camera [Leica LAS Core software]). (E) Plasma hepcidin in 6-month-old control, Hepc∆total, and Hepc∆liver mice. n = 4.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/23/10.1182_blood-2014-01-550467/4/m_3646f2.jpeg?Expires=1769082180&Signature=qDtJzmgL~1Ojwj8py3tHEceI8ysXVhejz2QFNn1FgA72xmada~i98eeiJnFOw6QW5q2vnoU6wd9oqvwH17mi2pGHo0GmKN9gRiRrQ118AYxqa2HX8ocHmwc4E1CIEIWfJX1ePdpcNlQRT8tNRD8FAUoNdGxIvGHLVktbsZhSwQeDMx0PCKr~g40T0~IAgn8euq6rIvkatjFSsd37ryL6rgybxuTX6qSZzTRtRIv6Uu9ZlNunw80G7DUd066hfi9hdrPZchQ7l1KJB-q6nVIkoJ91AGoBXcCdShVv7hSzil~qloCuEjPSqqcq5DHyJ7qMjEUVgDHOM9Fm2XVntU1lRQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal