Key Points
Tmod3 deletion leads to reduced erythroid progenitors and impaired erythroblast survival, cell-cycle exit, and enucleation.
Erythroblast-macrophage islands are reduced in the absence of Tmod3, which is required in both cell types for island formation.
Abstract
Tropomodulin (Tmod) is a protein that binds and caps the pointed ends of actin filaments in erythroid and nonerythoid cell types. Targeted deletion of mouse tropomodulin3 (Tmod3) leads to embryonic lethality at E14.5-E18.5, with anemia due to defects in definitive erythropoiesis in the fetal liver. Erythroid burst-forming unit and colony-forming unit numbers are greatly reduced, indicating defects in progenitor populations. Flow cytometry of fetal liver erythroblasts shows that late-stage populations are also decreased, including reduced percentages of enucleated cells. Annexin V staining indicates increased apoptosis of Tmod3−/− erythroblasts, and cell-cycle analysis reveals that there are more Ter119hi cells in S-phase in Tmod3−/− embryos. Notably, enucleating Tmod3−/− erythroblasts are still in the process of proliferation, suggesting impaired cell-cycle exit during terminal differentiation. Tmod3−/− late erythroblasts often exhibit multilobular nuclear morphologies and aberrant F-actin assembly during enucleation. Furthermore, native erythroblastic island formation was impaired in Tmod3−/− fetal livers, with Tmod3 required in both erythroblasts and macrophages. In conclusion, disruption of Tmod3 leads to impaired definitive erythropoiesis due to reduced progenitors, impaired erythroblastic island formation, and defective erythroblast cell-cycle progression and enucleation. Tmod3-mediated actin remodeling may be required for erythroblast-macrophage adhesion, coordination of cell cycle with differentiation, and F-actin assembly and remodeling during erythroblast enucleation.
Introduction
Biogenesis of mammalian red blood cells (RBCs) is a complex morphogenetic process of coordinated gene expression, proliferation, and terminal differentiation, with the final stages involving nuclear condensation and cell-cycle exit coordinated with nuclear polarization and expulsion.1,2 During nuclear polarization and expulsion, RBC membrane proteins are retained with the nascent reticulocyte, while unwanted proteins are sorted to the membrane overlying the expelled nucleus.3-5 Erythroblast enucleation has been likened to a form of asymmetric cell division, based on cell polarity signals contributed by microtubules,6,7 filamentous actin (F-actin), and myosin IIB assembly into a contractile ring-like structure at the constriction site between the extruding nucleus and the nascent reticulocyte.3,8-10 Similar to cytokinesis, F-actin assembly stimulated by Rac GTPases and a downstream effector, mDia2 (an F-actin–nucleating formin-homology protein), are required for enucleation of mouse erythroblasts.11-13 Inhibition of myosin IIB activity and filament assembly also reduces enucleation in mouse and human erythroblast cultures.6,13,14 Other studies implicate actin-regulated membrane trafficking of late endosomes and membrane remodeling as drivers of enucleation.4,15 With the exception of Rac GTPases, mDia2, and myosin II, the factors responsible for actin cytoskeleton remodeling during enucleation are not known.
Actin cytoskeleton regulation may also contribute to erythroblast enucleation via interactions of erythroblasts with macrophages.3,16 Erythroblasts differentiate in association with a large central macrophage to form erythroblastic islands in the bone marrow and fetal liver during embryonic development.16,17 Studies of fetal liver erythropoiesis show that erythroblast differentiation, survival, and enucleation depend on erythroblast-macrophage adhesion receptor interactions,18-20 and macrophage phagocytosis of extruded nuclei,21-24 processes that require the actin cytoskeleton. To date, palladin in macrophages is the only F-actin binding protein shown to be required for erythroblastic island formation.25
Tmods bind to tropomyosins and cap the pointed ends of actin filaments to regulate their length and stability.26 The Tmod family consists of four Tmods, with Tmod1 being the sole Tmod in human and mouse RBCs, where it caps the short actin filaments in the spectrin-actin network of the membrane skeleton.27 Tmod1-null mice display a mild compensated, sphero-elliptocytic anemia with osmotically fragile RBCs, due to misregulation of F-actin lengths and a disrupted spectrin-actin lattice.28 Tropomodulin3 (Tmod3) is unexpectedly present in Tmod1-null RBCs, suggesting that Tmod3 partially compensates for the absence of Tmod1.28 Here, we show that Tmod3 is expressed in human and mouse erythroblasts during erythroid terminal differentiation. Hence, we conclude that the presence of Tmod3 in Tmod1-null RBCs is likely due to aberrant persistence of Tmod3 in RBCs.
To investigate Tmod3 function in erythroid differentiation, we created global Tmod3 knockout mice, which were embryonic lethal at E14.5-E18.5, with anemia evident at ∼E13.5. Tmod3−/− mice are characterized by defective fetal liver erythropoiesis with impaired terminal differentiation via multiple mechanisms. Our results suggest that Tmod3-mediated actin remodeling plays critical roles in definitive erythropoiesis to promote erythroblast-macrophage adhesion, erythroblast survival, cell-cycle exit, F-actin assembly, and remodeling in enucleation.
Methods
Generation of Tmod3−/− mice
Mouse embryonic stem cells with a gene trap insertion in intron 1 of Tmod3 (RRF004) were obtained from BayGenomics (San Francisco, CA) (Figure 2). Blastocyst injections were performed in the Transgenic Mouse Core at The Scripps Research Institute. Resulting male chimeras were bred to C57BL/6J females to obtain germline transmission, and backcrossed to C57BL/6J mice for at least 3 generations. Timed matings of Tmod3+/− intercrosses were performed to obtain Tmod3−/− embryos. Wild-type and mutant Tmod3 allele primers for polymerase chain reaction (PCR) genotyping were as shown in Figure 2A and supplemental Table 1 on the Blood Web site. All experiments were performed according to the National Institutes of Health animal care guidelines, as approved and enforced by The Scripps Research Institute’s Institutional Animal Care and Use Committee.
Tmod1 expression is upregulated while Tmod3 is downregulated during erythroid differentiation. (A) mRNA levels of Tmods during in vitro erythroid differentiation from human CD34+ cells, determined by qRT-PCR on days 7, 10, and 14, normalized to ACTB mRNA. (B) Western blots of Tmod1 and Tmod3 proteins during in vitro erythroid differentiation of CD34+ cells. Cell pellets were collected on days 3, 6, 8, 10, 13, and 16 after initiation of differentiation, solubilized, and analyzed by western blotting (4.1R was used as positive control for erythroid differentiation, and total actin [C4 antibody] and glyceraldehyde-3-phosphate dehydrogenase proteins were used as loading controls). (C-D) qRT-PCR analysis of Tmod1 and Tmod3 mRNAs in highly pure erythroblasts at distinct developmental stages isolated by FACS from (C) in vitro CD34+cell-erythroid differentiation cultures, or (D) mouse bone marrow, normalized to ACTB mRNA. Stages were: proerythroblasts (Pro-E), early basophilic erythroblasts (early Baso), late basophilic erythroblasts (late Baso), basophilic erythroblasts (Baso-E), polychromatic erythroblasts (Poly-E), and orthochromatic erythroblasts (Ortho-E).
Tmod1 expression is upregulated while Tmod3 is downregulated during erythroid differentiation. (A) mRNA levels of Tmods during in vitro erythroid differentiation from human CD34+ cells, determined by qRT-PCR on days 7, 10, and 14, normalized to ACTB mRNA. (B) Western blots of Tmod1 and Tmod3 proteins during in vitro erythroid differentiation of CD34+ cells. Cell pellets were collected on days 3, 6, 8, 10, 13, and 16 after initiation of differentiation, solubilized, and analyzed by western blotting (4.1R was used as positive control for erythroid differentiation, and total actin [C4 antibody] and glyceraldehyde-3-phosphate dehydrogenase proteins were used as loading controls). (C-D) qRT-PCR analysis of Tmod1 and Tmod3 mRNAs in highly pure erythroblasts at distinct developmental stages isolated by FACS from (C) in vitro CD34+cell-erythroid differentiation cultures, or (D) mouse bone marrow, normalized to ACTB mRNA. Stages were: proerythroblasts (Pro-E), early basophilic erythroblasts (early Baso), late basophilic erythroblasts (late Baso), basophilic erythroblasts (Baso-E), polychromatic erythroblasts (Poly-E), and orthochromatic erythroblasts (Ortho-E).
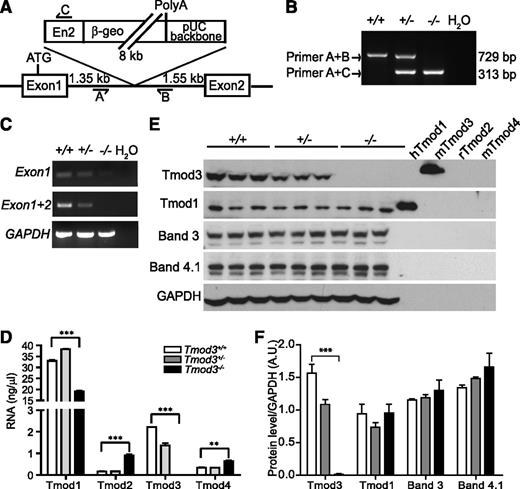
Targeted disruption of Tmod3 gene in mice. (A) Tmod3 gene targeting strategy, depicting insertion site of BayGenomics β-geo gene-trap vector into intron 1 of Tmod3, as confirmed by DNA sequencing. Primers A, B, and C were designed for genotyping wild-type and mutant alleles, as shown. (B) PCR analysis of genomic DNA from mouse embryonic yolk sacs derived from Tmod3+/− intercrosses. Primers A and B detect the expected 729 bp band in the wild-type allele, while primers A and C detect the predicted 313 bp band in the mutant allele. (C) RT-PCR of Tmod3 transcripts in E14.5 fetal liver mRNA, using Tmod3 exon 1 and 2 primers. (D) Tmod mRNA levels in Tmod3+/+, Tmod3+/−, and Tmod3−/− fetal livers from E14.5 embryos, determined by qRT-PCR. (E) Western blot analysis of Tmods and erythroid membrane proteins bands 3, and 4.1R in Tmod3+/+, Tmod3+/−, and Tmod3−/− fetal livers from E14.5 embryos. Each lane is from a single embryo. Purified Tmod proteins (5 ng/lane) were loaded as controls for antibody specificity (hTmod1, Tmod1 from Homo sapiens; mTmod3 and mTmod4, Tmod3 and Tmod4 from Mus musculus; rTmod2, Tmod2 from Rattus norvegicus).43 (F) Calculation of Tmod1, Tmod3, bands 3, and 4.1R protein levels, normalized to glyceraldehyde-3-phosphate dehydrogenase. Band intensities were analyzed by ImageJ. Values in panels D and F are means ± SD from triplicate individual fetal livers from different embryos. **P < .005; ***P < .001. β-geo, fusion of β-galactosidase and neomycin phosphotransferase II; En2, engrailed 2-splice acceptor sequence; PolyA, polyadenylation.
Targeted disruption of Tmod3 gene in mice. (A) Tmod3 gene targeting strategy, depicting insertion site of BayGenomics β-geo gene-trap vector into intron 1 of Tmod3, as confirmed by DNA sequencing. Primers A, B, and C were designed for genotyping wild-type and mutant alleles, as shown. (B) PCR analysis of genomic DNA from mouse embryonic yolk sacs derived from Tmod3+/− intercrosses. Primers A and B detect the expected 729 bp band in the wild-type allele, while primers A and C detect the predicted 313 bp band in the mutant allele. (C) RT-PCR of Tmod3 transcripts in E14.5 fetal liver mRNA, using Tmod3 exon 1 and 2 primers. (D) Tmod mRNA levels in Tmod3+/+, Tmod3+/−, and Tmod3−/− fetal livers from E14.5 embryos, determined by qRT-PCR. (E) Western blot analysis of Tmods and erythroid membrane proteins bands 3, and 4.1R in Tmod3+/+, Tmod3+/−, and Tmod3−/− fetal livers from E14.5 embryos. Each lane is from a single embryo. Purified Tmod proteins (5 ng/lane) were loaded as controls for antibody specificity (hTmod1, Tmod1 from Homo sapiens; mTmod3 and mTmod4, Tmod3 and Tmod4 from Mus musculus; rTmod2, Tmod2 from Rattus norvegicus).43 (F) Calculation of Tmod1, Tmod3, bands 3, and 4.1R protein levels, normalized to glyceraldehyde-3-phosphate dehydrogenase. Band intensities were analyzed by ImageJ. Values in panels D and F are means ± SD from triplicate individual fetal livers from different embryos. **P < .005; ***P < .001. β-geo, fusion of β-galactosidase and neomycin phosphotransferase II; En2, engrailed 2-splice acceptor sequence; PolyA, polyadenylation.
qRT-PCR analysis of Tmods
Fetal livers were dissected from mouse embryos at E14.5. Human CD34+ stem/early progenitor cells for erythroid cultures in Figure 1A-B were isolated from growth-factor-mobilized peripheral blood using the CliniMACS (Miltenyi Biotec) device, and cultured in vitro to promote terminal differentiation into erythroid cells.29 Pure populations of human erythroblasts from cord blood-derived CD34+ cell erythroid cultures (Figure 1C) or murine bone marrow erythroblasts (Figure 1D) at distinct developmental stages were obtained by fluorescence-activated cell sorting (FACS).30,31 The quantitative reverse-transcription polymerase chain reaction (qRT-PCR) protocol is available in the supplemental Methods, with primer and probe sequences in supplemental Table 1.
Western blotting
Cells from different days of in vitro erythropoiesis culture initiated from human CD34+ cells29 or fetal livers were homogenized in ice-cold immunoprecipitation assay buffer32 with protease inhibitor cocktail (1:100; Sigma-Aldrich). After centrifugation, the supernatant was solubilized in an equal volume of 2 × sodium dodecyl sulfate sample buffer, followed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and western blotting.32 Antibodies are presented in supplemental Table 2. Proteins were detected using enhanced chemiluminescence (Thermo Scientific) with radiograph film.
Fluorescence staining and confocal microscopy
To stain isolated erythroblasts, E14.5 fetal livers were dissociated into phosphate-buffered saline (PBS) by gentle trituration using a cutoff P200 tip, followed by 4 hours of fixation in 4% paraformaldehyde in PBS at room temperature (RT). Samples were centrifuged in an Eppendorf microfuge at 4000 rpm for 4 minutes and re-suspended in 0.3% Triton in PBS. After 30 minutes at RT, cells were centrifuged, resuspended in blocking buffer (3% BSA/1% goat serum in PBS) and stored overnight at 4°C. Cells were stained for 2 hours at RT with antibodies and dyes as shown in supplemental Table 2. After washing, cells were deposited onto coverslips using a Thermo-Fisher Cytospin 4 at 900 to 1000 rpm for 3 minutes, mounted on slides using Fluoro-gel (Electron Microscopy Sciences), and imaged by confocal fluorescence microscopy.
Erythroid colony analysis
Erythroid colony-forming units (CFU-E) and burst-forming units (BFU-E) were analyzed using E12.5 fetal liver cells, according to protocols from Stemcell Technologies. MethoCult M3231 and M3534 media with 2 U/mL rhepo (Life Technologies) were used for CFU-E and BFU-E colonies, respectively, and numbers of CFU-E and BFU-E colonies determined after 2 days and 7 days of culture, respectively.
Flow cytometry
Fetal liver cells were isolated by trituration with a 1 mL serological pipette, passed through a 70-µm cell strainer (BD Biosciences), and immunostained for 30 minutes at 4°C in PBS/2% fetal bovine serum with propidium iodide (PI) staining solution (BD Biosciences), Alexa Fluor 700-anti-Ter119, and phycoerythrin–anti-CD71 (Life Technologies). For apoptosis, cells were additionally stained for 5 minutes at RT with Pacific Blue-Annexin V (Life Technologies). To quantify dead cells, 7-AAD (Life Technologies) was added before flow cytometry. For enucleation, Syto-16 (nuclei) and SytoX (viability) (Life Technologies) were added to the cocktail of Ter119 and CD71 antibodies. For cell-cycle analysis, cells were fixed in 70% cold ethanol for at least 2 hours, and treated with DNase-free RNase (Roche) at 37°C for 1 hour, and stained with PI. Flow cytometry was performed with a BD FACS LSR II with BD FACSDiva software. Data were analyzed with FlowJo 9.2 software (Tree Star).
Erythroblastic islands
Native erythroblastic islands were isolated from E13.5 or E14.5 fetal livers.22 For island reconstitution, erythroblasts were stripped from adherent macrophages with Dulbecco’s PBS, and erythroid cells from other embryos (Tmod3+/+ or Tmod3−/−) were added to the stripped macrophages. Native or reconstituted clusters were dipped in RPMI medium (Life Technologies) to remove non-adherent cells. Islands were fixed in 4% paraformaldehyde in PBS for 20 minutes, stained with Hoechst 33258, Alexa Fluor-488-anti-Ter119 and Alexa Fluor-647-anti-F4/80, and imaged by confocal microscopy.
Statistical analysis
Results are means ± standard deviation (SD). Significance was evaluated with a 2-tailed unpaired Student t-test. Excel and Prism version 5.0 software (GraphPad Software) were used for statistical evaluation.
Results
Tmod1 expression increases while Tmod3 decreases during erythroid differentiation
To investigate Tmod1 and Tmod3 expression during erythroid differentiation, human erythroblasts were obtained by in vitro culture of CD34+ cells.29 qRT-PCR analysis showed that Tmod1 and Tmod3 messenger RNA (mRNA) expression levels were similar early in differentiation, with a fourfold increase in Tmod1 levels during differentiation, while Tmod3 levels decreased (Figure 1A). Tmod1 and Tmod3 proteins displayed the same pattern (Figure 1B), consistent with Tmod1 as the sole Tmod1 in mature RBCs.28 This analysis is supported by qRT-PCR analysis of highly purified erythroblast populations obtained by FACS from in vitro differentiated erythroid cultures of human CD34+ cells,30 and from mouse bone marrow31 (Figure 1C-D). Almost no Tmod2 and Tmod4 mRNA transcripts were detected in human or mouse erythroblasts (Figure 1A, data not shown). Downregulation of Tmod3 mRNA and protein expression during erythroid differentiation suggests that Tmod3 plays a role in erythropoiesis prior to assembly of the RBC membrane skeleton, which contains Tmod1.28
Generation of Tmod3−/− mice
Tmod3 knockout mice were created from embryonic stem cells in which intron 1 of Tmod3 was disrupted by a β-galactosidase/neomycin (β-geo) marker through retroviral-mediated insertion, creating a novel fusion transcript of the first exon with the β-geo marker33 (Figure 2A). Genotyping of progeny embryos from Tmod3+/− intercrosses showed expected PCR products of 729 bp and 313 bp from wild-type and mutant alleles, respectively (Figure 2B). To confirm insertion of the β-geo marker, LacZ staining was performed on Tmod3+/− E13.5 embryos, demonstrating bright blue staining of the whole embryo (supplemental Figure 1A), consistent with previous reports of ubiquitous Tmod3 expression.34
In fetal livers from Tmod3−/− embryos, Tmod3 mRNA was neither detected by RT-PCR using primers for exon 1 and 2 (Figure 2C), nor by qRT-PCR (Figure 2D). Western blotting showed that the Tmod3 ∼40kD polypeptide was absent in Tmod3−/− fetal livers (Figure 2E-F). Analysis of other Tmods revealed that Tmod1 mRNA was decreased but Tmod1 protein was unchanged (Figure 2D-F). Tmod2 and Tmod4 mRNAs were increased somewhat in absence of Tmod3 (Figure 2D), but Tmod2 and Tmod4 proteins were undetectable in all genotypes (data not shown). In addition, there were no significant changes in transcript or protein levels of other erythroid membrane proteins in fetal livers (Figure 2E-F and supplemental Figure 1B). Therefore, altered expression of other Tmods does not compensate for the absence of Tmod3.
Tmod3−/− mice are embryonic lethal with anemia at midgestation
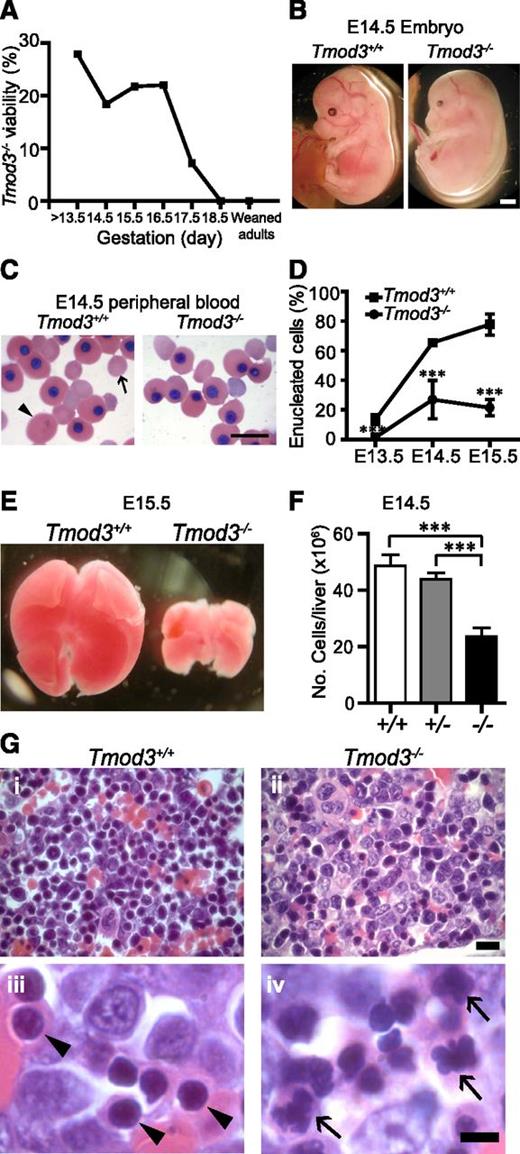
Tmod3+/− mice are viable and fertile, with no significant hematologic abnormalities (supplemental Table 3), RBC morphology, or osmotic fragility (supplemental Figure 2). In contrast, Tmod3−/− embryos begin to die starting at E14.5, with lethality increasing markedly after E16.5, and no viable Tmod3−/− embryos detected by E18.5 (Figure 3A and supplemental Table 4). The paleness of the surviving Tmod3−/− embryos at E14.5 suggests anemia (Figure 3B).
Tmod3−/−mice are embryonic lethal with anemia. (A) Viability of Tmod3−/− embryos. Timed matings of Tmod3+/− intercrosses were performed and embryos were harvested at stages E13.5 up to E18.5. The percentages reflect the numbers of live Tmod3−/− embryos with respect to all embryos harvested in litters at each gestational stage. (B) Gross morphology of Tmod3+/+ and Tmod3−/− embryos at E14.5. Scale bar, 1 mm. (C) Wright-Giemsa staining of peripheral blood cytospins from Tmod3+/+ and Tmod3−/− embryos at E14.5. Large primitive enucleated RBC (arrowhead); smaller definitive enucleated RBC (arrow). Note that sizes of enucleated cells from Tmod3−/− embryos are variable and do not fall into clear size categories. Scale bar, 20 µm. (D) The percentage of enucleated RBCs in peripheral blood cytospins from Tmod3+/+ and Tmod3−/− embryos at E13.5, E14.5, and E15.5. At least 300 cells were counted for each sample. Values are means ± SD (n = 5 to 13). ***P < .001. (E) Gross morphology of representative fetal livers from Tmod3+/+ and Tmod3−/− embryos at E15.5. (F) Total number of fetal liver cells from Tmod3+/+ (n = 7), Tmod3+/− (n = 18), and Tmod3−/− (n = 5) embryos at E14.5. Values are means ± SD. ***P < .001. (G) Hematoxylin and eosin staining of paraffin sections from Tmod3+/+ and Tmod3−/− fetal livers fixed in Bouin’s solution. Normal orthochromatic erythroblast nuclei in Tmod3+/+ fetal liver (arrowheads). Abnormal multilobular orthochromatic erythroblast nuclei in Tmod3−/− fetal liver (arrows). Scale bar, 10 µm (i-ii); 5 µm (iii-iv). Panels C and G were acquired with a Zeiss Axioskop microscope with AxioCam ICc3 color camera using a ×20 objective (N.A. 0.5) at a zoom of 2 (Gi-ii) or a ×100 oil-immersion objective (N.A. 1.3) at a zoom of 1 (C and Giii-iv).
Tmod3−/−mice are embryonic lethal with anemia. (A) Viability of Tmod3−/− embryos. Timed matings of Tmod3+/− intercrosses were performed and embryos were harvested at stages E13.5 up to E18.5. The percentages reflect the numbers of live Tmod3−/− embryos with respect to all embryos harvested in litters at each gestational stage. (B) Gross morphology of Tmod3+/+ and Tmod3−/− embryos at E14.5. Scale bar, 1 mm. (C) Wright-Giemsa staining of peripheral blood cytospins from Tmod3+/+ and Tmod3−/− embryos at E14.5. Large primitive enucleated RBC (arrowhead); smaller definitive enucleated RBC (arrow). Note that sizes of enucleated cells from Tmod3−/− embryos are variable and do not fall into clear size categories. Scale bar, 20 µm. (D) The percentage of enucleated RBCs in peripheral blood cytospins from Tmod3+/+ and Tmod3−/− embryos at E13.5, E14.5, and E15.5. At least 300 cells were counted for each sample. Values are means ± SD (n = 5 to 13). ***P < .001. (E) Gross morphology of representative fetal livers from Tmod3+/+ and Tmod3−/− embryos at E15.5. (F) Total number of fetal liver cells from Tmod3+/+ (n = 7), Tmod3+/− (n = 18), and Tmod3−/− (n = 5) embryos at E14.5. Values are means ± SD. ***P < .001. (G) Hematoxylin and eosin staining of paraffin sections from Tmod3+/+ and Tmod3−/− fetal livers fixed in Bouin’s solution. Normal orthochromatic erythroblast nuclei in Tmod3+/+ fetal liver (arrowheads). Abnormal multilobular orthochromatic erythroblast nuclei in Tmod3−/− fetal liver (arrows). Scale bar, 10 µm (i-ii); 5 µm (iii-iv). Panels C and G were acquired with a Zeiss Axioskop microscope with AxioCam ICc3 color camera using a ×20 objective (N.A. 0.5) at a zoom of 2 (Gi-ii) or a ×100 oil-immersion objective (N.A. 1.3) at a zoom of 1 (C and Giii-iv).
In Tmod3+/+ embryos at E14.5, circulating RBCs are predominantly enucleated, indicating definitive RBC production in the fetal liver, along with some larger nucleated primitive RBCs produced earlier in the yolk sac, a few of which are enucleated (Figure 3C-D).35,36 In contrast, in Tmod3−/− embryos, enucleated RBCs were rarely observed (Figure 3C), and quantification confirmed a significant reduction in percentage starting at E14.5 (Figure 3D). These results suggested a defect in fetal liver definitive erythropoiesis in the absence of Tmod3. The morphology and enucleation of the larger primitive circulating RBCs also appeared to be defective (supplemental Figure 3). However, in this study, we focused on definitive erythropoiesis, since Tmod3−/− embryos die after E14.5 when definitive erythropoiesis is dominant.
Erythroblast terminal differentiation is impaired in Tmod3−/− fetal livers
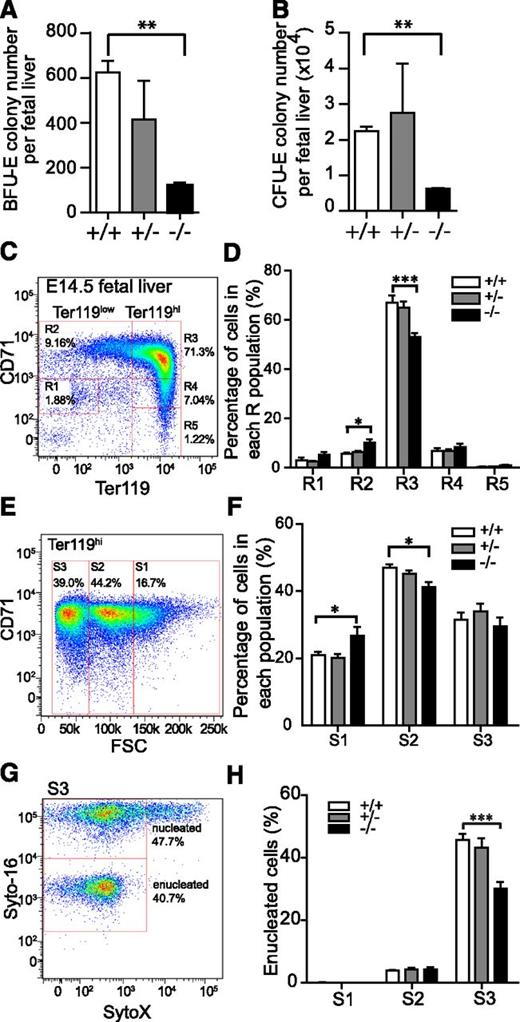
The fetal liver is the key organ for definitive erythropoiesis during midgestation, containing erythroblasts at various stages of differentiation. The Tmod3−/− fetal liver was noticeably smaller, starting at E12.5 (not shown) with reduced cell number (Figure 3E-F). Hematoxylin and eosin staining of Tmod3−/− fetal liver sections indicated abnormal cellular structure, with more abundant larger blast-type cells, and fewer late-stage orthochromatophilic erythroblasts (Figure 3Gi-ii). At high magnification, nuclei of Tmod3+/+ orthochromatophilic erythroblasts were round and condensed (Figure 3Giii), while those of Tmod3−/− erythroblasts often showed abnormal multilobular morphologies or “butterfly” shapes (Figure 3Giv). At E12.5, both BFU-E and CFU-E colony numbers per fetal liver were reduced in Tmod3−/− embryos (Figure 4A-B), consistent with the smaller liver size, and indicating fewer functional erythroid progenitors in Tmod3−/− fetal livers.
Tmod3−/−embryos are defective in definitive erythropoiesis with reduced progenitors, impaired terminal differentiation, and enucleation. (A) BFU-E per fetal liver. (B) CFU-E per fetal liver. Values in both are means ± SD of at least 3 independent embryos. (C) Representative flow cytometry profiles of R1-R5 erythroblast populations labeled with CD71/Ter119 in fetal livers from Tmod3+/+ mice at E14.5. The percentage of cells in each R population with respect to total PI-negative cells is indicated for a representative Tmod3+/+ fetal liver. (D) Percentage of cells in each R population, normalized to the number of total viable fetal liver cells from each embryo (Tmod3+/+, n = 6; Tmod3+/−, n = 19; Tmod3−/−, n = 8). (E) Representative CD71/FSC profile of Tmod3+/+ Ter119hi cells sorted into 3 populations (S1, S2, and S3) according to cell size. The percentage of cells in each S population with respect to total Ter119hi cells is indicated for a representative Tmod3+/+ fetal liver. (F) Percentage of cells in each S population, normalized to the number of total Ter119hi fetal liver cells from each embryo. (G) Representative flow cytometry of enucleated cells in the S3 population of Tmod3+/+ fetal livers, using Syto-16 for nuclei and SytoX for cell viability. The percentage of enucleated and nucleated cells with respect to total S3 cells is indicated for a representative Tmod3+/+ fetal liver. (H) Percentages of enucleated cells in S1, S2, and S3 populations. All values are means ± SD with at least 3 replicates. *P < .05, **P < .005; ***P < .001 for Tmod3−/− vs Tmod3+/+.
Tmod3−/−embryos are defective in definitive erythropoiesis with reduced progenitors, impaired terminal differentiation, and enucleation. (A) BFU-E per fetal liver. (B) CFU-E per fetal liver. Values in both are means ± SD of at least 3 independent embryos. (C) Representative flow cytometry profiles of R1-R5 erythroblast populations labeled with CD71/Ter119 in fetal livers from Tmod3+/+ mice at E14.5. The percentage of cells in each R population with respect to total PI-negative cells is indicated for a representative Tmod3+/+ fetal liver. (D) Percentage of cells in each R population, normalized to the number of total viable fetal liver cells from each embryo (Tmod3+/+, n = 6; Tmod3+/−, n = 19; Tmod3−/−, n = 8). (E) Representative CD71/FSC profile of Tmod3+/+ Ter119hi cells sorted into 3 populations (S1, S2, and S3) according to cell size. The percentage of cells in each S population with respect to total Ter119hi cells is indicated for a representative Tmod3+/+ fetal liver. (F) Percentage of cells in each S population, normalized to the number of total Ter119hi fetal liver cells from each embryo. (G) Representative flow cytometry of enucleated cells in the S3 population of Tmod3+/+ fetal livers, using Syto-16 for nuclei and SytoX for cell viability. The percentage of enucleated and nucleated cells with respect to total S3 cells is indicated for a representative Tmod3+/+ fetal liver. (H) Percentages of enucleated cells in S1, S2, and S3 populations. All values are means ± SD with at least 3 replicates. *P < .05, **P < .005; ***P < .001 for Tmod3−/− vs Tmod3+/+.
Next, we investigated whether the absence of Tmod3 also affected erythroid terminal differentiation using flow cytometry with CD71 and Ter119 markers to identify the R1-R5 erythroblast populations37,38 (Figure 4C). This showed that the normally abundant late-stage R3 erythroblast population (CD71hi /Ter119hi) was reduced substantially in the fetal livers of Tmod3−/− embryos, suggesting a late block in erythroid terminal differentiation (Figure 4D and supplemental Figure 4C). To further characterize terminal differentiation, we used FSC to separate Ter119hi cells into 3 populations: S1 (large), S2 (medium), and S3 (small), indicative of progressively more mature cells (Figure 4E).12 This showed that the percentage of Ter119hi cells in S1 increased, while in S2 it decreased (Figure 4F and supplemental Figure 4D), indicating a block in the progression from large- to medium-size cells, consistent with a defect at late stages of erythroblast maturation. Next, we analyzed erythroblast enucleation in S1-S3 populations using live-cell nuclear staining with Syto-16 (Figure 4G). As expected, most enucleation events occurred in the S3 population, and Tmod3−/− fetal livers showed a significant reduction of enucleation efficiency in this population (Figure 4H).
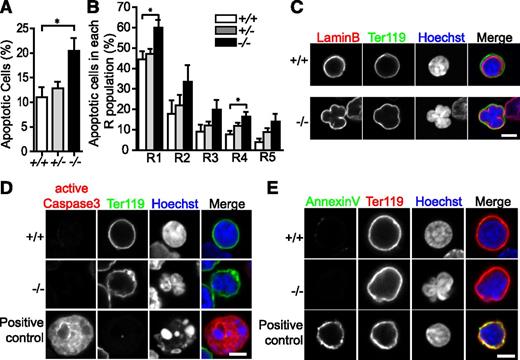
There are more apoptotic erythroblasts in Tmod3−/− fetal livers
To further explore the reasons for the smaller fetal liver and reduced late erythroblasts in Tmod3−/− embryos, we analyzed apoptosis by flow cytometry with Annexin V (Figure 5A), and by immunostaining of fetal liver sections for active caspase-3 (supplemental Figure 5A-B). These approaches demonstrated an ∼twofold increase of apoptotic cells in Tmod3−/− fetal livers, which may be due to apoptosis of erythroblasts and other cell types. To investigate apoptosis in erythroblasts, we analyzed each R population for the percentage of Annexin V-positive cells. All R populations showed modest increases, which only achieved statistical significance for R1 and R4 (Figure 5B), indicating that reduced erythroblast numbers in Tmod3−/− fetal livers may be partly explained by increases in apoptosis. However, confocal fluorescence microscopy of Tmod3−/− Ter119-positive erythroblasts with multilobular nuclei demonstrated continuous lamin B staining, indicative of an intact nuclear envelope (Figure 5C), negative staining for active caspase-3 and Annexin V (Figure 5D-E), positive staining for Ki67, and a cell proliferation marker (supplemental Figure 5C). Thus, we can deduce that multilobular nuclear morphologies of Tmod3−/− late erythroblasts are not due to apoptosis.
Tmod3−/−fetal liver erythroblasts display increased apoptosis throughout terminal differentiation. (A) Percentage of apoptotic cells based on Annexin V-positive cells as a proportion of total 7-AAD–negative fetal liver cells at E14.5. (B) Percentage of apoptotic cells within each R population. The 7-AAD–negative cells were first gated on CD71/Ter119 to identify R populations as in Figure 4C, and percentages for each R was calculated as the number of Annexin V-positive cells as a fraction of the total cell number in that R population. All values are means ± SD with at least 3 replicates with samples prepared from different individual embryos. *P < .05. (C) Cytospins of fetal liver cells stained with lamin B (red) for nuclear envelope, Ter119-Alexa488 (green) for a GPA-associated antigen to identify late erythroblasts, and Hoechst (blue) for nuclei. (D) Cytospins of fetal liver cells stained with active-caspase3 (red) for apoptosis, Ter119-Alexa488 (green), and Hoechst (blue). (E) Cytospins of fetal liver cells stained with Annexin V (green) for apoptosis, Ter119-PE (red), and Hoechst (blue). Panels C-E were acquired with a Zeiss LSM 780 laser scanning confocal fluorescence microscope using a Zeiss 100× oil-immersion objective (N.A. 1.4) at a zoom of 2. Scale bar, 4 µm.
Tmod3−/−fetal liver erythroblasts display increased apoptosis throughout terminal differentiation. (A) Percentage of apoptotic cells based on Annexin V-positive cells as a proportion of total 7-AAD–negative fetal liver cells at E14.5. (B) Percentage of apoptotic cells within each R population. The 7-AAD–negative cells were first gated on CD71/Ter119 to identify R populations as in Figure 4C, and percentages for each R was calculated as the number of Annexin V-positive cells as a fraction of the total cell number in that R population. All values are means ± SD with at least 3 replicates with samples prepared from different individual embryos. *P < .05. (C) Cytospins of fetal liver cells stained with lamin B (red) for nuclear envelope, Ter119-Alexa488 (green) for a GPA-associated antigen to identify late erythroblasts, and Hoechst (blue) for nuclei. (D) Cytospins of fetal liver cells stained with active-caspase3 (red) for apoptosis, Ter119-Alexa488 (green), and Hoechst (blue). (E) Cytospins of fetal liver cells stained with Annexin V (green) for apoptosis, Ter119-PE (red), and Hoechst (blue). Panels C-E were acquired with a Zeiss LSM 780 laser scanning confocal fluorescence microscope using a Zeiss 100× oil-immersion objective (N.A. 1.4) at a zoom of 2. Scale bar, 4 µm.
Cell-cycle progression is impaired during terminal differentiation in Tmod3−/− fetal liver
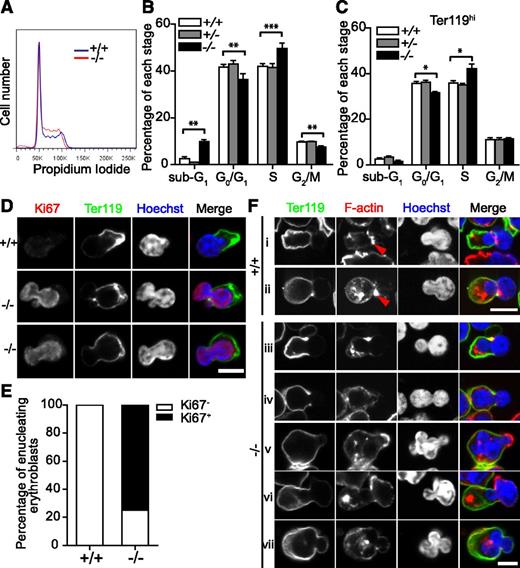
To investigate the basis for the late block in terminal differentiation, we analyzed the cell cycle of fetal liver cells using PI staining (Figure 6A-B). Consistent with the apoptosis analysis presented earlier, there were increased numbers of sub-G1 cells in Tmod3−/− fetal livers. Additionally, more cells were in S-phase, while cells at G0/G1 and G2/M were decreased, indicating impaired cell-cycle progression in Tmod3−/− fetal livers. Further analysis of the Ter119hi cell population showed an increase in cells at S-phase, but no increase of sub-G1 cells (Figure 6C), implying that impaired terminal differentiation in Tmod3−/− erythroblasts might be due to abnormal cell-cycle progression.
Cell-cycle progression and actin cytoskeleton structures are impaired during erythropoiesis in Tmod3−/−fetal liver. (A) Representative flow cytometry profiles of cell-cycle analysis of fetal liver cells at E14.5 after PI staining. (B) Percentages of fetal liver cells in sub-G1, G0/G1, S, and G2/M phase. (C) Percentages of Ter119hi fetal liver cells in sub-G1, G0/G1, S, and G2/M phase. All values are means ± SD with at least 3 replicates. **P < .005; ***P < .001 for Tmod3−/− vs Tmod3+/+. (D) Confocal fluorescence microscopy images of enucleating Tmod3+/+ or Tmod3−/− erythroblasts stained with Ter119-Alexa488 (green), Ki67 (red), and Hoechst (blue), and imaged in cytospins. Scale bar, 4 µm. (E) Percentages of enucleating Ki67+ and Ki67- erythroblasts in Tmod3+/+ or Tmod3−/− fetal livers. (F) Confocal fluorescence microscopy images of enucleating Tmod3+/+ (i-ii) and Tmod3−/− (iii-vii) erythroblasts, imaged in cytospins of fetal liver cells stained with Ter119-Alexa488 (green), rhodamine-phalloidin (red), and Hoechst (blue). Enucleating erythroblasts were identified in both genotypes by membrane sorting of Ter119 staining, and a nuclear constriction at the transition between the bright and dim Ter119 membrane staining. F-actin assembles into a contractile actin ring at the neck region in Tmod3+/+ enucleating erythroblasts and in bright foci in the cytoplasm of the incipient reticulocyte (i-ii). Tmod3−/− enucleating erythroblasts occasionally have an F-actin contractile ring (iii), but not always (iv-vii), and sometimes have F-actin enrichment on the dim Ter119 membrane overlying a protruding nuclear lobe (v-vi). Similar to wild-type, Tmod3−/− enucleating erythroblasts also contain F-actin foci in the cytoplasm (iii-vii). Scale bar, 6 µm. Panels D, Fiii-vii were acquired with a Zeiss LSM 780 laser scanning confocal microscope using a Zeiss 100× oil immersion objective (N.A. 1.4) at a zoom of 2, and panels Fi-ii with a Bio-Rad Radiance 2100 confocal microscope using a Zeiss 100× oil-immersion objective (N.A. 1.2) at a zoom of 3.
Cell-cycle progression and actin cytoskeleton structures are impaired during erythropoiesis in Tmod3−/−fetal liver. (A) Representative flow cytometry profiles of cell-cycle analysis of fetal liver cells at E14.5 after PI staining. (B) Percentages of fetal liver cells in sub-G1, G0/G1, S, and G2/M phase. (C) Percentages of Ter119hi fetal liver cells in sub-G1, G0/G1, S, and G2/M phase. All values are means ± SD with at least 3 replicates. **P < .005; ***P < .001 for Tmod3−/− vs Tmod3+/+. (D) Confocal fluorescence microscopy images of enucleating Tmod3+/+ or Tmod3−/− erythroblasts stained with Ter119-Alexa488 (green), Ki67 (red), and Hoechst (blue), and imaged in cytospins. Scale bar, 4 µm. (E) Percentages of enucleating Ki67+ and Ki67- erythroblasts in Tmod3+/+ or Tmod3−/− fetal livers. (F) Confocal fluorescence microscopy images of enucleating Tmod3+/+ (i-ii) and Tmod3−/− (iii-vii) erythroblasts, imaged in cytospins of fetal liver cells stained with Ter119-Alexa488 (green), rhodamine-phalloidin (red), and Hoechst (blue). Enucleating erythroblasts were identified in both genotypes by membrane sorting of Ter119 staining, and a nuclear constriction at the transition between the bright and dim Ter119 membrane staining. F-actin assembles into a contractile actin ring at the neck region in Tmod3+/+ enucleating erythroblasts and in bright foci in the cytoplasm of the incipient reticulocyte (i-ii). Tmod3−/− enucleating erythroblasts occasionally have an F-actin contractile ring (iii), but not always (iv-vii), and sometimes have F-actin enrichment on the dim Ter119 membrane overlying a protruding nuclear lobe (v-vi). Similar to wild-type, Tmod3−/− enucleating erythroblasts also contain F-actin foci in the cytoplasm (iii-vii). Scale bar, 6 µm. Panels D, Fiii-vii were acquired with a Zeiss LSM 780 laser scanning confocal microscope using a Zeiss 100× oil immersion objective (N.A. 1.4) at a zoom of 2, and panels Fi-ii with a Bio-Rad Radiance 2100 confocal microscope using a Zeiss 100× oil-immersion objective (N.A. 1.2) at a zoom of 3.
Previous studies showed that mammalian erythroblasts exit the cell cycle prior to enucleation.4 To test whether cell-cycle exit was affected by the absence of Tmod3, we stained cytospins of fetal liver cells for Ki67 (Figure 6D-E). Enucleating erythroblasts were identified by their Hoechst-stained nuclei on one side of the cell, together with a nuclear constriction located at the boundary between the bright and dim (or absent) rim of Ter119-membrane staining. As expected, all Tmod3+/+ enucleating erythroblasts were Ki67−. However, ∼75% of Tmod3−/− erythroblasts undergoing enucleation showed a strong Ki67+ signal. These results suggest that cell-cycle exit is not a prerequisite for enucleation in Tmod3−/− erythroblasts.
Enucleating erythroblasts show abnormal F-actin organization in Tmod3−/− fetal livers
The F-actin cytoskeleton plays key roles in erythroblast enucleation, and Tmod3 is an actin-regulatory protein.26,39 In Tmod3+/+ enucleating erythroblasts, confocal fluorescence microscopy revealed the expected contractile F-actin ring around the nuclear constriction site at the neck region, as well as additional bright spots of F-actin in the cytoplasm of the nascent reticulocyte (Figure 6Fi-ii). In Tmod3−/− enucleating erythroblasts, an F-actin ring appeared to form in the neck region of some, but not in most erythroblasts (Figure 6Fiii), although the cytoplasmic F-actin spots were still present (Figure 6Fiv-vii). In some cells, a prominent F-actin cap also appeared on the Ter119-negative membrane overlying one of the protruding nuclear lobes, which was not observed in wild-type cells (Figure 6Fv-vi). Thus, while abnormal Tmod3−/− erythroblasts with multilobular nuclei appeared to initiate enucleation, their F-actin organization was aberrant, likely impairing their ability to complete enulceation.
Erythroblast-macrophage island formation is defective in Tmod3−/− fetal livers
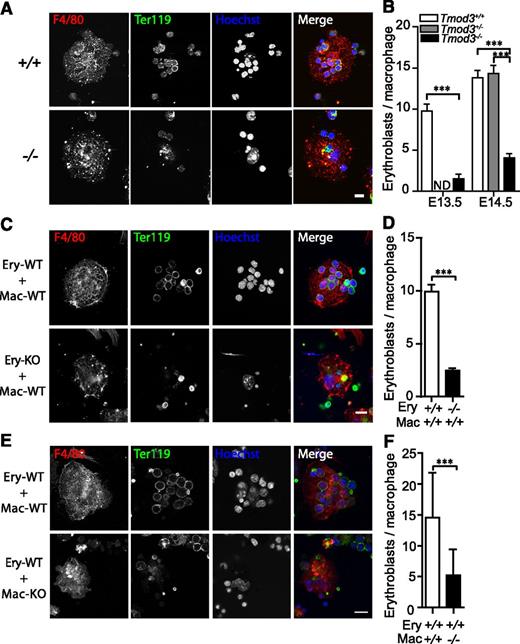
Fetal liver erythroblasts differentiate in association with central macrophages, forming islands, which play an important role in erythroblast enucleation.16,17,36 Tmod3 message and protein are expressed in both macrophages and erythroblasts, but macrophage morphologies and numbers appeared relatively normal in Tmod3−/− fetal liver sections (supplemental Figure 6A-B). To investigate a potential role for Tmod3 in island formation, native erythroblastic islands were isolated from fetal livers and the numbers of erythroblasts bound to each macrophage were counted.22 This showed that about three- to sixfold fewer erythroblasts were associated with each macrophage in native islands isolated from Tmod3−/− fetal livers (Figure 7A-B). In reconstitution experiments, Tmod3−/− erythroblasts were considerably less effective than Tmod3+/+ erythroblasts in forming islands with wild-type macrophages (Figure 7C-D). Conversely, Tmod3−/− macrophages were also less effective than Tmod3+/+ macrophages at supporting the attachment of wild-type erythroblasts (Figure 7E-F). Therefore, Tmod3 function is required in both erythroblasts and macrophages to form islands in the fetal liver.
Tmod3 is required in both erythroblasts and macrophages for erythroblast-macrophage island formation in the fetal liver. (A) Representative images of native islands isolated from fetal livers at E14.5. Cells were stained with F4/80-Alexa647 (red) for macrophages, Ter119-Alexa488 (green) for erythroblasts, and Hoechst (blue) for nuclei. (B) Numbers of erythroblasts bound per macrophage at E13.5 and E14.5. Islands were prepared from 3 independent litters and 30 to 90 islands were counted for each genotype. (C) Representative images of reconstituted islands from Tmod3+/+ or Tmod3−/− erythroblasts (Ery) incubated with Tmod3−/− macrophages (Mac). (D) Numbers of erythroblasts bound per macrophage. For each cell combination, 3 independent litters with a total of 80 to 100 islands were counted (9.9 ± 0.7 Tmod3+/+ erythroblasts, but only 2.5 ± 0.2 Tmod3−/− erythroblasts bound to each wild-type macrophage). (E) Representative images of reconstituted islands from Tmod3+/+ erythroblasts (Ery) incubated with Tmod3+/+ or Tmod3−/− macrophages (Mac). (F) Numbers of erythroblasts bound per macrophage. For each cell combination, 3 independent litters with a total of 70 to 80 islands were counted; 14.6 ± 0.9 wild-type erythroblasts were attached to each Tmod3+/+ macrophage, while only 5.2 ± 0.5 were attached to each Tmod3−/− macrophage. All values are means ± SD. ***P < .001. Panels A, C, and E were acquired using a Bio-Rad Radiance 2100 confocal microscope with a Zeiss 63× oil immersion objective (N.A. 1.4). Scale bar, 10 µm. ND, not determined.
Tmod3 is required in both erythroblasts and macrophages for erythroblast-macrophage island formation in the fetal liver. (A) Representative images of native islands isolated from fetal livers at E14.5. Cells were stained with F4/80-Alexa647 (red) for macrophages, Ter119-Alexa488 (green) for erythroblasts, and Hoechst (blue) for nuclei. (B) Numbers of erythroblasts bound per macrophage at E13.5 and E14.5. Islands were prepared from 3 independent litters and 30 to 90 islands were counted for each genotype. (C) Representative images of reconstituted islands from Tmod3+/+ or Tmod3−/− erythroblasts (Ery) incubated with Tmod3−/− macrophages (Mac). (D) Numbers of erythroblasts bound per macrophage. For each cell combination, 3 independent litters with a total of 80 to 100 islands were counted (9.9 ± 0.7 Tmod3+/+ erythroblasts, but only 2.5 ± 0.2 Tmod3−/− erythroblasts bound to each wild-type macrophage). (E) Representative images of reconstituted islands from Tmod3+/+ erythroblasts (Ery) incubated with Tmod3+/+ or Tmod3−/− macrophages (Mac). (F) Numbers of erythroblasts bound per macrophage. For each cell combination, 3 independent litters with a total of 70 to 80 islands were counted; 14.6 ± 0.9 wild-type erythroblasts were attached to each Tmod3+/+ macrophage, while only 5.2 ± 0.5 were attached to each Tmod3−/− macrophage. All values are means ± SD. ***P < .001. Panels A, C, and E were acquired using a Bio-Rad Radiance 2100 confocal microscope with a Zeiss 63× oil immersion objective (N.A. 1.4). Scale bar, 10 µm. ND, not determined.
Based on microarray data, the inability to form normal islands is not due to altered expression of erythroblast-macrophage adhesion molecules in Tmod3−/− fetal livers (supplemental Figure 6C). However, phalloidin staining and confocal microscopy indicate abnormal F-actin organization in Tmod3−/− macrophages (supplemental Figure 6D). Tmod3-mediated actin remodeling in macrophages and erythroblasts may be required for macrophage-erythroblast adhesion, and hence, island formation. The inability to form normal islands may contribute to the abnormal morphologies and reduced enucleation frequency of erythroblasts in the absence of Tmod3.
Discussion
In this study, we showed that mice missing the actin pointed-end-capping protein, Tmod3, are embryonic lethal starting at E14.5, with anemia caused by defective fetal liver definitive erythropoiesis. Tmod3 is required for multiple aspects of erythroid differentiation, namely, erythroid progenitor function at the BFU-E and CFU-E stages, erythroblast survival, cell-cycle exit, enucleation of late erythroblasts, and erythroblastic island formation with macrophages. Tmod3 function is required in both erythroblasts and macrophages, with the absence of Tmod3 leading to defects in F-actin contractile ring formation in enucleating erythroblasts, and to abnormal F-actin organization in macrophages. Thus, Tmod3 appears to promote erythroblast enucleation via cell-cycle exit and actin cytoskeletal remodeling, and by enabling erythroblast-macrophage adhesion to form islands, it promotes erythroblast cell-cycle progression and enucleation.16,40
As a member of the Tmod family, Tmod3 binds tropomyosins and caps the pointed ends of actin filaments, similar to Tmod1 in mature RBCs.26,41 However, Tmod1 and Tmod3 display differences in recognition of TM isoforms, and in TM-regulated capping of F-actin pointed ends.26,42 In addition, Tmod3 has a greater ability to form a 1:1 complex with actin monomers compared with Tmod1,39,43 and can also nucleate actin filament assembly using both its monomer-binding and pointed-end-capping sites.43 Thus, in contrast to Tmod1 stabilizing F-actin through binding pointed ends, Tmod3 may regulate actin assembly and turnover in dynamic cell contexts, such as erythroid proliferation and differentiation.26,41,44 This is supported by the differential expression patterns of Tmods, with Tmod3 decreasing and Tmod1 increasing in erythroid terminal differentiation (Figure 1). Loss of Tmod3 function in erythroid differentiation is not compensated for by other Tmods, because Tmod1 protein levels are unchanged, and Tmod2 and Tmod4 protein levels are undetectable in Tmod3−/− fetal liver (Figure 2D-F).
Actin filament dynamics play an important role in cell morphology and cell-cycle machinery.45,46 In the absence of Tmod3, there were more erythroblasts in S-phase and fewer in G0/G1 (Figure 6A-C). Moreover, ∼75% of late erythroblasts undergoing enucleation were Ki67+ (Figure 6D-E and supplemental Figure 5C), implying an inability to exit the cell cycle without Tmod3. This suggests that the G1/S checkpoint might be impaired in Tmod3−/− erythroblasts, possibly leading to apoptosis in proliferating erythroblasts (Figure 5B). However, apoptosis is apparently less significant for Tmod3−/− late erythroblasts, as observed by immunostaining and confocal microscopy for apoptotic markers (Figure 5C-E) and by cell-cycle analysis (Figure 6C). Thus, Tmod3−/− late stage erythroblasts may be able to initiate enucleation despite remaining in S-phase (Figure 6D-E).
Another possibility for the increased numbers of Tmod3−/− erythroblasts in S-phase is that S-phase is extended in the absence of Tmod3. Microarray analysis shows that several DNA replication-related genes (such as Rfc4 and Orc2l) are downregulated in Tmod3−/− fetal livers. DNA in Tmod3−/− erythroblasts might be inefficiently replicated, leading to prolongation of S-phase. A previous study of cell-cycle checkpoint kinase function showed that nuclear expression of Chk induced a prolonged S-phase, characterized morphologically by multilobular nuclei,47 similar to Tmod3−/− late erythroblasts, which have Ki67+ multilobular nuclei (supplemental Figure 5C). Tmod3 would be the first actin regulatory protein reported to participate in the process of cell-cycle exit during erythropoiesis, and may coordinate the cell cycle with erythroid terminal differentiation.
Tmod3 also functions to promote erythroblast enucleation.11,13 Our confocal microscopy studies show that absence of Tmod3 leads to aberrant F-actin ring formation with abnormal membrane sites of F-actin assembly in Ter119-positive, late-stage erythroblasts (Figure 6F). Tmod3-mediated actin dynamics may assist in erythroblast polarization and nuclear displacement, along with assembly of the F-actin ring required for nuclear expulsion. On the other hand, membrane domain sorting appears to be less affected, based on the presence of a Ter119-negative membrane overlying the protruding nuclear lobe(s) in Tmod3−/− erythroblasts attempting to enucleate (Figure 6F). A potential role for Tmod3-mediated actin dynamics in cell polarization is supported by our previous knockdown and overexpression experiments in human microvascular endothelial cells.41
The central macrophages of erythroblastic islands in the mouse fetal liver play an important role in promoting erythroblast survival, terminal differentiation, and cell-cycle progression, as well as enucleation of late-stage erythroblasts.16,21,22 In Tmod3−/− embryos, fetal liver erythroblastic island formation is impaired due to reduced adhesion of erythroblasts to macrophages, with Tmod3 required in both cell types (Figure 7). Moreover, Tmod3−/− macrophages also displayed abnormal F-actin organization (supplemental Figure 6D). Several adhesion proteins are critical for erythroblastic island formation, including erythroblast macrophage protein in erythroblasts and macrophages, and α4β1 integrin/vascular cell adhesion molecule-1 or intercellular adhesion molecule-4/αv receptors in erythroblasts and macrophages, respectively.16,17,20 While disruption of Tmod3 did not affect transcript levels of these receptors in the fetal liver (supplemental Figure 6C), their localizations or interactions could be affected by disturbances in F-actin assembly in the absence of Tmod3, as F-actin assembly is well-known to regulate integrin receptor clustering and adhesion.48,49 Evidence that F-actin stability in macrophages is important for island formation comes from previous studies of palladin, an F-actin-binding and stabilizing protein that functions in macrophages to promote island formation in the fetal liver.25 Defective adhesive interactions in erythroblastic islands in the Tmod3−/− fetal liver could partly explain impaired terminal erythroid differentiation with impaired survival, cell-cycle progression, and reduced enucleation of late erythroblasts (Figures 5 and 6).
In summary, our studies show that Tmod3 affects mouse fetal liver erythropoiesis at multiple points: numbers of functional erythroid progenitors, promotion of erythroblast adhesion to macrophages, erythroblast survival, coordination of cell cycle with erythroblast enucleation, and regulation of F-actin assembly in both erythroblasts and macrophages. To determine if Tmod3 is required for erythropoiesis in adult mice, future studies using conditional knockout mice in which Tmod3 is selectively deleted in erythroblasts or macrophages, are needed.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Aaron Sy for genotyping, Heather Paik for osmotic fragility, and Xin Du at UCSD for CBCs.
This work was supported by grants from the National Institutes of Health (R01-HL083464) (V.M.F.), (DK26263 and DK32094) (X.A), and by the Leukemia and Lymphoma Society and Giving Tree Foundation (A.W).
Authorship
Contribution: Z.S. analyzed mRNA and protein expression, erythroid colonies, cell cycle, erythroblast-macrophage islands, and wrote the manuscript; R.B.N. performed embryo analysis and erythroblast confocal microscopy; A.B. performed qRT-PCR and flow cytometry; N.E.K. conducted cell counts and blood smears; Z.S., R.B.N., and A.B. also analyzed data, and created figures and tables; H.L. and A.W. provided cells and RNA from CD34+ cultures; J.L. and X.A. analyzed Tmod expression in FACS purified erythroblasts; and V.M.F. designed experiments, interpreted the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Velia M. Fowler, Department of Cell and Molecular Biology, CB163, The Scripps Research Institute, 10550 N. Torrey Pines Rd, La Jolla, CA 92037; e-mail: velia@scripps.edu.
References
Author notes
Z.S., R.B.N., and A.B. contributed equally to this study.

![Figure 1. Tmod1 expression is upregulated while Tmod3 is downregulated during erythroid differentiation. (A) mRNA levels of Tmods during in vitro erythroid differentiation from human CD34+ cells, determined by qRT-PCR on days 7, 10, and 14, normalized to ACTB mRNA. (B) Western blots of Tmod1 and Tmod3 proteins during in vitro erythroid differentiation of CD34+ cells. Cell pellets were collected on days 3, 6, 8, 10, 13, and 16 after initiation of differentiation, solubilized, and analyzed by western blotting (4.1R was used as positive control for erythroid differentiation, and total actin [C4 antibody] and glyceraldehyde-3-phosphate dehydrogenase proteins were used as loading controls). (C-D) qRT-PCR analysis of Tmod1 and Tmod3 mRNAs in highly pure erythroblasts at distinct developmental stages isolated by FACS from (C) in vitro CD34+cell-erythroid differentiation cultures, or (D) mouse bone marrow, normalized to ACTB mRNA. Stages were: proerythroblasts (Pro-E), early basophilic erythroblasts (early Baso), late basophilic erythroblasts (late Baso), basophilic erythroblasts (Baso-E), polychromatic erythroblasts (Poly-E), and orthochromatic erythroblasts (Ortho-E).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/5/10.1182_blood-2013-03-492710/4/m_758f1.jpeg?Expires=1769081638&Signature=C-av7bqO805HRlwQ17sJaw0-R~IEc-KwTX2M21wGZQhTIMT7ccViO-CkxaaCO7Kg21pBU0X2IRooW~XQVSHSlQ5HFi4loHvR5CQXLAUo0r5BrB3h1hv8V48SpiA2WbLTIb0jME8P2KC8jBU7OUvfAQRQd0MkhYH1AjIox0kUm3b-q3BhgFa~jpOhWtSNS1R-~dUEI-bPRldqO~a3a--UVdo~-TAhxnDa6RkIWCqpdlSSp90r~4Af41YL4ip4LyUNOHmn05v51-YhqKuSKCotOluFUi5LX1RS3oyxdv-OROS8i4ov-p07TpT9zmC8vEpIXryhpNzusQASqL4iCW94IQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)