Key Points
Aggf1 is required for both primitive and definitive hematopoiesis.
Aggf1 is the earliest known regulator for differentiation of hemangioblasts.
Abstract
The hemangioblast is a multipotential progenitor, which is derived from the mesoderm and can further differentiate into hematopoietic and endothelial lineages. The molecular mechanism governing the specification of hemangioblasts is fundamental to regenerative medicine based on embryonic stem cells for the treatment of various hematologic and vascular diseases. Here we show that aggf1 acts at the top of the genetic regulatory hierarchy in the specification of hemangioblasts in zebrafish. Knockdown of aggf1 expression decreases expression of endothelial cell–specific markers (cdh5, admr) and disrupts primitive hematopoiesis as shown by a decreased number of erythroid cells and reduced expression of gata1 (marker for erythroid progenitors) and pu.1 (myeloid progenitors). Aggf1 knockdown also decreases expression of runx1 and c-myb, indicating that it is required for specification of hematopoietic stem cells (definitive hematopoiesis). Aggf1 knockdown led to dramatically reduced expression of hemangioblast markers fli1, etsrp, lmo2, and scl, and hematopoietic/endothelial defects in aggf1 morphants were rescued by messenger RNA for scl, fli-vp16, or etsrp. Taken together, these data indicate that aggf1 is involved in differentiation of both hematopoietic and endothelial lineages and that aggf1 acts upstream of scl, fli1, and etsrp in specification of hemangioblasts.
Introduction
Hemangioblasts are the multipotent progenitors that can differentiate into both hematopoietic and endothelial cells (ECs).1,2 Defining the molecular mechanisms underlying specification of hemangioblasts may have great clinical potential for regenerative medicine because it may lead to new strategies for treating a variety of hematologic and vascular disorders.3 The existence of hemangioblasts has been demonstrated in human and mouse embryos by the finding that single-cell–derived colonies can generate both hematopoietic and ECs in in vitro differentiation assays of mouse or human embryonic stem cells.2,4,5 Single-cell–resolution fate mapping of late zebrafish blastula and gastrula demonstrated the existence of hemangioblasts that produced both hematopoietic and ECs in vivo.6 Emerging evidence suggests that hemangioblasts may exist as circulating stem cells in the peripheral blood in the adult.7
The bipotential hemangioblasts are developed from the mesoderm in the early vertebrate embryo or the ventral mesoderm from zebrafish embryos.1,8 Hemangioblasts can then differentiate into either hematopoietic or EC lineages.1,2 For specification of the EC lineage, hemangioblasts first differentiate into angioblasts, which then give rise to vascular ECs, including arterial ECs and venous ECs.9 The hematopoietic lineage will generate blood cells, including erythroid cells, myeloid cells, and lymphoid cells.10,11
Early vertebrate embryonic hematopoiesis consisted of 2 stages or waves, the primitive hematopoiesis and definitive hematopoiesis, at different anatomical sites.12 The first stage of primitive hematopoiesis occurs in the intermediate cell mass in zebrafish.11,13 The second wave of definitive hematopoiesis produces adult-type hematopoietic stem cells (HSCs) in major vessels or highly vascularized structures, that is, the aorta-gonad-mesonephros region, umbilical/vitelline arteries, yolk sac, and placenta at the E10-11 stage.14 In zebrafish, the functionally equivalent structure of the aorta-gonad-mesonephros region is found in the ventral wall of the dorsal aorta marked by expression of c-myb and runx1.8,15,16
The differentiation of hemangioblasts from the mesoderm is regulated by a number of transcriptional factor genes, including several members of the E-twenty six family (etsrp, fli1), scl, and lmo2. Scl/tal1 encodes a basic helix-loop-helix transcription factor and plays an essential role in hematopoiesis and development of the dorsal aorta during zebrafish embryogenesis.17-19 Fli1 was shown to act upstream of scl to drive blood and EC differentiation.20 Etsrp was shown to act upstream of the full-length isoform of scl (scl-α) and fli1 to specify angioblasts or the N-terminal truncated isoform of scl-β to specify HSCs.21,22 Lmo2 encodes an LIM-containing protein that forms a complex with Scl/Tal1 to specify the development of hemangioblasts.23,24
A number of transcription factors are expressed in the anterior lateral mesoderm and/or the posterior lateral mesoderm and play a critical role in primitive and definitive hematopoiesis. Gata1 and pu.1 encode 2 early transcription factors regulating primitive hematopoiesis. Gata1 is a critical gene in dictating the erythroid lineage fate.25 In contrast, pu.1 plays a potential role in promoting myeloid cell fate and repressing erythroid differentiation.26 C-myb and runx1 are 2 genes that are involved in definitive hematopoiesis to generate HSCs that further differentiate into erythroid, myeloid, and lymphoid lineages.8,15,16
Aggf1 encodes an interesting angiogenic factor with G-Patch and FHA domains and is associated with increased susceptibility for the vascular disease Klippel-Trenaunay syndrome.27-29 Similar to the vascular endothelial growth factor (VEGF) A, purified wild-type AGGF1 protein promoted strong angiogenesis.27 Recently, we demonstrated that aggf1 plays an important role in the specification of veins and angiogenesis by activating the AKT signaling pathway in zebrafish embryogenesis.30
As discussed previously, several regulators have been found to specify differentiation of hemangioblasts, but the molecular mechanisms underlying hemangioblast differentiation are largely unknown, and the master genes controlling hemangioblast development remain to be identified. In this study, we found that aggf1 is a master molecular determinant for hemangioblast specification and is involved in the specification of both hematopoietic and endothelial lineages during zebrafish embryogenesis.
Methods
Zebrafish and microinjection
Wild-type AB strain zebrafish and clom39+/− mutant line (kindly provided by Dr Jing-Wei Xiong from Peking University) were used in the study. Embryos were raised at 28.5°C, treated with 0.003% 1-phenyl-2-thiourea (Sigma-Aldrich) to prevent pigment formation, and collected at different stages for analysis.31,32 The study was approved by the ethics committee of Huazhong University of Science and Technology.
Two aggf1-specific morpholinos (MO1, MO2) and a standard morpholino (Std-MO) were tested and used as described in our previous study.30 Different doses of aggf1 MOs were analyzed, and doses of 2 ng for aggf1 MO1 and 4 ng for MO2 were selected to avoid toxicity and lethality (supplemental Figure 1; available on the Blood Web site).
In vitro transcription was used to generate messenger RNA (mRNA) for aggf1, scl, etsrp, egfp, and lycat-egfp using the mMessage mMachine SP6 kit (Ambion) and for fli-vp16 using the T3 kit as described previously.20
Whole mount in situ hybridization
o-Dianisidine staining
Staining of hemoglobin was carried out in zebrafish by o-dianisidine as described previously.35
Results
Aggf1 is required for primitive hematopoiesis
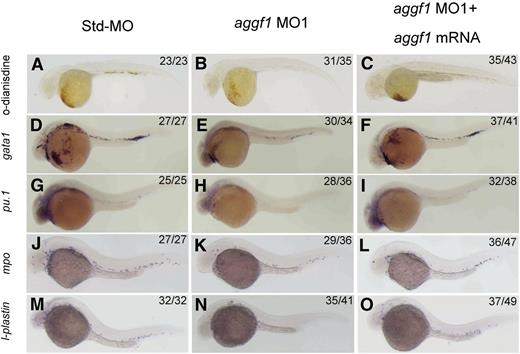
To characterize the important physiological functions of aggf1 during embryogenesis, we used 2 different aggf1 MOs to knock down the expression of aggf1 in zebrafish embryos. The first MO, MO1, targets the junction between the first exon and the first intron, whereas the second MO, MO2, targets the aggf1 ATG region. The effectiveness of aggf1 MO1 and MO2 has been demonstrated previously.30 Std-MO was used as control. During the study, careful examinations of MO-injected embryos under a phase-contrast microscope revealed an interesting morphologic abnormality in blood circulation. Zebrafish embryos injected with aggf1 MO1 showed fewer red cells than embryos injected with Std-MO (supplemental Video 1). Furthermore, aggf1 morphants showed no apparent circulation at 48 hpf compared with control embryos (supplemental Video 1). Similar results were obtained for aggf1 MO2 (data not shown). Thus, aggf1 may be involved in hematopoiesis. To further verify this finding, we used o-dianisidine staining (binding to heme) to detect erythroid cells. At 48 hpf, 2 ng of aggf1 MO1 caused a marked decrease in the number of red blood cells (Figure 1A-B). In order to test whether the observed result was specific to aggf1 or off target, we coinjected aggf1 MO1 (2 ng) and in vitro synthesized, capped zebrafish aggf1 mRNA (100 pg) into 1- to 2-cell embryos. At 48 hpf, coinjection of aggf1 mRNA rescued the decreased number of erythroid cells caused by aggf1 MO1 (compare Figure 1C with Figure 1B). Similar results were obtained for aggf1 MO2 (data not shown). These results suggest that aggf1 plays an important role in hematopoiesis.
Knockdown of aggf1 expression results in abnormal primitive hematopoiesis during zebrafish embryogenesis. (A-C) Dramatic reduction of the number of erythroid cells by an aggf1 morpholino (MO1). Zebrafish embryos in the 1- to 2-cell stage were injected with 2 ng of Std-MO (A) or 2 ng of MO1 (B). Erythroid cells in the circulatory system were detected with o-dianisidine at 48 hpf. A similar experiment with injection of 2 ng of MO1 together with 100 pg of zebrafish aggf1 mRNA (C) into embryos was performed to determine whether overexpression of aggf1 can rescue the defects in aggf1 morphants. MO1 was designed in a manner to knock down endogenous zebrafish aggf1 mRNA only and does not knock down the in vitro synthesized aggf1 mRNA used for injection. (D-F) Knockdown of aggf1 reduces the gata1 signal, a marker for erythroid progenitors. Zebrafish embryos (1- to 2-cell stage) were injected with 2 ng of Std-MO (D), 2 ng of aggf1 MO1 (E), or 2 ng of aggf1 MO1 together with 100 pg of aggf1 mRNA (F) and used for WISH at 26 hpf with an antisense probe for gata1. (G-I) Knockdown of aggf1 reduces the pu.1 signal, a marker for myeloid progenitors. The embryos were injected and processed as in panels D-F, but with an antisense probe for pu.1. (J-O) Knockdown of aggf1 expression results in a reduced number of granulocytes and macrophages during zebrafish early embryogenesis. (J-L) Knockdown of aggf1 expression reduces the mpo signal, a marker for granulocytes. Zebrafish embryos (1- to 2-cell stage) were injected with 2 ng of Std-MO (J), 2 ng of aggf1 MO1 (K), or 2 ng of aggf1 MO1 together with 100 pg of aggf1 mRNA (L) and used for WISH at 28 hpf with an antisense probe for mpo. Knockdown of aggf1 expression reduces the l-plastin signal at 28 hpf, a marker for macrophages (M-O). All embryos are shown in a lateral view with the anterior on the left.
Knockdown of aggf1 expression results in abnormal primitive hematopoiesis during zebrafish embryogenesis. (A-C) Dramatic reduction of the number of erythroid cells by an aggf1 morpholino (MO1). Zebrafish embryos in the 1- to 2-cell stage were injected with 2 ng of Std-MO (A) or 2 ng of MO1 (B). Erythroid cells in the circulatory system were detected with o-dianisidine at 48 hpf. A similar experiment with injection of 2 ng of MO1 together with 100 pg of zebrafish aggf1 mRNA (C) into embryos was performed to determine whether overexpression of aggf1 can rescue the defects in aggf1 morphants. MO1 was designed in a manner to knock down endogenous zebrafish aggf1 mRNA only and does not knock down the in vitro synthesized aggf1 mRNA used for injection. (D-F) Knockdown of aggf1 reduces the gata1 signal, a marker for erythroid progenitors. Zebrafish embryos (1- to 2-cell stage) were injected with 2 ng of Std-MO (D), 2 ng of aggf1 MO1 (E), or 2 ng of aggf1 MO1 together with 100 pg of aggf1 mRNA (F) and used for WISH at 26 hpf with an antisense probe for gata1. (G-I) Knockdown of aggf1 reduces the pu.1 signal, a marker for myeloid progenitors. The embryos were injected and processed as in panels D-F, but with an antisense probe for pu.1. (J-O) Knockdown of aggf1 expression results in a reduced number of granulocytes and macrophages during zebrafish early embryogenesis. (J-L) Knockdown of aggf1 expression reduces the mpo signal, a marker for granulocytes. Zebrafish embryos (1- to 2-cell stage) were injected with 2 ng of Std-MO (J), 2 ng of aggf1 MO1 (K), or 2 ng of aggf1 MO1 together with 100 pg of aggf1 mRNA (L) and used for WISH at 28 hpf with an antisense probe for mpo. Knockdown of aggf1 expression reduces the l-plastin signal at 28 hpf, a marker for macrophages (M-O). All embryos are shown in a lateral view with the anterior on the left.
Zebrafish hematopoiesis includes primitive hematopoiesis and definitive hematopoiesis.36 To assess the role of aggf1 in primitive hematopoiesis, we performed WISH for gata1, a marker for erythroid progenitors23 in aggf1 morphants. The expression of gata1 was downregulated by 2 different aggf1 MOs compared with control Std-MO (Figure 1D-E; supplemental Figure 2A-B). The decreased expression of gata1 was partially rescued by injection of in vitro synthesized aggf1 mRNA in aggf1 morphants (Figure 1D-F). Similarly, the expression of pu.1 (marker for myeloid progenitors)37 was reduced in aggf1 morphants compared with control embryos injected with Std-MO (Figure 1G-H; supplemental Figure 2C-D). Coinjection of in vitro synthesized aggf1 mRNA (100 pg) rescued the decreased expression of pu.1 in aggf1 morphants (compare Figure 1I with Figure 1H). These results indicate that aggf1 is involved in primitive hematopoiesis during zebrafish embryogenesis.
We also investigated the development of myeloid cells in aggf1 morphants by WISH. The expression of mpo (marker for granulocytes)38 and l-plastin (marker for macrophages)38 was reduced in aggf1 morphants compared with control embryos injected with Std-MO at 28 hpf (Figure 1J-K,M-N; supplemental Figure 3A-D). Coinjection of in vitro synthesized aggf1 mRNA (100 pg) rescued the decreased expression of myeloid cell markers in aggf1 morphants (compare Figure 1L with Figure 1K; compare Figure 1O with Figure 1N).
Aggf1 is required for the definitive hematopoiesis
To assess the role of aggf1 in definitive hematopoiesis and the emergence of HSCs in zebrafish embryogenesis, we analyzed the expression of runx1 and c-myb in the ventral wall of the dorsal aorta in aggf1 morphants by WISH.8,15,16 In aggf1 morphants, expression of runx1 and c-myb was dramatically reduced in the ventral wall of the dorsal aorta at 30 hpf (Figure 2A-C,E-G), which suggests that the definitive hematopoiesis is defective when aggf1 is knocked down. Injection of aggf1 mRNA (100 pg) rescued the downregulated expression of runx1 and c-myb in aggf1 morphants, which suggests that the defect of definitive hematopoiesis is caused by specific aggf1 knockdown. Together, these data show that zebrafish aggf1 is involved in the definitive hematopoiesis and specification of HSCs in zebrafish embryogenesis. This finding was further confirmed by the defective development of lymphoid cells in aggf1 morphants. The expression of rag1 (marker for lymphoid cells)36 was nearly absent in aggf1 morphants (Figure 2I-J; supplemental Figure 3E-F). Coinjection of in vitro synthesized aggf1 mRNA (100 pg) rescued the decreased expression rag1 in aggf1 morphants (compare Figure 2K with Figure 2J).
Knockdown of aggf1 expression results in abnormal definitive hematopoiesis during zebrafish embryogenesis. Knockdown of aggf1 expression reduces the runx1 signal (A-D) or c-myb signal (E-H) in the ventral wall of the dorsal aorta, 2 independent markers for HSCs. Zebrafish embryos (1- to 2-cell stage) were injected with 4 ng of Std-MO (A,E), 2 ng of aggf1 MO1 (B,F), 4 ng of aggf1 MO2 (C,G), or 2 ng of aggf1 MO1 together with 100 pg of aggf1 mRNA (D,H) and used for WISH at 30 hpf with an antisense probe for runx1 or c-myb. Knockdown of aggf1 expression abolishes the rag1 signal, a marker for lymphoid cells, at 5 dpf (I-K). (A-H) Embryos are shown in a lateral view with the anterior on the left. (I-K) Embryos are shown as a dorsal view with the anterior at the top.
Knockdown of aggf1 expression results in abnormal definitive hematopoiesis during zebrafish embryogenesis. Knockdown of aggf1 expression reduces the runx1 signal (A-D) or c-myb signal (E-H) in the ventral wall of the dorsal aorta, 2 independent markers for HSCs. Zebrafish embryos (1- to 2-cell stage) were injected with 4 ng of Std-MO (A,E), 2 ng of aggf1 MO1 (B,F), 4 ng of aggf1 MO2 (C,G), or 2 ng of aggf1 MO1 together with 100 pg of aggf1 mRNA (D,H) and used for WISH at 30 hpf with an antisense probe for runx1 or c-myb. Knockdown of aggf1 expression abolishes the rag1 signal, a marker for lymphoid cells, at 5 dpf (I-K). (A-H) Embryos are shown in a lateral view with the anterior on the left. (I-K) Embryos are shown as a dorsal view with the anterior at the top.
Aggf1 is involved in specification of vascular ECs
There is a close lineage relationship between hematopoiesis and development of ECs.1,2 As such, aggf1 is also involved in specification of EC.30 This is further demonstrated by WISH with several new EC-specific markers. The cadherin 5 (cdh5, vascular endothelium–cadherin) and adrenomedullin receptor (admr) genes are expressed in all vascular ECs in early zebrafish embryos.39,40 We found that expression of cdh5 and admr were both dramatically reduced in axial vessels and nearly absent in intersegmental vessels at 26 hpf after injection of aggf1 MO1 or MO2 (supplemental Figure 4). The reduced expression of cdh5 and admr was restored to nearly normal levels by coinjection of aggf1 mRNA in aggf1 morphants. The data indicate that aggf1 is involved in development of ECs.
Aggf1 is involved in specification of hemangioblasts
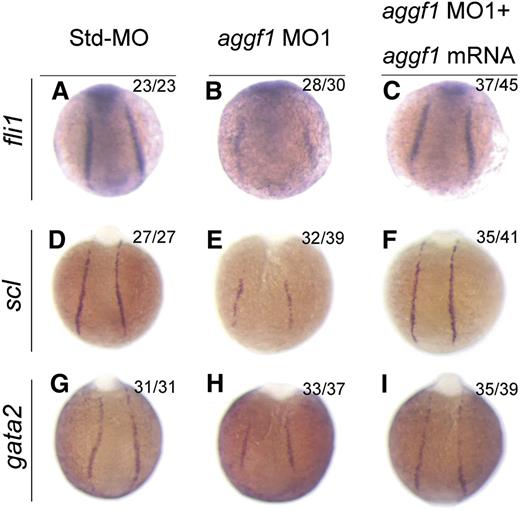
Because HSCs and ECs are derived from the same progenitor, the hemangioblast,2 and aggf1 is involved in specification of both types of cells, we hypothesized that aggf1 was involved in specification of hemangioblasts during early somitogenesis stages. To test the hypothesis, we performed WISH with several markers for hemangioblasts, HSCs, and angioblasts. Fli1 was shown to be at the top of the transcriptional network for specification of hemangioblasts.20 The expression of fli1 was reduced in aggf1 morphants compared with control embryos injected with Std-MO (Figure 3A-B; supplemental Figure 5A). This result was confirmed by quantitative reverse-transcription polymerase chain reaction analysis (supplemental Figure 6).
Specification of hemangioblasts and hematopoietic cells is defective in aggf1 morphants. (A-C) Zebrafish embryos injected with Std-MO (2 ng; A,D,G), aggf1 MO1 (2 ng; B,E,H), or MO1 (2 ng) together with 100 pg of aggf1 mRNA (C,F,I), and used for WISH with probes fli1 and scl (markers for hemangioblasts) and gata2 (marker for hematopoietic cells) at 8 somites. Expression of scl, fli1, and gata2 is severely reduced in the posterior lateral plate mesoderm (PLPM) by knockdown of aggf1 expression. Embryos are shown at a dorsal view with the anterior at the top.
Specification of hemangioblasts and hematopoietic cells is defective in aggf1 morphants. (A-C) Zebrafish embryos injected with Std-MO (2 ng; A,D,G), aggf1 MO1 (2 ng; B,E,H), or MO1 (2 ng) together with 100 pg of aggf1 mRNA (C,F,I), and used for WISH with probes fli1 and scl (markers for hemangioblasts) and gata2 (marker for hematopoietic cells) at 8 somites. Expression of scl, fli1, and gata2 is severely reduced in the posterior lateral plate mesoderm (PLPM) by knockdown of aggf1 expression. Embryos are shown at a dorsal view with the anterior at the top.
Scl is a marker for hemangioblasts, HSCs, and angioblasts.21 The expression of scl was reduced in aggf1 morphants compared with control embryos injected with Std-MO (Figure 3D-E; supplemental Figure 5B). The expression levels of scl and another hemangioblast marker, etsrp, were dependent on the doses of aggf1 MO (supplemental Figure 7). This result was confirmed by quantitative reverse-transcription polymerase chain reaction analysis (supplemental Figure 6).
Gata2 binds to scl and lmo2 to regulate the development of HSCs.41 The expression of gata2 and lmo2 was reduced in aggf1 morphants compared with control embryos injected with Std-MO (Figure 3G-H; supplemental Figure 5C-D).
Coinjection of in vitro synthesized full-length aggf1 mRNA rescued the reduced expression of hemangioblast markers fli1, scl, and lmo2 (compare Figure 3C with Figure 3B; compare Figure 3F with Figure 3E; compare Figure 3I with Figure 3H). Together, these data suggest that the zebrafish aggf1 gene is involved in the specification of hemangioblasts, HSCs, and angioblasts.
Knockdown of aggf1 expression does not affect expression of endoderm-specific markers, ectoderm markers, or nonhemangioblast-related mesoderm markers
To investigate whether the function of aggf1 is limited to the specification of hemangioblasts derived from the mesoderm, we examined the expression of 1 endoderm-specific marker, sox17, and 2 ectoderm markers, krox20 and otx1, in aggf1 morphants by WISH.42,43 The expression of sox17, krox20, or otx1 was not affected in aggf1 morphants compared with control embryos injected with standard MO (supplemental Figure 8A-F,Q-R).
To further investigate whether aggf1 knockdown affects the expression of the mesoderm markers not related to specification of hemangioblasts, we performed WISH for ntl, papc, eve1, fgf8, and nkx2.5.44 The expression of these markers was not changed in aggf1 morphants compared with control embryos (supplemental Figure 8G-P).
Together with our previous results showing lack of effects of aggf1 knockdown on mesoderm markers myoD and pax2.1,30 these data suggest that the function of aggf1 is likely limited to specification of hemangioblasts in zebrafish embryogenesis.
Aggf1 acts upstream of scl to regulate hematopoiesis
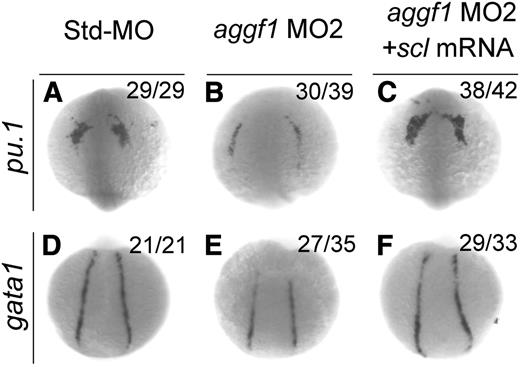
Scl has been implicated in the hematopoietic and vascular development in zebrafish.17-19 Because knockdown of aggf1 expression decreased scl expression, which might result in reduced expression of gata1 (erythroid progenitor marker) and pu.1 (myeloid progenitor marker), aggf1 may regulate hematopoiesis through scl. To further verify this hypothesis, we performed rescue studies with scl mRNA. As shown in Figure 4, aggf1 MO2 reduced the expression of gata1 in the PLPM and pu.1 in the anterior lateral plate mesoderm at the 8-somite stage, but the effect was rescued by coinjection of scl mRNA. These results suggest that aggf1 acts upstream of scl to regulate hematopoiesis during zebrafish embryogenesis.
Overexpression of scl rescues defects in hematopoiesis in aggf1 morphants. Zebrafish embryos were injected with Std-MO (4 ng; A,D), aggf1 MO2 (4 ng; B,E), or aggf1 MO2 (4 ng) together with in vitro synthesized full-length scl mRNA (75 pg) (C,F) and used for WISH at 8 somites with antisense probes for pu.1 (A-C) and gata1 (D-F). Reduced expression of pu.1 (marker for myeloid progenitor) in anterior lateral plate mesoderm by knockdown of aggf1 expression was rescued by overexpression of scl mRNA (A-C). Coinjection of scl mRNA rescued decreased expression of gata1 (marker for erythroid progenitor) in PLPM caused by aggf1 MO2 (D-F). (A-C) Anterior view with the ventral side at the top. (D-F) Dorsal view with the anterior at the top.
Overexpression of scl rescues defects in hematopoiesis in aggf1 morphants. Zebrafish embryos were injected with Std-MO (4 ng; A,D), aggf1 MO2 (4 ng; B,E), or aggf1 MO2 (4 ng) together with in vitro synthesized full-length scl mRNA (75 pg) (C,F) and used for WISH at 8 somites with antisense probes for pu.1 (A-C) and gata1 (D-F). Reduced expression of pu.1 (marker for myeloid progenitor) in anterior lateral plate mesoderm by knockdown of aggf1 expression was rescued by overexpression of scl mRNA (A-C). Coinjection of scl mRNA rescued decreased expression of gata1 (marker for erythroid progenitor) in PLPM caused by aggf1 MO2 (D-F). (A-C) Anterior view with the ventral side at the top. (D-F) Dorsal view with the anterior at the top.
Aggf1 acts upstream of fli1 and etsrp to regulate specification of hemangioblasts
Fli1 has been reported to act at the top of the transcriptional network to drive development of blood cells and ECs.20 Because of reduced fli1 expression in aggf1 morphants, we hypothesized that aggf1 acted upstream of fli1 to regulate development of both hematopoietic and endothelial lineages. To test the hypothesis, we coinjected fli-vp16 mRNA together with aggf1 MO2 into embryos at the 1- to 2-cell stage. WISH showed reduced expression of lmo2 (hemangioblast marker), scl (hemangioblast marker), and gata2 (hematopoietic factor)22 at the 10-somite stage in aggf1 morphants, but the effect was rescued by fli-vp16 mRNA (Figure 5A-I). These results suggest that aggf1 acts upstream of fli1 to regulate specification of hemangioblasts.
Etsrp is a marker for hemangioblasts.22 We previously showed that knockdown of aggf1 expression significantly reduced expression of etsrp in the PLPM.30 Here we performed rescue studies with etsrp mRNA to conclusively demonstrate that aggf1 acts upstream of etsrp, which is necessary and sufficient for initiating vasculogenesis and specification of hemangioblasts.22 We coinjected etsrp mRNA together with aggf1 MO1 into embryos at the 1- to 2-cell stage. WISH showed that reduced expression of scl (hemangioblast marker) and lmo2 (hemangioblast marker) at the 6-somite stage by aggf1 MO1 was rescued by etsrp mRNA (Figure 5J-O). These results suggest that aggf1 acts upstream of etsrp to regulate specification of hemangioblasts.
Overexpression of fli-vp16 or etsrp rescues defects in specification of hemangioblasts and hematopoietic cells in aggf1 morphants. (A-I) Zebrafish embryos (1- to 2-cell stage) were injected with Std-MO (4 ng; A,D,G), aggf1 MO2 (4 ng; B,E,H), or aggf1 MO2 (4 ng) together with fli-vp16 mRNA (75 pg) (C,F,I) and used for WISH at 10 somites with antisense probes for scl (A-C), lmo2 (D-F), and gata2 (G-I). Reduced expression of hemangioblast markers (scl, lmo2) in PLPM by aggf1 MO2 was restored by overexpression of fli-vp16 mRNA (A-F). Similarly, reduced expression of hematopoietic marker gata2 in PLPM by knockdown of aggf1 expression was restored by overexpression of fli-vp16 mRNA (G-I). (J-O) Zebrafish embryos (1- to 2-cell stage) were injected with Std-MO (2 ng; J,M), aggf1 MO1 (2 ng; K,N), or aggf1 MO1 (2 ng) together with etsrp mRNA (100 pg) (L, O) and used for WISH at 6 somites with antisense probes for scl (J-L) and lmo2 (M-O). Reduced expression of scl and lmo2 in PLPM by aggf1 MO1 was restored by overexpression of etsrp mRNA, suggesting that etsrp is downstream of aggf1 during differentiation of hemangioblasts. Embryos are shown in a dorsal view with the anterior at the top.
Overexpression of fli-vp16 or etsrp rescues defects in specification of hemangioblasts and hematopoietic cells in aggf1 morphants. (A-I) Zebrafish embryos (1- to 2-cell stage) were injected with Std-MO (4 ng; A,D,G), aggf1 MO2 (4 ng; B,E,H), or aggf1 MO2 (4 ng) together with fli-vp16 mRNA (75 pg) (C,F,I) and used for WISH at 10 somites with antisense probes for scl (A-C), lmo2 (D-F), and gata2 (G-I). Reduced expression of hemangioblast markers (scl, lmo2) in PLPM by aggf1 MO2 was restored by overexpression of fli-vp16 mRNA (A-F). Similarly, reduced expression of hematopoietic marker gata2 in PLPM by knockdown of aggf1 expression was restored by overexpression of fli-vp16 mRNA (G-I). (J-O) Zebrafish embryos (1- to 2-cell stage) were injected with Std-MO (2 ng; J,M), aggf1 MO1 (2 ng; K,N), or aggf1 MO1 (2 ng) together with etsrp mRNA (100 pg) (L, O) and used for WISH at 6 somites with antisense probes for scl (J-L) and lmo2 (M-O). Reduced expression of scl and lmo2 in PLPM by aggf1 MO1 was restored by overexpression of etsrp mRNA, suggesting that etsrp is downstream of aggf1 during differentiation of hemangioblasts. Embryos are shown in a dorsal view with the anterior at the top.
Aggf1 and cloche act independently during differentiation of hemangioblasts
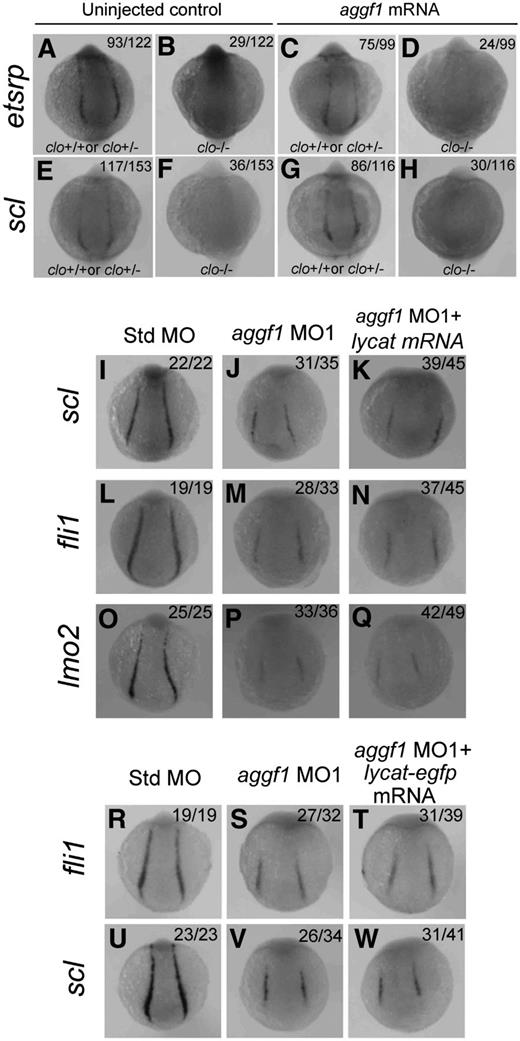
The zebrafish cloche mutant showed defects in endothelial and hematopoietic lineage development as well as lack of endocardium at the homozygous state.45 Similar to aggf1, cloche has been shown to act upstream of etsrp, fli1, scl, lmo2, and other markers.18,22,40,45 Thus, both aggf1 and cloche are the earliest known players in the generation of both hematopoietic and endothelial lineages. In this study, we tried to determine the relationship between aggf1 and cloche. First, we determined whether aggf1 mRNA could rescue the phenotypes of the clom39 mutant. We injected in vitro synthesized, capped aggf1 mRNA into 1- to 2-cell embryos (F1) from clom39+/− × clom39+/− crosses and analyzed expression of etsrp and scl by WISH at the 10-somite stage. The F1 embryos are expected to contain 25% of wild-type embryos, 50% of heterozygous embryos, and 25% of homozygous embryos. We found that 23.8% (29/122) and 23.5% (36/153) of uninjected control F1 embryos, which represent homozygous clom39−/− embryos, displayed no detectable expression of etsrp and scl, respectively (Figure 6A-B,E-F). After injection, 24.2% (24/99) of F1 embryos displayed no detectable etsrp expression (Figure 6C-D), and similarly, 25.8% (30/116) of homozygous F1 embryos showed no expression of scl (Figure 6G-H). The data suggest that aggf1 mRNA is not able to rescue the phenotype of the clom39 mutant, and aggf1 does not act downstream of cloche in specification of hemangioblasts.
Zebrafish aggf1 and cloche act independently during differentiation of hemangioblasts. (A-H) Heterozygous clochem39 mutant zebrafish were mated with each other. The offspring embryos were injected with 100 pg of aggf1 mRNA and used for WISH at 10 somites with antisense probes for etsrp (A-D) and scl (E-H). (A-B) Uninjected control embryos (23.8%, 29/122) showed no detectable etsrp expression, whereas 76.2% (93/122) embryos showed nearly normal expression of etsrp at 10 somites. (C-D) No detectable etsrp expression was displayed in 24.2% (24/99) of the embryos, whereas 75.8% (75/99) embryos showed nearly normal expression of etsrp at 10 somites. (E-F) Uninjected control embryos (76.5%, 117/153) showed normal expression of scl, whereas 23.5% (36/153) of embryos did not display expression of scl at 10 somites. (G-H) No detectable scl expression was displayed in 25.8% (30/116) of the embryos, whereas 74.2% (86/116) of the embryos showed nearly normal expression of scl at 10 somites. The ratios are consistent with the autosomal recessive inheritance pattern (3:1), suggesting that injection of aggf1 mRNA did not alter the 3:1 ratio and did not rescue the expression of hemangioblast markers in clochem39 mutant embryos. (I-Q) Overexpression of lycat mRNA did not rescue abnormal hemangioblast phenotypes in aggf1 morphants. Zebrafish embryos (1- to 2-cell stage) were injected with Std-MO (2 ng; I,L,O), aggf1 MO1 (2 ng; J,M,P), or aggf1 MO1 (2 ng) together with lycat mRNA (100 pg) (K,N,Q) and used for WISH at 6 somites with antisense probes for fli1 (I-K), scl (L-N), and lmo2 (O-Q). Injection of lycat mRNA failed to rescue the reduced expression of fli1, scl, and lmo2 in PLPM by aggf1 MO1. Embryos are shown in a dorsal view with the anterior at the top. (R-W) Overexpression of lycat-egfp mRNA did not rescue abnormal hemangioblast phenotypes in aggf1 morphants, that is, reduced expression of fli1 (R-T) or scl (U-W).
Zebrafish aggf1 and cloche act independently during differentiation of hemangioblasts. (A-H) Heterozygous clochem39 mutant zebrafish were mated with each other. The offspring embryos were injected with 100 pg of aggf1 mRNA and used for WISH at 10 somites with antisense probes for etsrp (A-D) and scl (E-H). (A-B) Uninjected control embryos (23.8%, 29/122) showed no detectable etsrp expression, whereas 76.2% (93/122) embryos showed nearly normal expression of etsrp at 10 somites. (C-D) No detectable etsrp expression was displayed in 24.2% (24/99) of the embryos, whereas 75.8% (75/99) embryos showed nearly normal expression of etsrp at 10 somites. (E-F) Uninjected control embryos (76.5%, 117/153) showed normal expression of scl, whereas 23.5% (36/153) of embryos did not display expression of scl at 10 somites. (G-H) No detectable scl expression was displayed in 25.8% (30/116) of the embryos, whereas 74.2% (86/116) of the embryos showed nearly normal expression of scl at 10 somites. The ratios are consistent with the autosomal recessive inheritance pattern (3:1), suggesting that injection of aggf1 mRNA did not alter the 3:1 ratio and did not rescue the expression of hemangioblast markers in clochem39 mutant embryos. (I-Q) Overexpression of lycat mRNA did not rescue abnormal hemangioblast phenotypes in aggf1 morphants. Zebrafish embryos (1- to 2-cell stage) were injected with Std-MO (2 ng; I,L,O), aggf1 MO1 (2 ng; J,M,P), or aggf1 MO1 (2 ng) together with lycat mRNA (100 pg) (K,N,Q) and used for WISH at 6 somites with antisense probes for fli1 (I-K), scl (L-N), and lmo2 (O-Q). Injection of lycat mRNA failed to rescue the reduced expression of fli1, scl, and lmo2 in PLPM by aggf1 MO1. Embryos are shown in a dorsal view with the anterior at the top. (R-W) Overexpression of lycat-egfp mRNA did not rescue abnormal hemangioblast phenotypes in aggf1 morphants, that is, reduced expression of fli1 (R-T) or scl (U-W).
The lycat gene was located within the clom39 deletion interval and suggested as the candidate gene for the cloche mutant.46 Therefore, we performed a rescue study with lycat mRNA to determine whether it can rescue defects in aggf1 morphants. We coinjected aggf1 MO1 together with in vitro synthesized lycat mRNA into 1- to 2-cell embryos and examined the expression of hemangioblast markers by WISH at 6-somite embryos (Figure 6I-Q). Injection of lycat mRNA failed to rescue the reduced expression of scl, fli1, and lmo2 in PLPM caused by aggf1 MO1 (Figure 6I-Q). To ensure that the lycat protein is translated in the rescue study, we created a lycat-egfp fusion expression construct. Injection of lycat-egfp mRNA into 1- to 2-cell embryos resulted in successful synthesis of the lycat–enhanced green fluorescent protein fusion protein at 28 hpf (green signal, supplemental Figure 9). As with lycat mRNA, lycat-egfp mRNA failed to rescue the reduced expression of fli1 and scl in aggf1 morphants (Figure 6R-W).
Together, all these data suggest that aggf1 and cloche act independently in the differentiation of hemangioblasts during zebrafish embryogenesis (Figure 747-49 ).
Working model for differentiation of hemangioblasts in zebrafish with the acting position of aggf1 indicated. Genes involved in the generation of both hematopoietic and endothelial lineages in zebrafish embryos are indicated. This study suggests that aggf1 acts as the earliest gene that affects the differentiation of hemangioblasts. Modified from H. Yamauchi et al,47 Zhang et al,48 and Bertrand et al.49
Working model for differentiation of hemangioblasts in zebrafish with the acting position of aggf1 indicated. Genes involved in the generation of both hematopoietic and endothelial lineages in zebrafish embryos are indicated. This study suggests that aggf1 acts as the earliest gene that affects the differentiation of hemangioblasts. Modified from H. Yamauchi et al,47 Zhang et al,48 and Bertrand et al.49
Discussion
In this study, we established angiogenic factor AGGF1 as the earliest molecular determinant for differentiation of hemangioblasts from the mesoderm (Figure 7). Downregulation of aggf1 expression dramatically decreased the expression of key hemangioblast genes, such as fli1, etsrp, scl, and lmo2 (Figure 3; supplemental Figure 5). In 2008, Liu et al showed that a constitutively active fli1, fli1-VP16, induced expression of key hemangioblast genes scl, lmo2, gata2, and etsrp and failed to rescue the phenotype of scl morphants or etsrp morphants (reduced flk1 expression).20 Liu et al argued that fli1 acted upstream of scl and etsrp in hemangioblast specification.20 Our data showed that injection of fli-vp16 mRNA rescued the phenotype of reduced expression of lmo2, scl, and gata2 in aggf1 morphants (Figure 5A-I), indicating that aggf1 acts upstream of fli1 to regulate specification of hemangioblasts. In 2010, Ren et al showed that injection of either fli1 RNA or scl RNA rescued the flk signal in etsrp morphants, suggesting that etsrp acted upstream of scl and fli1 to specify angioblasts.21 Our data showed that injection of etsrp RNA rescued the phenotype of reduced expression of scl and lmo2 in aggf1 morphants (Figure 5J-O), indicating that aggf1 acts upstream of etsrp to regulate specification of hemangioblasts. Thus, although there exists a controversy with regard to whether it is fli1 or etsrp that acts at the top of the regulatory hierarchy for hemangioblast specification, our study indicates that aggf1 acts upstream of both fli1 and etsrp during this developmentally important process (Figure 7).
The etsrp gene was initially identified in a microarray study for gene expression changes in the cloche mutant embryos40 and was later found to be able to rescue the abnormal gene expression phenotype in cloche embryos.22 Other studies also found that cloche acted upstream of fli1, scl, and lmo2.8,18,50 A candidate cloche gene has been identified as lycat encoding a lysocardiolipin acyltransferase.46 Considering that aggf1 morphants showed similar phenotypes in hematopoietic and EC development markers, we determined the relationship between aggf1 and cloche in hemangioblast specification. Capped aggf1 mRNA failed to rescue the reduced expression of hemangioblast markers in the cloche embryos, and likewise, lycat-egfp mRNA could not rescue the reduced hemangioblast signals in aggf1 MO morphants (Figure 6). These data suggest that aggf1 and cloche act independently or in parallel in hemangioblast differentiation. It is interesting to note that zebrafish evolved 2 independent upstream factors for specification of hemangioblasts, but the underlying advantages are unknown and warrant further investigation. Our rescue experiments used the m39 cloche mutant, which carries a large chromosomal deletion. In the future, it may be interesting to perform similar studies with other types of cloche mutants such as the s5 allele with a presumed point mutation,51 which might provide more specificity.
Hemangioblasts are the common bipotential progenitors shared by both endothelial and blood cells.1,2 Indeed, we found that aggf1 is an important gene required for the generation of both hematopoietic and endothelial lineages in zebrafish embryos. Expression of endothelial lineage markers kdrl, cdh5, and admr was also dramatically reduced in axial vessels and nearly absent in intersegmental vessels at 26 hpf by aggf1 MO1 or MO2 (supplemental Figure 4).30 Previously, we also reported that aggf1 played an important role in specification of veins by activating the AKT signaling pathway in zebrafish. The present study significantly expanded the physiological roles of aggf1 by identifying a novel function of aggf1 in specification of both HSCs and angioblasts, as well as their precursor, the hemangioblast.
Hemangioblasts are derived from mesodermal cells localized in the ventral mesoderm.1 Our previous study showed that aggf1 was a maternally and ubiquitously expressed gene in zebrafish.30 Furthermore, expression of aggf1 did not overlap with hemangioblast markers etsrp, fli, and scl (data not shown). These results are consistent with the role of aggf1 as a maternal factor acting at a very early stage in specification and differentiation of hemangioblasts from the ventral mesoderm. We found that knockdown of aggf1 did not affect the expression of endoderm-specific marker sox17 and ectoderm markers krox20 and otx1 (supplemental Figure 8). Aggf1 MOs did not have any effect on expression of mesoderm markers, such as ntl, papc, eve1, fgf8, and nkx2.5 (supplemental Figure 8) and myoD and pax2.1.30 Therefore, aggf1 may be specifically involved in specification of hemangioblasts.
For arterial EC specification, VEGFA was found to be the key molecular determinant, which binds to its receptors VEGFR2 and/or Nrp1 and activates the extracellular signal-regulated kinase/mitogen-activated protein kinase signaling pathway (ERK1/2/MAPK), leading to activation of Notch/Δ-like 4 (Dll4) signaling and arterial differentiation.52 We have recently shown that the specification of the venous EC fate is mediated by AGGF1 through activating the AKT signaling pathway.30 Considering the important role of AGGF1 in early hemangioblast specification unraveled here, we speculate that ECs committed to arterial or venous fate may be molecularly distinct extremely early. It is commonly thought that hemangioblasts are differentiated into angioblasts, which then become restricted to either an arterial or venous EC fate. Our data here may suggest that hemangioblasts have already become restricted to either an arterial or venous cell fate, a process regulated at least partly by aggf1. Aggf1 may drive differentiation of specific hemangioblasts that specify venous cell fate. Knockdown of aggf1, therefore, leads to defective specification of venous EC fate but does not affect specification of arterial EC fate as described previously.30 Extensive future studies are needed to further test this hypothesis.
One limitation of this study is that it solely relies on MO-mediated knockdown of aggf1 to investigate the physiological function of aggf1. The results from this study should be validated in targeted and germline-transmissible knockout fish in the future. In addition, our in situ hybridization of runx1 and c-myb showed that aggf1 was involved in differentiation of HSCs. This finding may be further validated by future studies with in vitro differentiation studies53 and embryonic transplantation studies as previously described.54
In conclusion, the data presented here demonstrate that aggf1 plays an important role in the differentiation of both hematopoietic and ECs in vivo and acts at the top of the genetic regulatory hierarchy in the specification of hemangioblasts. The study provides a fundamental understanding of differentiation of hemangioblasts and HSCs, which has general implications in regenerative medicine for hematologic and vascular diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Bo Zhang for providing the pu.1, mpo, and l-plastin plasmids; Aldo Ciau-Uitz for the Fli-VP16 plasmid; Jing-Wei Xiong for zebrafish clom39+/− mutant line and a lycat plasmid; and Zhan Yin for assistance.
This work was supported by grants from the China National Basic Research Programs (Program 973: 2013CB531101 and 2012CB517801), the “Innovative Development of New Drugs” Key Scientific Project (2011ZX09307-001-09), the National Institutes of Health, National Heart, Blood, and Lung Institute (R01 HL094498) (Q.K.W.), the Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education, and a grant from the State Key Laboratory of Freshwater Ecology and Biotechnology (2011FB16).
Authorship
Contribution: L.L. designed and performed experiments, analyzed data, and drafted the manuscript; D.C. performed some experiments and analyzed data; J.L., X.W., and N.W. constructed some plasmids and performed some experiments; C.X. provided valuable assistance; and Q.K.W. designed and supervised the project, assisted with data analysis and interpretation, and critically revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qing Kenneth Wang, Center for Human Genome Research and College of Life Science and Technology, Huazhong University of Science and Technology, 1037 Luoyu Rd, Wuhan 430074, People’s Republic of China; e-mail: qkwang@mail.hust.edu.cn or wangq2@ccf.org.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal