Key Points
First comprehensive and time-resolved characterization of platelet cAMP/PKA signaling upon iloprost treatment.
More than 2700 phosphorylation sites quantified between 4 time points and from 3 individual healthy donors.
Abstract
One of the most important physiological platelet inhibitors is endothelium-derived prostacyclin which stimulates the platelet cyclic adenosine monophosphate/protein kinase A (cAMP/PKA)–signaling cascade and inhibits virtually all platelet-activating key mechanisms. Using quantitative mass spectrometry, we analyzed time-resolved phosphorylation patterns in human platelets after treatment with iloprost, a stable prostacyclin analog, for 0, 10, 30, and 60 seconds to characterize key mediators of platelet inhibition and activation in 3 independent biological replicates. We quantified over 2700 different phosphorylated peptides of which 360 were significantly regulated upon stimulation. This comprehensive and time-resolved analysis indicates that platelet inhibition is a multipronged process involving different kinases and phosphatases as well as many previously unanticipated proteins and pathways.
Introduction
In the blood flow, a delicate equilibrium between activating and inhibiting stimuli prevents spontaneous platelet aggregation and occlusion of blood vessels while allowing steady plug formation in case of blood vessel lesions. One of the compounds keeping platelets in their resting state is prostacyclin I2 (PGI2), derived from endothelial cells through conversion of prostaglandin H2 by prostacyclin synthase.1 PGI2 binds specifically to the Gs-coupled prostacyclin receptor on the platelet plasma membrane,2 thus stimulating the cyclic adenosine monophosphate/protein kinase A (cAMP/PKA)–signaling cascade. This leads to downstream phosphorylation events resulting in the inactivation of small G proteins of the Ras and Rho families, inhibition of Ca2+ release, and modulation of actin cytoskeleton dynamics.3 Due to their central role in platelet inhibition, defects in cAMP and guanosine 3′,5′-cyclic monophosphate (cGMP) pathways might contribute to cardiovascular diseases,4 whereas prostacyclin and modulators of platelet cyclic nucleotide levels are highly interesting pharmacologic targets.5 In the clinic, drugs modulating cAMP or cAMP-regulating pathways, such as the phosphodiesterase 3 (PDE3) inhibitor cilostazol or the P2Y12 receptor inhibitors clopidogrel or prasugrel, are used for antiplatelet treatment.5,6
Due to the short half-life of prostacyclin, more stable analogs such as iloprost with a half-life time of over 20 minutes were developed.7 In this study, we analyzed platelets prepared from fresh blood donations to investigate molecular mechanisms underlying platelet inhibition upon stimulation of the PGI2 receptor. Using quantitative mass spectrometry (MS), we analyzed thousands of phosphorylation events after stimulation with 2 or 5nM iloprost (3 biological replicates each), respectively, for 0, 10, 30 or 60 seconds.
This is the first study to elucidate time-resolved changes in phosphorylation patterns of human platelets upon stimulation of the cAMP/PKA pathway. We quantified a total of 2738 phosphorylated peptides, corresponding to 2598 and 1457 phosphorylation sites of high (probability of >0.9 for correct phosphorylation site localization within the peptide sequence) and lower (<0.9) confidence from 1223 unique proteins (see supplemental Tables 3-5, available on the Blood Web site). Among those are almost 300 proteins with differential phosphorylation patterns of which 137 are potential PKA targets, compared with the 15 PKA targets established in platelets so far.3 Our data provide unprecedented insight into the crosstalk between the inhibiting cAMP/PKA pathway and downstream targets such as ubiquitin, Rho-Rac, and Ca2+/inositol phosphate–signaling pathways.
In addition, we assessed the biological variance of the basal phosphoproteome in human platelets, relatively quantifying 700 phosphopeptides between 4 different healthy donors. As we previously demonstrated for the complete human platelet proteome,8 the variance of basal phosphorylation levels in healthy donors seems to be surprisingly low as well–with a median coefficient of variation (CV) of 16% from the expected 1:1:1:1 ratios.
In summary, our novel quantitative data on platelet phosphorylation provide a rich source for further functional research and represent another step toward a deeper understanding of the mechanisms which contribute to platelet physiology as well as pathophysiology. Some of the signaling proteins detected in this study are so-far unknown key players of platelet activation and inhibition; thus, they might be novel candidates for monitoring functional defects or impaired responsiveness of human platelets to antiplatelet treatment, imposing an increased risk for severe cardiovascular or other adverse secondary effects.
Materials and methods
Platelet preparation
Blood was obtained from healthy volunteers according to our Institute’s guidelines and the Declaration of Helsinki. Our studies with human platelets were approved by the ethics committee of the University of Würzburg (study numbers: 67/92 and 114/04).
Full blood (40 mL) was immediately collected into sample tubes containing 10 mL of citrate buffer (CCD, 100mM Na2HPO4 × 2 H2O, 7mM citric acid, 140mM glucose; pH 6.5) and 10 U/mL apyrase. Platelet-rich plasma (PRP) was obtained by centrifugation at 300g for 20 minutes at room temperature (RT). The PRP layer was removed and diluted 1:1 with HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer (145mM NaCl, 5mM KCl, 1mM MgCl2, 10mM HEPES, and 10mM d-glucose; pH 7.4). The diluted PRP was centrifuged at 200g for 10 minutes at RT to reduce leukocyte contamination. Subsequently, the platelets were pelleted from the supernatant by centrifugation at 400g for 20 minutes at RT. Each pellet was finally diluted in Tyrode's buffer (2.2mM Na2HPO4 × 2 H2O, 7.8mM NaH2PO4 × H2O, 10mM NaHCO3, 150mM NaCl, 5mM KCl, 5mM glucose, pH 6.8) and incubated for 20 minutes at 37°C. Meanwhile, a small aliquot of each pellet was used for cell counting on a cell-sorting machine (Sysmex; Norderstedt, Germany). Aliquots of the platelet suspension were either incubated for 10, 30, or 60 seconds with 2 or 5nM iloprost or vehicle at 37°C. For western blot analysis, 50 µL of platelet suspension was mixed with 100 µL of 3× sodium dodecyl sulfate (SDS) gel-loading buffer. For proteome analysis, the remaining samples were stopped by adding the lysis buffer (50mM Tris, 1% SDS, 150mM NaCl, 1 tablet PhosStop/10 mL, pH 7.8) in a ratio of 1:2 to each platelet sample. Lysed samples were immediately shock-frozen in liquid nitrogen and stored at −80°C until further usage.
Sample preparation and liquid chromatography-mass spectrometry analysis
All steps of the analytical workflow, including sample preparation, proteolytic digestion, quality control, iTRAQ labeling, global proteome analysis, phosphopeptide enrichment, mass spectrometry, spectrum processing, database searching, and quantification are detailed in the supplemental Materials and methods. Lysed samples (200 µg each) were processed separately and digested using trypsin. Digest efficiency and reproducibility were quality controlled9 prior to labeling samples with 4 different isobaric iTRAQ 4plex labels. Samples were pooled and 3 µg each were used for global proteome quantification. Afterward, samples were subjected to TiO2-based phosphopeptide enrichment, as described previously.10 Data interpretation was conducted using the Proteome Discoverer software (Thermo Scientific). A false discovery rate of <1% was applied and phosphorylation site localization probabilities were determined by using the phosphoRS11 algorithm. Raw data and search results can be accessed using the ProteomeXchange accession: PXD000242.
Western blot analysis
For western blot analysis the samples were added directly to SDS gel loading buffer, then analyzed by SDS polyacrylamide gel electrophoresis (PAGE). Separated proteins were transferred to nitrocellulose membrane, blocked with 3% milk in Tris-buffered saline (TBS, 150mM NaCl, 50mM Tris) containing 0.1% Tween and incubated with primary antibodies against phosphorylated GRP2Ser587 (CalDAG-GEF1), SrcTyr530, VASPSer157, VASPSer239,12 GSK3αSer21, GSK3βSer9, ZyxinSer142/143, Filamin-ASer2152, and LASPSer146, as well as total GRP2, Src, Filamin-A, Zyxin and PKA substrate antibody overnight at 4°C (see supplemental Methods). For visualization of the signal, goat anti-rabbit or anti-mouse immunoglobulin G (IgG)–conjugated with horseradish peroxidase was used as a secondary antibody followed by ECL detection (Amersham; Pharmacia Biotech). Blots were scanned using SilverFast software and analyzed densitometrically by NIH Image J software for uncalibrated optical density.
Results
Quality control of samples
To ensure that the detected phosphopeptide regulations do not originate from variations of protein levels (eg, due to technical variation in sample processing, digestion, etc9 ), we additionally compared protein levels between the different time points for each biological replicate, using 1% of the labeled samples prior to phosphopeptide enrichment. The CV was below 12% for all replicates over all quantified proteins (∼1000 per replicate), which is close to the expected technical error of iTRAQ experiments. In addition, we subjected a single 5nM iloprost-stimulated time course to label-free quantitation, as detailed in the supplemental Material and methods, as an alternative and unbiased strategy to also identify phosphorylation-independent regulation events.
As we have demonstrated recently, intrasubject as well as intersubject variances of the global protein expression patterns are low in human platelets, as 85% of the relatively quantified proteins showed no significant variation.8 After purification of platelets of the required quality without preactivation was established,8 we addressed the intersubject variance of basal phosphorylation patterns to determine whether reproducible phosphorylation time courses can be obtained from different donors. Quantifying 700 phosphopeptides between “resting” platelets from 4 healthy donors, we assessed a CV of 16%. Even for low abundant membrane receptors such as P2X1 with ∼1400 copies per platelet, the CV for the phosphopeptide 382DLAApTSSTLGLQENMR397 was only 15%. These data indicate that intersubject comparisons of time-resolved phosphoproteomic profiles in human platelets are feasible.
Classification of phospho-regulated proteins
To determine which phosphopeptides are significantly regulated, for each replicate iTRAQ, ratios obtained from the Proteome Discoverer software were normalized against the corresponding global analysis, hence compensating for pipetting imprecisions and other systematic errors based on the assumption that the majority of peptides remain stable. Then, peptide-spectrum matches of each replicate were grouped according to protein accessions and the positions of phosphorylation sites in the respective protein sequence, as determined by the phosphoRS algorithm. For each of those groups, the corresponding medians of ratios and SDs were calculated. Grouping and median calculation were repeated to combine all 3 biological replicates. Subsequently, in order to evaluate the significance of regulations, the distribution of the logarithmically transformed ratios was modeled for every iTRAQ channel using a skewed normal distribution centered on the median and normalized to the upper and lower 34th percentiles. Thus, a peptide was considered as regulated with >99% confidence if the corresponding ratio had a probability >99% to deviate from the normal distribution. Finally, only those peptide groups presenting either (1) a regulation confidence >99% in at least 2 replicates or (2) more than twofold upregulation or downregulation in a single replicate were considered to generate a final set of reliably regulated phosphopeptides.
In total, 299 unique proteins show a regulation of phosphorylation upon stimulation with iloprost. Notably, these proteins are henceforth referred to as “regulated proteins.” According to our previous data on human platelets,8 this corresponds to ∼6% of the platelet proteome. Comparing this set of 299 regulated proteins with the human proteome based on GO annotation13 (see Table 1 and supplemental Table 1 for details) reveals an enrichment of proteins involved in “cytoskeleton rearrangement and organization” (26 proteins) and “vesicle-mediated transport” (54 proteins). According to the GPS 2.1 algorithm for kinase consensus sequence prediction,14 many of these regulations occur within a PKA consensus sequence. Furthermore, many of these are early responses (first 10 seconds) as well, indicating an early inhibition of degranulation and actin rearrangement by PKA phosphorylation.
Selection of significantly enriched biological processes
| Enriched biological process (P value of enrichment) . | All proteins . | Potentially PKA regulated . | Early responders (10 s), 2nM/5 nM . |
|---|---|---|---|
| Vesicle-mediated transport (1.1 × 10−20) | 54 | 19 | 14/14 |
| Platelet activation (4.4 × 10−12) | 22 | 6 | 5/7 |
| Actin cytoskeleton organization (1.5 × 10−12) | 26 | 15 | 11/10 |
| Regulation of small GTPase–mediated signal transduction (5.5 × 10−12) | 26 | 13 | 10/9 |
| Platelet degranulation (1.5 × 10−8) | 12 | 2 | 3/3 |
| Negative regulation of cytoskeleton organization (3.7 × 10−7) | 10 | 6 | 2/3 |
| Blood coagulation (1.5 × 10−6) | 28 | 8 | 7/9 |
| Regulation of Ras protein signal transduction (4.8 × 10−5) | 21 | 8 | 8/4 |
| Cell-surface receptor linked signaling pathway (1.9 × 10−2) | 42 | 24 | 11/12 |
| Enriched biological process (P value of enrichment) . | All proteins . | Potentially PKA regulated . | Early responders (10 s), 2nM/5 nM . |
|---|---|---|---|
| Vesicle-mediated transport (1.1 × 10−20) | 54 | 19 | 14/14 |
| Platelet activation (4.4 × 10−12) | 22 | 6 | 5/7 |
| Actin cytoskeleton organization (1.5 × 10−12) | 26 | 15 | 11/10 |
| Regulation of small GTPase–mediated signal transduction (5.5 × 10−12) | 26 | 13 | 10/9 |
| Platelet degranulation (1.5 × 10−8) | 12 | 2 | 3/3 |
| Negative regulation of cytoskeleton organization (3.7 × 10−7) | 10 | 6 | 2/3 |
| Blood coagulation (1.5 × 10−6) | 28 | 8 | 7/9 |
| Regulation of Ras protein signal transduction (4.8 × 10−5) | 21 | 8 | 8/4 |
| Cell-surface receptor linked signaling pathway (1.9 × 10−2) | 42 | 24 | 11/12 |
Selection of significantly enriched biological processes, according to gene ontology (GO) annotation,13 within the regulated proteins. Given is the number of proteins which can be assigned to the respective biological process from the share of all regulated proteins, the potentially PKA-regulated proteins as well as the early responders which show significant regulation already 10 seconds after stimulation.
Among the regulated proteins are 16 kinases, which can be classified in those which show an early (10 seconds) or later (≥30 seconds) response to platelet inhibition as summarized in Table 2.
Summary of regulated kinases
| Accession . | Gene . | Site . | Regulation . |
|---|---|---|---|
| Kinase early responders, 10 s | |||
| Q8TD19 | NEK9 | Ser827 | ↓105nM |
| Q9UKE5 | TNIK | Ser1021/1028 | ↑105nM |
| Ser764/Ser766/Ser769 | ↑105nM | ||
| P07948 | LYN | Thr489/Tyr492/Ser495/Tyr501/Thr502/Thr504 | ↑60/302/5nM |
| Q00537 | CDK17 | Ser9 | ↑105nM |
| Ser180 | ↑105nM | ||
| Ser182 | ↑102nM | ||
| Q02750 | MAP2K1 | Ser377/Thr378/Ser385/Thr386/Thr388 | ↓105nM |
| Ser385/Thr386 | ↑105nM | ||
| Q93100 | PHKB | Ser27 | ↑102/5nM |
| Ser29 | ↑102/5nM | ||
| Q8N4C8 | MNK1 | Ser699 | ↑102/5nM |
| Ser701 | ↑102/5nM | ||
| Ser732 | ↓105nM | ||
| Ser993 | ↑105nM | ||
| Q9BZ23 | PANK2 | Ser189 | ↑30/102/5nM |
| O95819 | MAP4K4 | Ser900/Thr907/Thr909/Ser913 | ↑102/5nM |
| Q15746 | MLCK | Ser1438 | ↓105nM |
| Ser1773 | ↑30/102/5nM | ||
| P12931 | SRC | Tyr530 | ↑105nM |
| Kinase late responders, >10 s | |||
| Q12851 | MAP4K2 | Ser365/367 | ↑305nM |
| P46019 | PHKA2 | Ser729 | ↑302/5nM |
| P52333 | JAK3 | Ser17 | ↑605nM |
| P49840 | GSK3A | Thr19/Ser20/Ser21/Ser39 | ↑105nM |
| P27448 | MARK3 | Tyr441/Ser442 | ↑305nM |
| Accession . | Gene . | Site . | Regulation . |
|---|---|---|---|
| Kinase early responders, 10 s | |||
| Q8TD19 | NEK9 | Ser827 | ↓105nM |
| Q9UKE5 | TNIK | Ser1021/1028 | ↑105nM |
| Ser764/Ser766/Ser769 | ↑105nM | ||
| P07948 | LYN | Thr489/Tyr492/Ser495/Tyr501/Thr502/Thr504 | ↑60/302/5nM |
| Q00537 | CDK17 | Ser9 | ↑105nM |
| Ser180 | ↑105nM | ||
| Ser182 | ↑102nM | ||
| Q02750 | MAP2K1 | Ser377/Thr378/Ser385/Thr386/Thr388 | ↓105nM |
| Ser385/Thr386 | ↑105nM | ||
| Q93100 | PHKB | Ser27 | ↑102/5nM |
| Ser29 | ↑102/5nM | ||
| Q8N4C8 | MNK1 | Ser699 | ↑102/5nM |
| Ser701 | ↑102/5nM | ||
| Ser732 | ↓105nM | ||
| Ser993 | ↑105nM | ||
| Q9BZ23 | PANK2 | Ser189 | ↑30/102/5nM |
| O95819 | MAP4K4 | Ser900/Thr907/Thr909/Ser913 | ↑102/5nM |
| Q15746 | MLCK | Ser1438 | ↓105nM |
| Ser1773 | ↑30/102/5nM | ||
| P12931 | SRC | Tyr530 | ↑105nM |
| Kinase late responders, >10 s | |||
| Q12851 | MAP4K2 | Ser365/367 | ↑305nM |
| P46019 | PHKA2 | Ser729 | ↑302/5nM |
| P52333 | JAK3 | Ser17 | ↑605nM |
| P49840 | GSK3A | Thr19/Ser20/Ser21/Ser39 | ↑105nM |
| P27448 | MARK3 | Tyr441/Ser442 | ↑305nM |
Kinases showing an early and late response upon iloprost stimulation. ↓105nM corresponds to downregulation of phosphorylation after 10 seconds in the 5nM iloprost samples. If the phosphorylation site localization was not confident (eg, Lyn, MAPKK1, MEKKK4, and GSK3α), all potentially phosphorylated residues are listed according to phosphoRS.
In addition, 3 phosphatases, namely PTPRJ (Ser1311: ↑30/102/5nM), CTDSPL (Ser32: ↓302nM), and PTPN12 (Ser332: ↓602nM; 351EEILQPPEPHPVPPILp(TPSPPSAFPT)VTTVWQDNDR386: ↑30/105nM; 565TVSLp(TPSPTTQVET)PDLVDHDNTSPLFR591: ↓605nM) and 3 phosphatase regulatory subunits, namely PPP2R5D (Ser573: ↑105nM), PPP1R14A (Ser26: ↓302nM; Ser128 and Ser130: ↓605nM), PPP1R3D (Ser77 and Ser78: ↓302nM) were phospho-regulated upon platelet inhibition.
Analyzing all regulated phosphopeptides for the potential presence of consensus motifs of all classes of kinases using GPS 2.1 clearly suggests the additional involvement of CK1/2 and Akt1 kinases in the processes of platelet inhibition and activation. GO-term enrichment analysis with regard to protein function (see Table 3, supplemental Table 2 for details) reveals 6 proteins with Src-homology (SH)3/SH2 adaptor activity. Furthermore, among the regulated proteins, 16 are calmodulin binding and 22 small GTPase regulator proteins. Of the latter, many are connected to Rho-Rac and Rab pathways. In principle, GTPase regulators represent interesting key players in platelet regulation, as they are essential switches of cellular changes and responses upon stimulation.15
Protein functions which are significantly enriched within the 299 regulated proteins
| Enriched protein function (P value of enrichment) . | All proteins . | Potentially PKA regulated . | Early responders, 10 s, 2nM/5 nM . |
|---|---|---|---|
| Actin binding (1.6 × 10−11) | 26 | 17 | 7/8 |
| Small GTPase regulator activity (1.7 × 10−10) | 22 | 10 | 12/7 |
| Calmodulin binding (8.6 × 10−9) | 16 | 7 | 3/3 |
| SH3/SH2 adaptor activity (1.9 × 10−4) | 6 | 3 | 1/1 |
| Ubiquitin ligase binding (1.8 × 10−3) | 5 | 3 | 0/2 |
| Serine/threonine kinase activity (4.4 × 10−3) | 12 | 5 | 2/4 |
| Ubiquitin protein ligase activity (8.2 × 10−2) | 4 | 2 | 3/2 |
| Tyrosine kinase activity (9.0 × 10−2) | 4 | 0 | 0/2 |
| Enriched protein function (P value of enrichment) . | All proteins . | Potentially PKA regulated . | Early responders, 10 s, 2nM/5 nM . |
|---|---|---|---|
| Actin binding (1.6 × 10−11) | 26 | 17 | 7/8 |
| Small GTPase regulator activity (1.7 × 10−10) | 22 | 10 | 12/7 |
| Calmodulin binding (8.6 × 10−9) | 16 | 7 | 3/3 |
| SH3/SH2 adaptor activity (1.9 × 10−4) | 6 | 3 | 1/1 |
| Ubiquitin ligase binding (1.8 × 10−3) | 5 | 3 | 0/2 |
| Serine/threonine kinase activity (4.4 × 10−3) | 12 | 5 | 2/4 |
| Ubiquitin protein ligase activity (8.2 × 10−2) | 4 | 2 | 3/2 |
| Tyrosine kinase activity (9.0 × 10−2) | 4 | 0 | 0/2 |
Protein functions which are significantly enriched within the 299 regulated proteins, according to GO annotation.13 Given is the number of (1) proteins which can be assigned to the respective function from all regulated proteins, (2) potentially PKA-phosphorylated proteins (according to GPS 2.1 consensus motif prediction) as well as (3) early responders which show significant regulation already 10 seconds after stimulation.
Validation using western blot
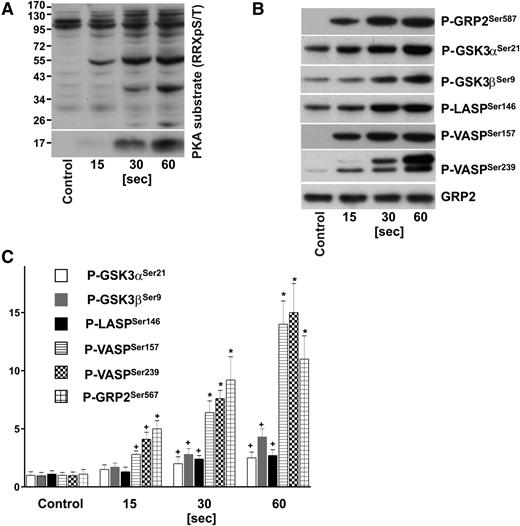
We used site-specific antibodies against phosphorylated GRP2Ser587, SrcTyr530, VASPSer157, VASPSer239, GSK3αSer21, GSK3βSer9, ZyxinSer142/143, Filamin-ASer2152, LASPSer146, and against total GRP2, Src, Filamin-A, Zyxin to verify some of our results by immunoblotting. As depicted in Figure 1 (and supplemental Figure 1) our MS-based findings could be validated using platelets obtained from additional healthy donors. Indeed, in some cases, the western blot data indicate an even higher increase in phosphorylation, whereas in accordance with the quantitative MS data, phosphorylation of Filamin-ASer2152 remains unchanged upon iloprost treatment, however, increases upon platelet activation with thrombin (supplemental Figure 1).
Time course of PKA substrate phosphorylation in iloprost-stimulated human platelets. Washed human platelets (3 × 108/mL) were incubated with iloprost (2nM) for the indicated time and processed for western blot analysis with antibodies directed against (A) Phospho-PKA substrates (RRXpS/T), (B) P-GRP2Ser587, P-GSK3αSer21, P-GSK3βSer9, P-LASPSer146, P-VASPSer157, P-VASPSer239, or total GRP2 (CalDAG-GEFI) antibodies. (C) Immunoblots were scanned and intensities of the phospho-specific antibodies were normalized to the total GRP2 signal. All images were quantified by the Image J program and expressed as fold changes, where control samples were taken as 1. Results are mean ± SEM, n = 4. +P < .05 compared with the control; *P < .05 compared with 15-second samples.
Time course of PKA substrate phosphorylation in iloprost-stimulated human platelets. Washed human platelets (3 × 108/mL) were incubated with iloprost (2nM) for the indicated time and processed for western blot analysis with antibodies directed against (A) Phospho-PKA substrates (RRXpS/T), (B) P-GRP2Ser587, P-GSK3αSer21, P-GSK3βSer9, P-LASPSer146, P-VASPSer157, P-VASPSer239, or total GRP2 (CalDAG-GEFI) antibodies. (C) Immunoblots were scanned and intensities of the phospho-specific antibodies were normalized to the total GRP2 signal. All images were quantified by the Image J program and expressed as fold changes, where control samples were taken as 1. Results are mean ± SEM, n = 4. +P < .05 compared with the control; *P < .05 compared with 15-second samples.
Novel putative targets of PKA
In his comprehensive review article about cAMP/cGMP-dependent signaling in platelets, Smolenski recently published a list of 17 PKA/PKG targets.3 Our data not only confirm these findings, but substantially expand our knowledge about putative PKA targets in human platelets. From the 12 PKA substrates described by Smolenski, we could detect 9 with putative PKA sites; in addition, we could confirm phosphorylation of the established PKG substrate IRAG at Ser657 and Ser670, both upregulated 10 seconds after iloprost stimulation. However, direct crosstalk between cAMP and cGMP are unlikely.16 From that list, 2 PKA substrates are missing in our data due to the peptide sequence which is rather too short or too long for confident identification after tryptic digestion.
In total about half of all regulated proteins (137) show a regulation of potential PKA sites. Moreover, 22 proteins have >1 regulated peptide containing a potential PKA site, for example, PDE3A and MRVI1/IRAG. Interestingly, phosphorylation of PDE3A (Ser311 and 312: ↑102/5nM; Ser428: ↑30/102/5nM) is upregulated 10 seconds after iloprost treatment, suggesting an immediate negative feedback loop.17 In contrast, AMP deaminase 2 is ↑305nM on Ser76 whereas another potential PKA target Ser168 is ↓102nM, suggesting a positive feedback loop to maintain cAMP levels.18 In total, 30 proteins are potential early PKA targets (changes after 10 seconds) after both 2nM and 5nM iloprost stimulation (see Table 4). This list includes known mediators of platelet activation/inhibition but also indicates an important role for so-far unknown proteins such as the endosulfine family (ENSA, ARPP19) or BIN2.
Potential PKA targets showing early responses 10 seconds after both 2nM and 5nM iloprost treatment
| Accession . | Gene . | Protein name . | Putative PKA site . | Accession . | Gene . | Protein name . | Putative PKA site . |
|---|---|---|---|---|---|---|---|
| O14828 | SCAMP3 | Secretory carrier–associated membrane protein 3 | S32 | Q8N4C8 | MNK1 | Misshapen-like kinase 1 | S699 and S701 |
| O00501 | CLDN5 | Claudin-5 | T207 | Q8N5J2 | FAM63A | Protein FAM63A | S441 |
| O43312 | MTSS1 | Metastasis suppressor protein 1 | S272 | Q92974 | ARHGEF2 | Rho guanine nucleotide exchange factor 2 | S886 |
| O43768 | ENSA | α-endosulfine | S109 | Q93100 | PHKB | Phosphorylase b kinase regulatory subunit β | S27 |
| O94929 | ABLIM3 | Actin-binding LIM protein 3 | S277 | Q96MK2 | FAM65C | Protein FAM65C | S340* |
| O95644 | NFATC1 | Nuclear factor of activated T cells, cytoplasmic 1 | S245 | Q99490 | AGAP2 | Arf-GAP with GTPase, ANK repeat and PH domain–containing protein 2 | T647 |
| O95819 | MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | S900* | Q9BZ72 | PTTPNM2 | Membrane-associated phosphatidylinositol transfer protein 2 | S1277 |
| P16150 | SPN | Leukosialin | T341 | Q9C0C9 | UBE2O | Ubiquitin-conjugating enzyme E2 O | S515 |
| P56211 | ARPP19 | cAMP-regulated phosphoprotein 19 | S104 | Q9P270 | SLAIN2 | SLAIN motif-containing protein 2 | S391 |
| Q14162 | SCARF1 | Endothelial cells scavenger receptor | S589 and S605 | Q9UBW5 | BIN2 | Bridging integrator 2 | S259 |
| Q14432 | PDE3A | cGMP-inhibited 3′,5′-cyclic phosphodiesterase A | S312 | Q9UJU6 | DBNL | Drebrin-like protein | S269* |
| Q15149 | PLEC | Plectin | S4386 | Q9UKE5 | TNIK | TRAF2 and NCK-interacting protein kinase | S1021* |
| Q684P5 | RAP1GAP2 | Rap1 GTPase-activathg protein 2 | S7 | Q9Y2J2 | EPB41L3 | Band 4.1-like protein 3 | S420 |
| Q7LDG7 | RASGRP2 | RAS guanyl-releasing protein 2 | S116, 117, and S587 | Q9Y385 | UBE2J1 | Ubiquitin-conjugating enzyme E2 J1 | S266 |
| Q8IWB9 | TEX2 | Testis-expressed sequence 2 protein | S295 | Q9Y6F6 | MRVI1 | Protein MRVI1 | S657 and S670 |
| Accession . | Gene . | Protein name . | Putative PKA site . | Accession . | Gene . | Protein name . | Putative PKA site . |
|---|---|---|---|---|---|---|---|
| O14828 | SCAMP3 | Secretory carrier–associated membrane protein 3 | S32 | Q8N4C8 | MNK1 | Misshapen-like kinase 1 | S699 and S701 |
| O00501 | CLDN5 | Claudin-5 | T207 | Q8N5J2 | FAM63A | Protein FAM63A | S441 |
| O43312 | MTSS1 | Metastasis suppressor protein 1 | S272 | Q92974 | ARHGEF2 | Rho guanine nucleotide exchange factor 2 | S886 |
| O43768 | ENSA | α-endosulfine | S109 | Q93100 | PHKB | Phosphorylase b kinase regulatory subunit β | S27 |
| O94929 | ABLIM3 | Actin-binding LIM protein 3 | S277 | Q96MK2 | FAM65C | Protein FAM65C | S340* |
| O95644 | NFATC1 | Nuclear factor of activated T cells, cytoplasmic 1 | S245 | Q99490 | AGAP2 | Arf-GAP with GTPase, ANK repeat and PH domain–containing protein 2 | T647 |
| O95819 | MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | S900* | Q9BZ72 | PTTPNM2 | Membrane-associated phosphatidylinositol transfer protein 2 | S1277 |
| P16150 | SPN | Leukosialin | T341 | Q9C0C9 | UBE2O | Ubiquitin-conjugating enzyme E2 O | S515 |
| P56211 | ARPP19 | cAMP-regulated phosphoprotein 19 | S104 | Q9P270 | SLAIN2 | SLAIN motif-containing protein 2 | S391 |
| Q14162 | SCARF1 | Endothelial cells scavenger receptor | S589 and S605 | Q9UBW5 | BIN2 | Bridging integrator 2 | S259 |
| Q14432 | PDE3A | cGMP-inhibited 3′,5′-cyclic phosphodiesterase A | S312 | Q9UJU6 | DBNL | Drebrin-like protein | S269* |
| Q15149 | PLEC | Plectin | S4386 | Q9UKE5 | TNIK | TRAF2 and NCK-interacting protein kinase | S1021* |
| Q684P5 | RAP1GAP2 | Rap1 GTPase-activathg protein 2 | S7 | Q9Y2J2 | EPB41L3 | Band 4.1-like protein 3 | S420 |
| Q7LDG7 | RASGRP2 | RAS guanyl-releasing protein 2 | S116, 117, and S587 | Q9Y385 | UBE2J1 | Ubiquitin-conjugating enzyme E2 J1 | S266 |
| Q8IWB9 | TEX2 | Testis-expressed sequence 2 protein | S295 | Q9Y6F6 | MRVI1 | Protein MRVI1 | S657 and S670 |
Phosphorylation could not be confidently localized within the peptide sequence.
Discussion
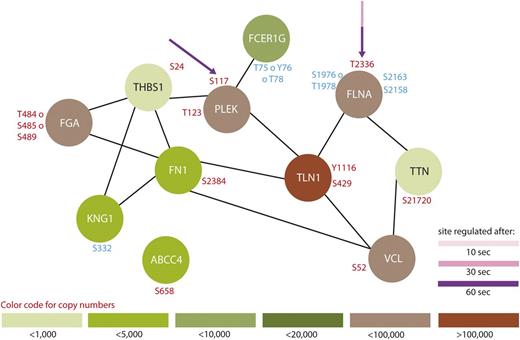
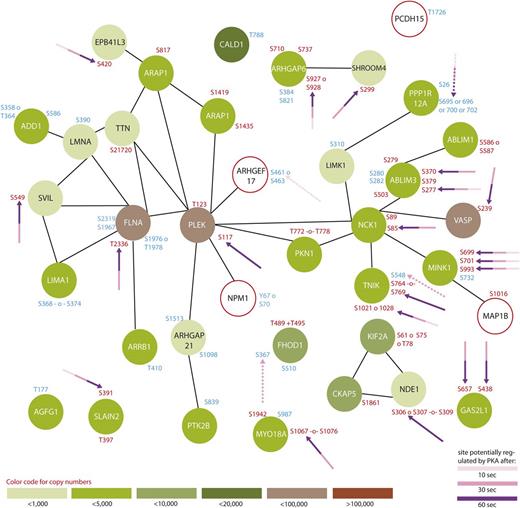
For the first time, our data provide a time-resolved and comprehensive insight into phosphorylation-dependent signaling upon cAMP-mediated inhibition of human platelets stimulated with the stable prostacyclin analog iloprost. It is obvious that platelet inhibition is a highly complex and concerted process, involving a variety of kinases and phosphatases with substantial crosstalk to activatory signaling pathways. All 3 major steps of platelet inhibition, (1) inactivation of small G proteins of the Ras/Rho family, (2) inhibition of Ca2+ release from intracellular stores, and (3) modulation of actin cytoskeleton dynamics,3 are represented by a variety of regulated proteins in the present study. Our quantitative phosphoproteomic data not only confirm known and expected time courses of proteins known to be involved in platelet inhibition, such as VASP, LASP, CalDAG-GEF119 (Figure 1), and Rap1GAP2,20 but also provide a detailed overview of many more phosphopeptides 10, 30, and 60 seconds after iloprost treatment, compared with untreated controls. Besides activation of PKA, platelet inhibition also seems to directly interfere with a variety of signaling pathways, such as Wnt,21 MEK/ERK, p38 MAPK, JNK,22 Src. We could detect a phosphorylation-dependent regulation of many proteins involved in known Gq-, Gi-, and G12/13-signaling pathways in platelets,23 such as phospholipase C (PLC)β, MLCK, Myosin, cytosolic phospholipase A2 (cPLA2), CalDAG-GEF1, Talin, IP3R, LIM-K. For assessing and summarizing the global data, we selected 3 GO terms which are important for platelets, namely (1) platelet activation, (2) platelet degranulation, (3) cytoskeleton organization, and created protein interaction networks based on STRING24 (Figures 2-4).
Protein interaction network summarizing all regulated proteins belonging to the GO-term “platelet activation”. From all 299 regulated proteins, only members of the GO-term “platelet activation” were searched for interactions using STRING,24 using a medium threshold confidence. Potential PKA sites are highlighted with arrows and the corresponding colors indicate the time points which are regulated compared with 0-second iloprost treatment (dashed lines for downregulation). Upregulated phosphorylation sites are labeled in red and downregulated in blue. Estimated protein copy numbers are taken from Burkhart et al.8
Protein interaction network summarizing all regulated proteins belonging to the GO-term “platelet activation”. From all 299 regulated proteins, only members of the GO-term “platelet activation” were searched for interactions using STRING,24 using a medium threshold confidence. Potential PKA sites are highlighted with arrows and the corresponding colors indicate the time points which are regulated compared with 0-second iloprost treatment (dashed lines for downregulation). Upregulated phosphorylation sites are labeled in red and downregulated in blue. Estimated protein copy numbers are taken from Burkhart et al.8
Protein interaction network summarizing all regulated proteins belonging to the GO-term “platelet degranulation.” From all 299 regulated proteins, only members of the GO-term “platelet degranulation” were searched for interactions using STRING,24 using a medium threshold. Potential PKA sites are highlighted with arrows and the corresponding colors indicate the time points which are regulated compared with 0-second iloprost treatment (dashed lines for downregulation). Upregulated phosphorylation sites are labeled in red and downregulated in blue. Estimated protein copy numbers are taken from Burkhart et al.8
Protein interaction network summarizing all regulated proteins belonging to the GO-term “platelet degranulation.” From all 299 regulated proteins, only members of the GO-term “platelet degranulation” were searched for interactions using STRING,24 using a medium threshold. Potential PKA sites are highlighted with arrows and the corresponding colors indicate the time points which are regulated compared with 0-second iloprost treatment (dashed lines for downregulation). Upregulated phosphorylation sites are labeled in red and downregulated in blue. Estimated protein copy numbers are taken from Burkhart et al.8
Protein interaction network summarizing all regulated proteins belonging to the GO-term “cytoskeleton organization.” From all 299 regulated proteins, only members of the GO-term “platelet degranulation” were searched for interactions using STRING,24 using a medium threshold. Potential PKA sites are highlighted with arrows and the corresponding colors indicate the time points which are regulated compared with 0-second iloprost treatment (dashed lines for downregulation). Upregulated phosphorylation sites are labeled in red and downregulated in blue. Estimated protein copy numbers are taken from Burkhart et al.8
Protein interaction network summarizing all regulated proteins belonging to the GO-term “cytoskeleton organization.” From all 299 regulated proteins, only members of the GO-term “platelet degranulation” were searched for interactions using STRING,24 using a medium threshold. Potential PKA sites are highlighted with arrows and the corresponding colors indicate the time points which are regulated compared with 0-second iloprost treatment (dashed lines for downregulation). Upregulated phosphorylation sites are labeled in red and downregulated in blue. Estimated protein copy numbers are taken from Burkhart et al.8
The Rho/Rac pathway and cytoskeletal reorganization
According to our data, the inhibitory cAMP/PKA pathway affects proteins connected to Rho-Rac pathways. During platelet activation, RhoA leads to various downstream effects such as an increase of actin polymerization, stress fiber, or focal adhesion formation.15 PKA phosphorylation could be involved in modulating the Rho-signaling pathway by phosphorylation (see Figure 4). For instance, several RhoGAP proteins which influence the hydrolysis of GTP to GDP show a regulation upon iloprost treatment, such as RhoGAP1 (site not confident –44p(SDDSKSSS)PELVTHLK58: ↓605nM), RhoGAP6 (Ser710: ↑605nM; Ser927 or 928: ↑305nM; Thr737: ↑602nM), and RhoGAP18 (Ser66: ↓105nM; site not confident –44p(SISQDSLDELS)MEDYWIELENIK66: ↑102nM). Furthermore, our results are in accordance with Nishikawa et al who have reported that MLCK is phosphorylated by PKA in vitro25 because we could identify 2 regulated phosphorylation sites at Ser1759 (↑305nM) and Ser1773 (↑302nM, ↑105nM) of which the latter is a potential PKA target. Moreover, we identified RhoGEF2 (Ser886: ↑102/5nM) and RhoGEF6 (Ser640: ↑302/5nM and Ser684: ↑305nM) as potential PKA substrates. RhoGEF2 mediates crosstalk between microtubules and activates RhoA by exchanging GDP to GTP, indicating a potentially similar mechanism as recently demonstrated for CalDAG-GEF1.20 According to Zenke et al, Ser886 is within a potential inhibiting domain of RhoGEF2 phosphorylated by PAK1,26 however, the sequence PRRRSL represents a classical PKA consensus sequence. RhoGEF6 is a nucleotide exchange factor for Rac1, which is involved in actin cytoskeleton reorganization27 and also interacts with BIN2. Indeed, BIN2 seems to be among the most regulated proteins with several phosphorylation sites responding after 10 seconds: Ser259 (↑102/5nM), Ser263 (↓105nM), 288SESEp(SVSAT)EDLAPDAAQGEDNSEIK313 (↑102nM), and 483ENENIHNQNPEELCp(TSPT)LMTSQVASEPGEAK514 (↓102nM). It contains a Bin–Amphiphysin–Rvs (BAR) domain (Val28-Asn244) which can mediate interactions with phosphatidylinositide-enriched membranes and seems to be involved in cell motility and migration.28
Further downstream, our data elucidate a change in phosphorylation of Talin at Ser429 (↑60/302/5nM) and Tyr1116 (↓102nM; ↑105nM). Filamin A is another cytoskeletal protein strongly regulated at several phosphorylation sites: Ser1976 or Thr1978 (↓10/302/5nM), Ser2163 (↓302nM) and Ser2158 (↓305nM). Although we could not detect changes at Ser2152 (confirmed by western blot, supplemental Figure 1), a known PKA site preventing Filamin from degradation,29 we identified a novel potential PKA site at Thr2336 (↑302/5nM). Further structural proteins such as Titin (Ser21720: ↑305nM) and Fibrinogen α chain (Thr484 or Ser485 or Ser489: ↑30/102/5nM; S485: ↓605nM) are also regulated upon iloprost treatment, as illustrated in Figures 2-4.
Different Myosin isoforms are also clearly regulated upon iloprost stimulation: we identified Myosin 9 regulated at Ser1943 (↓102nM) and Myosin 9B at 5 confident phosphorylation sites (Thr1346: ↓602nM; Ser1993: ↑102nM; Ser1353: ↑105nM; Ser1354: ↑102nM; Ser1356: ↑102/5nM). Finally, Myosin 18A phosphorylation is found to be regulated at Ser987 (↓105nM), Ser1942 (↑102/5nM), and 1065RVp(SSSSELDLPS)GDHCEAGLLQLDVPLLR1093 (↑302/5nM).
Kinases and phosphatases involved in platelet inhibition
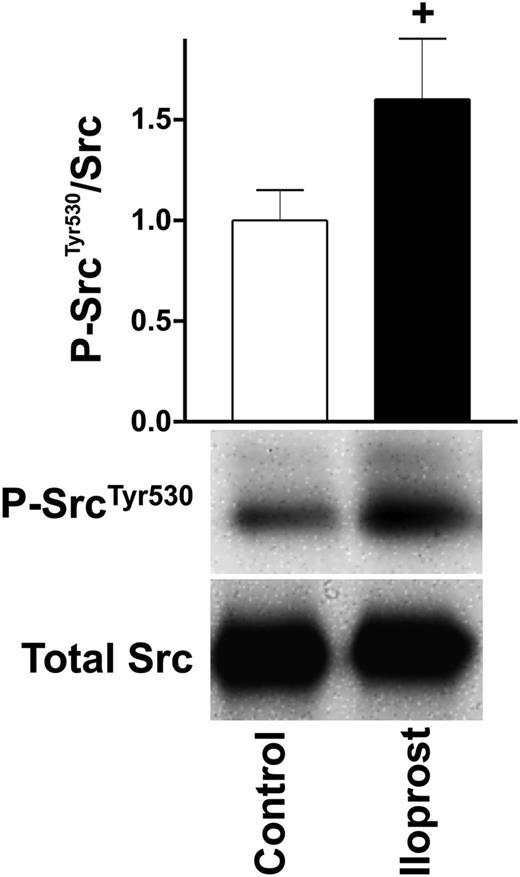
Regarding platelet activation (Figure 2) and aggregation, SH2 domain–containing proteins play an important role, serving as kinases as well as adaptor proteins.30 After iloprost treatment, 3 important SH2 domain–containing, nonreceptor tyrosine kinases involved in various downstream signaling cascades such as actin dynamics (Src),31 regulation of Ca2+ (Fyn),32 and degranulation (Lyn)33 are regulated as illustrated in Figures 2 and 4. The first, Src, is increasingly phosphorylated at Tyr530 (↑105nM), a site known to inhibit kinase activity,34 indicating a negative regulation of Src upon platelet inhibition. We confirmed these interesting MS-based findings by western blot analysis using a site-directed antibody against P-SrcTyr530, as depicted in Figure 5. The second, Lyn, is increasingly phosphorylated on the peptide 484AEERPp(TFDYLQSVLDDFYTAT)EGQYQQQP512 (↑60/302/5nM) and it has been reported that megakaryocyte-associated tyrosine-protein kinase inhibits Lyn by phosphorylation at Tyr508.35 Whether phosphorylation of any of these Ser/Thr/Tyr residues has a negative regulatory effect on kinase activity and subsequent platelet glycoprotein VI (GPVI) signaling in platelets requires further validation. At last, Fyn phosphorylation at Ser21 is decreased (↓302nM). While Fyn is involved in GPVI/integrin signaling and interacts with the phosphatidylinositol pathway,36 the function of Ser21 in Fyn is still unknown.
Western blot validation of increased Src Tyr530 phosphorylation. Washed human platelets (3 × 108/mL) were incubated with iloprost (5nM) for the indicated time and processed for western blot analysis with antibodies directed against P-SrcTyr530 and total Src. Signals were quantified as described in the Figure 1B legend. Shown are representatives of 3 independent experiments.
Western blot validation of increased Src Tyr530 phosphorylation. Washed human platelets (3 × 108/mL) were incubated with iloprost (5nM) for the indicated time and processed for western blot analysis with antibodies directed against P-SrcTyr530 and total Src. Signals were quantified as described in the Figure 1B legend. Shown are representatives of 3 independent experiments.
Ca2+ and phosphatidylinositol signaling
During platelet activation, phosphatidylinositol and mainly Ca2+ signaling regulate most activating pathways in platelets such as integrin activation, thrombus formation, vesicle degranulation, and cytoskeletal rearrangement. Stimulation of the Thromboxane A2 receptor (TPα) leads to activation of PLC, producing PIP2 and PIP3.37 We found TPα (Ser329/Ser331: ↑105nM) regulated on a potential PKA site which could lead to receptor desensitization.37-39 Additionally, we found PITPNM2 (Ser399 or Ser400: ↑102nM; Ser1277: ↑102/5nM; Ser1326: ↑305nM) as phosphatidylinositol transporter and PIKFYVE (Ser307: ↑102nM) as a PIP2-converting kinase to be potentially phosphorylated by PKA. Further downstream on the level of effector proteins, we identified several PIP3-signaling–dependent proteins to be directly regulated by (de)phosphorylation, among those IRS-2 (Ser560: ↑305nM), GSK3α (site not confident, 19p(TSSFAEPGGGGGGGGGGPGGS)ASGPGGTGGGK50: ↑305nM), GSK3β (Thr8/Ser9/Ser13: ↑302nM), centraurin-delta2 (Ser1419: ↑605nM; Ser1435: ↑305nM) and centaurin-gamma1 (Thr647: ↑102/5nM). We could confirm an increase in phosphorylation of GSK3αSer21 and GSK3βSer9 by immunoblotting (Figure 1). For GSK3α, it was demonstrated that phosphorylation of Ser21 by PKA can lead to its enzymatic inhibition.40
With regard to Ca2+ signaling, pleckstrin phosphorylation (Thr123: ↑305nM; Ser117: ↑605nM, a major PKC target in platelets41 ) is regulated, whereas CalDAG-GEF1 phosphorylation is regulated at Ser576 (↑102/5nM) and potentially PKA phosphorylated at Ser116 and Ser117 (both: ↑102/5nM) as well as Ser587 (↑30/102/5nM), which is in accordance with our own recent data20 demonstrating that phosphorylation of CalDAG-GEF1 inhibits Rap1b activation in human platelets. Moreover, cPLA2 phosphorylation is upregulated at Ser727 (↑302/5nM). Phosphorylation at this site might promote the dissociation of a heterotetramer of cPLA2, Annexin A2, and p11 and increases cPLA2 enzymatic activity.42 As direct regulators of intracellular calcium, we found the calcium release–activated calcium channel Orai1 being dephosphorylated (Thr295: ↓602nM) and the inositol phosphate receptor regulator MRVI1 (IRAG) increasingly phosphorylated at Ser657 and Ser670 (↑102/5nM). Both sites are known to be PKG targets preventing the release of Ca2+ from inositol phosphate sensitive stores and subsequent platelet aggregation.43 The role of Orai1 phosphorylation though is yet unclear.
In addition, we found novel putative PKA phosphorylation sites in TRPC63 at Ser839/840 and Ser92/Thr93/94, however, not regulated under the observed conditions.
A potential role for protein ubiquitination in platelet regulation
Our data also reveal differential phosphorylation upon iloprost stimulation in 7 proteins which are related to protein ubiquitination. Rad23B (Thr155/Thr159/Ser160: ↓102nM) is assumed to deliver polyubiquitinated substrates to the 26S proteasome and to participate in endoplasmic reticulum–associated degradation (ERAD) of misfolded glycoproteins, whereas Cullin4A and DCAF8 are part of the cullin-RING–based E3 ubiquitin-protein ligase.44 Cullin4A is potentially PKA phosphorylated at Ser10 (↑302/5nM) and DCAF8 phosphorylation is significantly downregulated at Ser99 (↓605nM). CYLD, a ubiquitin thiolesterase, is dephosphorylated at Ser418 (↓605nM) as well. MARCH-2 and NEDD-4 are E3 ubiquitin-protein ligases which are potentially PKA phosphorylated at Ser49 (↑302/5nM) and Ser747 (↑305nM), respectively. The latter regulates membrane-bound receptors by ubiquitination.45 Finally, TOM-like protein 1 phosphorylation is significantly regulated at Ser314/Ser321/Ser323 (↑102nM).
The reasons for phosphoregulation of components of the ubiquitin pathway after platelet inhibition–dependent stimulation remain unclear. However, there is evidence that Syk is ubiquitinated upon platelet activation by collagen, leading to a substantial increase of its enzymatic activity.46 Furthermore, cAMP/PKA-mediated ubiquitination leads to an increased proteasome and ubiquitin ligase activity in rat neurons47 and PKA phosphorylation of β-catenin prevents its ubiquitination and degradation in mouse fibroblasts, thus retaining Wnt-signaling activity.48
Additional phosphorylation-independent regulation
To identify further iloprost-mediated processes with an alternative approach which is not solely focused on phosphopeptides, we analyzed a time course of 1 donor using label-free quantitative proteomics, as described in supplemental Material and methods. Hereby, we quantified global changes within the complete samples to identify regulation events which are not primarily based on phosphorylation, but could be mediated by further posttranslational modifications such as (de)acetylation, (de)palmitoylation,49 degradation/shedding, and formation or cleavage of disulfide bonds. To improve the robustness and reduce the number of candidates considered, we filtered all proteins for the occurrence of at least 3 significantly regulated peptides, leading to a final list of 44 proteins of which 29 were identified as regulated on the phosphopeptide level too (see supplemental Table 6). Among the 15 additional candidates are proteins which are known to be involved in platelet activation, such as APP, Calpain-1 catalytic subunit, coagulation factor V, coagulation factor XIII A chain, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β-1, Hic-5, integrin αIIb, protein disulfide-isomerase A3, glycogen phosphorylase, kindlin-3, and vWF. These results indicate that, besides phosphorylation, further post-translational modification (PTM) such as degradation/processing, ubiquitination, and disulfide bond rearrangement/formation might play an important role in platelet activation and inhibition.
Novel key regulators of platelet activity?
ENSA (Ser109) and ARPP19 (Ser104) are low-molecular-weight proteins of the endosulfine family which have been neglected in platelets so far; however, both show a striking increase in phosphorylation already 10 seconds after stimulation with iloprost. It is known that these proteins are involved in the regulation of phosphatase PP2A which in turn regulates cPLA2 and thus the release of arachidonate.21 Moreover, the early responding ENSA phosphorylation Ser109 disrupts ENSA interaction with α-synuclein (SNCA),23 which was reported as potential negative regulator of α-granule release.50 Thus, these 2 proteins represent novel substrates which might have an important role in platelet activation and inhibition. We speculate that the present data set contains many more interesting targets for functional research, of which many have been completely unknown to the field.
In conclusion, our study represents the first global approach to elucidate time-resolved phosphorylation changes after cAMP-dependent PKA stimulation of human platelets using iloprost. In total, we quantified >2700 phosphopeptides thus yielding unprecedented insights into fundamental processes leading to platelet inhibition. Almost 300 proteins show regulated phosphopeptides, 137 of which are putative PKA targets. Our data clearly indicate that cAMP-dependent platelet inhibition involves the crosstalk of different signaling pathways, obviously through the concerted regulation of at least 16 kinases and 7 phosphatases, and thus is much more complex than originally expected.
Besides known players and substrates of platelet inhibition, we identified many so-far unknown players. We thus hypothesize that these data generated through a discovery-driven characterization of signaling events in human platelets will be the basis to identify and establish novel key players and target proteins in platelet signaling. Moreover, it seems obvious that besides phosphorylation, further dynamic PTM might also play an important role in regulating platelet activation, even in the early stages of cAMP/PKA signaling. Future experiments will have to characterize these novel PKA substrates on the functional level to define their roles in platelet inhibition.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
We thank Claudia Schütz for excellent technical assistance.
This work was supported by the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, by the Bundesministerium für Bildung und Forschung (MedSys Project SARA 31P5800: J.G., P.N., O.K., U.W., A.S., and R.Z., 01E010003: U.W.), and Deutsche Forschungsgemeinschaft (SFB688/TPA2 [U.W., S.G.], SPP1335 [O.K.]).
F.B. is candidate at the University of Dortmund, Germany. J.V. is candidate at the University of Tübingen, Germany, and this work is submitted in partial fulfillment of the requirement of the PhD.
Authorship
Contribution: F.B. designed and performed research, analyzed the data, and wrote the paper; J.G. analyzed data, collected the platelets from donors, and wrote the paper; S.G. performed western blots and contributed to study design and writing the paper; M.V., P.N., J.V., O.K., and L.M. analyzed the data; U.W. contributed to study design; A.S. contributed to study design and writing the paper; and R.P.Z. conceived of the study, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: René P. Zahedi, Leibniz-Institut für Analytische Wissenschaften–ISAS– e.V., Otto-Hahn-Strasse 6b, D-44227 Dortmund, Germany; e-mail: rene.zahedi@isas.de; and Albert Sickmann, Leibniz-Institut für Analytische Wissenschaften–ISAS–e.V., Otto-Hahn-Strasse 6b, D-44227 Dortmund, Germany; e-mail: albert.sickmann@isas.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal