Key Points
Acquired pathogenic mutations in SAMHD1 are found in up to 11% of relapsed/refractory patients with CLL.
SAMHD1 is mobilized to sites of DNA damage.
SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase and a nuclease that restricts HIV-1 in noncycling cells. Germ-line mutations in SAMHD1 have been described in patients with Aicardi-Goutières syndrome (AGS), a congenital autoimmune disease. In a previous longitudinal whole genome sequencing study of chronic lymphocytic leukemia (CLL), we revealed a SAMHD1 mutation as a potential founding event. Here, we describe an AGS patient carrying a pathogenic germ-line SAMHD1 mutation who developed CLL at 24 years of age. Using clinical trial samples, we show that acquired SAMHD1 mutations are associated with high variant allele frequency and reduced SAMHD1 expression and occur in 11% of relapsed/refractory CLL patients. We provide evidence that SAMHD1 regulates cell proliferation and survival and engages in specific protein interactions in response to DNA damage. We propose that SAMHD1 may have a function in DNA repair and that the presence of SAMHD1 mutations in CLL promotes leukemia development.
Introduction
Chronic lymphocytic leukemia (CLL) is a disease of the elderly, demonstrating both clinical and biological heterogeneity. Complications include the development of autoimmunity and secondary immunodeficiency. Recurrent copy number aberrations of prognostic significance in CLL have been identified by fluorescence in situ hybridization,1 including deletions of 17p13.1 involving TP53 and deletions of 11q22.3 involving ATM. Recent data from whole genome and exome sequencing (WGS and WES, respectively) have confirmed the heterogeneous nature of CLL, with all recurrent mutations described occurring at low frequency.2,,-5
Previously, we used WGS to perform longitudinal studies of CLL patients undergoing treatment and described likely pathogenic variants in candidate founder genes that might initiate and/or maintain leukemia.6 These mutations were somatic and defined by their presence at all 5 time points in all leukemia cells assayed.6 Of particular interest was an acquired uniparental isodisomy event with a homozygous nonsynonymous mutation in SAMHD1 (supplemental Figure 1 on the Blood Web site), a gene involved in the regulation of innate immune response and autoimmune disease.
Germ-line mutations in SAMHD1 have been described in 17% of patients diagnosed with a congenital autoimmune disease called Aicardi-Goutieres syndrome (AGS).7,8 AGS is an autosomal recessive disorder that is characterized by an autoimmune encephalopathy and chronic activation of the immune system. It occurs mainly in patients of consanguineous background. Most patients with AGS succumb to the disease in early childhood.
More recently, SAMHD1 has been identified as an HIV-1 restriction factor operating in nondividing cells including dendritic cells, macrophages, and resting CD4+ T cells.9,,-12 SAMHD1 is a deoxyribonucleotide triphosphate (dNTP) triphosphohydrolase and a nuclease13,14 that regulates the intracellular dNTP pool.15,16 It is expressed widely across most tissue types including in the hemopoietic system and normal B cells.17,18 In the case of HIV, SAMHD1 is responsible for maintaining the cellular dNTP pool at a level that is inadequate for replication of HIV-1 and other viruses,19,-21 thereby preventing viral propagation.
Importantly, there is increasing evidence that an imbalance in the cellular dNTP pool is also associated with DNA replication stress and an alteration in replication fork velocity. Perturbations of the replication machinery in turn have been shown to lead to increased mutagenesis, genomic instability, and cancer development.22,23
Taken together, (1) the potential role of a SAMHD1 mutation as a founder event in CLL, (2) the established function of SAMHD1 in maintenance of the intracellular dNTP pools, and (3) the involvement of SAMHD1 in the innate immune response and AGS encouraged us to further explore the role of SAMHD1 in CLL pathogenesis.
To this, we present a detailed molecular characterization of clinical trial samples from CLL patients in the United Kingdom together with functional experiments interrogating the role of SAMHD1 in the presence of DNA damage.
Materials and methods
Samples
Informed consent from CLL patients was obtained in line with the Declaration of Helsinki. Ethics approval for this project is covered under the Haematology Collection Protocol, HTA License Number 12217, Oxfordshire C REC: 09/H0606/5. We initially chose CLL patients from our institution (Oxford University Hospitals National Health Service [NHS] Trust) to evaluate whether SAMHD1 mutations were recurrent in CLL (cohort 1 = 100 samples). Overall, the patient cohort was that of a tertiary referral center with a high proportion of unmutated IGVH and treatment refractory cases. Chemotherapy refractoriness was defined by failure to respond to a purine analog or relapsing within 6 months of treatment.
To evaluate the frequency of SAMHD1 mutations in a pretreatment cohort, 200 samples from 2 United Kingdom (UK) National Cancer Research Network (NCRN) clinical trials were sequenced (cohort 2: Does the ADdition of Mitoxantrone Improve REsponse (AdMIRe) trial, n = 108; Attenuated dose Rituximab with ChemoTherapy In CLL (ARCTIC) trial, n = 92). A further 63 samples from 2 UK NCRN relapsed and refractory trials (cohort 3: CLL 201, n = 37; CLL 202, n = 26) were also evaluated for SAMHD1 mutations. All clinical trial samples were sequenced using our in-house next-generation sequencing (NGS) targeted panel (supplemental Table 1)
Peripheral blood mononuclear cells (PBMCs) were isolated from CLL blood samples by ficoll gradient centrifugation. DNA was extracted from peripheral blood CLL cells using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
Samples from cohorts 1, 2, and 3 were tested using genome-wide single nucleotide polymorphism (SNP) arrays and NGS, as well as Sanger sequencing. The B-allele frequencies (BAFs) from the array data and the variant allele frequencies (VAFs) from the NGS data showed that the contribution of CLL genomes was ≥80% in all samples.
Sanger sequencing
We designed primers to amplify the coding exons of SAMHD1 (supplemental Table 2). Each primer had an M13 tail: forward, 5′-GTAAAACGACGGCCAGT-3′; reverse, 5′-CAGGAAACAGCTATGAC-3′. Purified amplicons were sequenced from both strands using BigDye terminator chemistry and an ABI 3130 DNA sequencer. Mutation Surveyor DNA variant analysis software was used to compare sequences with germ-line reference sequence.
Genome-wide SNP platform hybridizations and analysis
Hybridization to Illumina SNP chips (HumanOmni1-Quad and HumanOmniS-8 platforms) was performed according to manufacturer’s protocols found on registration at http://www.illumina.com/products. The data were processed using GenomeStudioV2009.2 (Illumina, San Diego, CA) and analyzed using Nexus 6.1 Discovery Edition (BioDiscovery, El Segundo, CA). We excluded from further analyses all losses and gains that had been noted previously in the Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home) and all copy neutral loss of heterozygosity (cnLOH) <2 Mb.
To determine the genomic complexity of a sample, we performed 2 calculations. (1) The sum of the length of all losses and gains (>20 kb) and cnLOH (>2 Mb). A total length of >100 Mb was used to define genomic complexity. (2) The sum of the number of losses and gains (>20 kb) and cnLOH (>2 Mb). A total of ≥4 was used as the second criterion toward defining genomic complexity. A sample was defined as having genomic complexity only if both criteria were fulfilled.
We defined mosaic cnLOH as the occurrence of a region of allelic imbalance in the absence of a copy number change. Allelic imbalance was detected using the BAF data and defined as present when the heterozygous (AB) allele frequency deviated from the expected 0.5 value but did not reach full AA or BB homozygosity (at 0 and 1 BAF, respectively). We were guided by Nexus Discovery Edition v6.1 calls and reported only changes that were visible on manual inspection of the data plots.
Targeted NGS
A TruSeq Custom Amplicon panel (TSCA; Illumina Cambridge Ltd., Saffron Walden, UK) was designed, targeting mutational hotspots in 11 genes involved in the pathogenesis of CLL (supplemental Table 2). Dual-barcoded TSCA libraries were created from 250 ng of DNA, before undergoing 2 × 150-bp paired-end sequencing on the MiSeq platform (Illumina Cambridge Ltd.). Initial alignment and variant calling analysis was performed with the BaseSpace online analysis tool.24 The sequence data were also analyzed using the Stampy25 and Platypus26 tools to screen for large insertions/deletions. High confidence functional sequence variants were identified by removing any that failed the BaseSpace filter, had a quality score <60, had no predicted functional consequence (intergenic, synonymous), or were present in dbSNP (build 135). All variants with dbSNP identifiers were checked in dbSNP for clinical association.
Cell purification
PBMCs were stained and sorted by fluorescence-activated cell sorter Aria (BD-Pharmingen) as described previously.27 Cells were stained with anti-CD19 phycoerythrin (PE) (BD Pharmingen) and anti-CD5 allophycocyanin (APC) (BD Pharmingen). Postsort purity was >95%.
WES
The tumor sample was purified as described above with the CD19+, CD5+ fraction selected for WES. The sample was quantified using the High Sensitivity Qubit system (Invitrogen), and sample integrity was assessed using 1% E-Gel EX (Invitrogen). Library preparation was performed using the Illumina TruSeq DNA HT Sample Preparation kit and enriched for the exome using the Illumina TruSeq Exome Enrichment kit. Sequencing was performed using a HiSeq2000 as 100-bp paired end reads.
The sequence reads were mapped to the human genome assembly hg19/NCBI37 using Stampy,25 and variants were detected using Platypus.26 High-quality reads scoring 200, with sequence variations found within coding exons and a total coverage of ≥50 reads, were analyzed further. The database for normal variants (dbSNP)28 and the 1000 Genomes database29 were used to exclude common variants. We searched for any published CLL-associated mutations2,4,-6,30,31 present in the leukemia cells and excluded their presence in the germ line by Sanger sequencing.
RNA extraction and reverse transcription
Total RNA was isolated from sorted cell populations using RNAeasy kits according to the manufacturer’s protocol (Qiagen, Hilden, Germany) or purified with Trizol reagent (Invitrogen). Complementary DNA was prepared from 1 to 12 µL total RNA using SuperScript VILO kits (Invitrogen, Carlsbad, CA) or SuperScript II enzyme (Invitrogen) and oligo-dT primers according to the manufacturer’s protocol.
RNA isoforms
Reverse transcription (RT) products were used for the polymerase chain reaction (PCR) reaction using Phusion HSII enzyme (Finnzymes) and the following SAMHD1-specific primers: CGGCGCCGAGGTTCTTGACT (forward) and TTTGTAAACAACTGACTACAGACA (reverse). The PCR products were resolved on a 1% agarose gel containing 1× SYBR Safe DNA gel stain (Life Technologies).
Quantitative real-time RT-PCR of purified cells
To generate Figure 4A, absolute quantification of cDNA was performed in duplicate using real-time PCR and a transcript quantification TaqMan assay kit for SAMHD1 (Hs00210019_m1; Applied Biosystems, Foster City, CA). Amplifications were performed using Qiagen Rotor-Gene 6000. Following normalization using the control gene, c-abl, relative quantification was calculated using the ΔCt method. Quantification of SAMHD1 mRNA shown in Figure 4B was performed in triplicate using QuantiTect SYBR Green (Qiagen) according to manufacturer’s instructions. Further details in are in supplemental Methods.
cDNA sequencing
Primers were designed to amplify the complementary DNA of particular exons of SAMHD1 (supplemental Table 3). Purified amplicons were sequenced as detailed in the Sanger sequencing method to confirm the presence of the mutations found in the corresponding DNA samples.
Cell lines and expression constructs
Adherent and suspension cells were cultured in Dulbecco’s modified Eagle medium or RPMI-1640 respectively, supplemented with 10% fetal calf serum, ultraglutamine, and antibiotics. HeLa cells stably expressing Flag-HA–tagged SAMHD1 have been described previously.9 HeLa cells stably expressing inducible SAMHD1 wild-type (WT) and mutants were generated by transducing HeLa cells with lentiviral pTRIPZ vector (Thermo Scientific, Waltham, MA), in which the SAMHD1 open reading frame (ORF) was inserted. Transduced cells were selected with 2 µg/mL puromycin. SAMHD1 expression was induced with 2 µg/mL doxycycline (Clontech, Saint-Germain-en-Laye, France).
Plasmids
Cell extracts preparation and western blot analysis
For western blotting, cells were lysed for 30 minutes in a buffer containing 0.5% Triton X-100, 150 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, 10 mM β-mercaptoethanol, and 0.5 mM phenylmethylsulfonyl fluoride. Equal amount of proteins were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were transferred to nitrocellulose membranes (GE Healthcare, Little Chalfont, Buckinghamshire, UK). When specified, protein load was assessed by Ponceau S staining (Thermo Scientific). Membranes were blocked with 5% milk diluted in phosphate-buffered saline/0.1% Tween, incubated with primary antibody for 1 hour at room temperature, and incubated with horseradish peroxidase–conjugated secondary antibody for 1 hour at room temperature. Proteins were visualized with SuperSignal West pico or femto (Thermo Scientific).
Immunofluorescence
HeLa cells constitutively expressing Flag-HA–tagged SAMHD1 were seeded on cover glasses and treated with or without 25 nM of camptothecin (CPT) for 18 hours. Cells were then fixed with 2% paraformaldehyde and permeabilized using 0.1% Triton X-100. Immunodetection of 53BP1 and HA-tagged SAMHD1 was performed overnight at 4°C using specific antibodies. Images were acquired using the Zeiss confocal microscope system LSM780.
Antibodies
Anti-SAMHD1 antibodies used were purchased from Abcam (Cambridge, UK; ab96768 and ab67820) and Bethyl/Euromedex (Schiltigheim, France; A303-690A). Anti-phosphoSAMHD1Thr592 antibody has been described previously.33 Anti-CyclinA2 antibody was purchased from Cell Signaling, anti-DCAF1 antibody from Bethyl/Euromedex, anti-HA from Covance/Eurogentec, and anti-53BP1 antibody was from Millipore (Billerica, MA). Tubulin and actin antibodies were from Sigma-Aldrich Chemical (St Louis, MO). Mouse (NA931) and rabbit (NA934) secondary antibodies were purchased from GE Healthcare.
Drug treatment and cell proliferation
HeLa WT or expressing SAMHD1 were seeded in 96-well plates at a density of 1000 cells per well. Etoposide, camptothecin, mitomycin C, and methyl methanesulfonate (Sigma-Aldrich Chemical) were applied to the cells 6 hours after seeding in a total volume of 100 µL. Three days after treatment, the Cell Titer-Glo kit (Promega, Madison, WI) was used to measure cell survival according to the manufacturer’s instructions. Results are shown as percentage of survival compared with mock-treated cells.
For the cell proliferation experiment, the cell count was performed at the indicated time using MACSQuant Analyzer (Miltenyi Biotec, Bergisch Gladbach, Germany).
Cell fractionation and glycerol gradient
HeLa stably expressing SAMHD1 were seeded in 15-cm dishes and treated with 5 µM etoposide for 4 and 18 hours or mock treated. Cells were then harvested, washed with phosphate-buffered saline, and lysed successively in 5 volumes of CSK buffer (10 mM piperazine-N,N′-bis[2-ethanesulfonic acid] [PIPES], pH 7, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM β-glycerophosphate, 0.2 mM Na vanadate, and 5 mM Na fluoride) containing 0.5% and 0.1% of Triton to purify nuclear extracts. For density gradient sedimentation, 400 µg of nuclear extracts was loaded onto a glycerol gradient (15-35% in buffer containing 20 mM Tris, pH 7.5, 0.15 M KCl, 2.5 mM MgCl2, 0.05% NP40, 0.05% Tween, 1 mM dithiothreitol, and protease inhibitors; Roche) and centrifuged for 4 hours at 55 000 rpm. Two hundred–microliter fractions were collected from the top of the gradient.
Statistical methods
We wished to ascertain whether SAMHD1 mutations were associated with common CLL-described mutations (Figure 3B). We analyzed genomic data on 18 SAMHD1 mutated samples (Figure 3B) and 229 WT SAMHD1 samples (clinical trial samples with whole genome array and NGS data available). The sample proportions of described CLL mutations were calculated for the mutated and WT SAMHD1 patients, and the difference was tested for significance using the 2-sided pooled hypothesis z-test, which provides a P value. Unadjusted P values can be compared with an adjusted α level of 0.05/7 = 0.0071 using a conservative Bonferroni adjustment to reflect that 7 comparisons were made.
We compared the clinical outcome for the mutated and WT SAMHD1 patients on samples with clinical outcome available (AdMIRe Trial).34 The 2 populations were compared with respect to the complete remission (CR) and minimal residual disease (MRD), using for both comparisons the standard 2-sided pooled z-test for the difference in proportions.
For patients with both overall survival data and SAMHD1 mutation status available (CLL 201 and CLL 202 clinical trials, n = 65), a survival analysis was performed. Survival times for the populations of WT SAMHD1 and mutated SAMHD1 patients were compared by creating the corresponding Kaplan-Meier survival curves. The statistical difference between the 2 survival curves was quantified by performing the standard log-rank test.35
Results
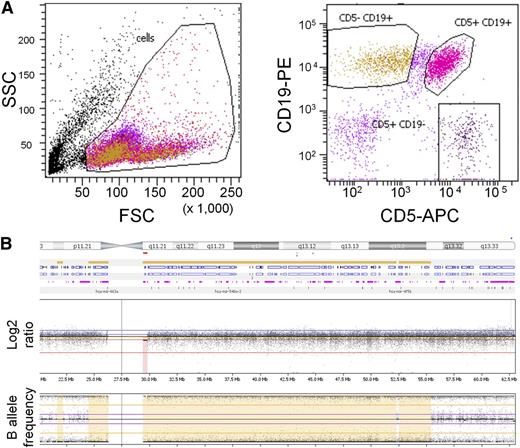
Initially, we reviewed 32 patients with congenital AGS and germ-line SAMHD1 mutations. One of these patients was a boy born to consanguineous parents who presented with features typical of AGS.7 This patient has a homozygous germ-line canonical splice-acceptor site (1609-1G>C) mutation in intron 14, leading to an alternative splice variant.7 Interestingly, at the age of 24 years, he presented with cervical and axillary lymphadenopathy and a lymphocytosis. Flow cytometry demonstrated the typical hallmark of CD19+CD5+ mature lymphocytes (Figure 1A), confirming CLL. High-resolution genome-wide array analysis of DNA from his CLL cells revealed a large region of homozygosity including the SAMHD1 locus on chromosome 20 (Figure 1B). This was not associated with a copy number change but was consistent with a germ-line inheritance pattern arising from consanguineous parents carrying shared haplotypes across this region. No other chromosomal aberrations associated with CLL and no novel aberrations were detected.
Analysis of an AGS patient. (A) Flow cytometry plot from whole peripheral blood of an AGS patient showing (left) forward-sideward scatter and (right) CD19 PE/CD5 APC scatter gated on lymphoid cells. (B) Whole genome array of chromosome 20 from leukemia cells of an AGS patient, visualized in Nexus (BioDiscovery). The log2 ratio indicates copy number changes, and the B-allele frequency plot indicates loss of heterozygosity.
Analysis of an AGS patient. (A) Flow cytometry plot from whole peripheral blood of an AGS patient showing (left) forward-sideward scatter and (right) CD19 PE/CD5 APC scatter gated on lymphoid cells. (B) Whole genome array of chromosome 20 from leukemia cells of an AGS patient, visualized in Nexus (BioDiscovery). The log2 ratio indicates copy number changes, and the B-allele frequency plot indicates loss of heterozygosity.
To establish whether acquired mutations were present, we tested DNA from the patient’s CLL cells using a targeted approach to sequence genes reported as mutated recurrently in CLL (supplemental Table 2). None of these genes were mutated in the patient’s sample. Next, we performed WES to identify other acquired mutations that might contribute to CLL pathogenesis, including recently described pathogenic mutations5 (Materials and methods). Again, no acquired mutations seen previously in CLL or other hematological malignancies were observed. Although these results do not rule out a contribution of other currently unknown genes in CLL pathogenesis, they support an involvement of SAMHD1.
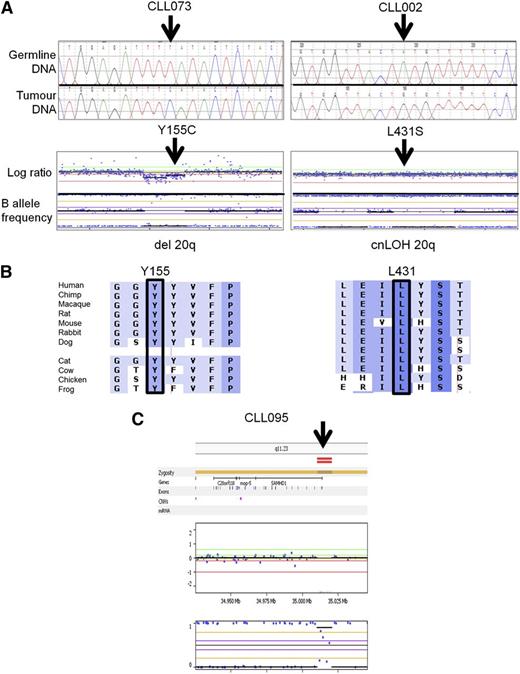
We therefore set out to establish whether acquired SAMHD1 mutations were recurrent in CLL patients. In our single institution cohort of 100 consecutive CLL patients,36 we identified 5 patients, carrying between them 6 mutations in SAMHD1 (Figure 2A; Table 1, cohort 1). Germ-line analysis using sorted CD5+CD19− cells could be performed for 5 of the mutations and confirmed that they were acquired (supplemental Table 5). Furthermore, 4 of the 5 patients carried a mutation associated with either a mono-allelic deletion or cnLOH as revealed by genome-wide array data (Figure 2A; Table 1) and reflected in the predominantly high VAF (Table 1). Mutations affected mainly conserved residues and were predicted to be deleterious for protein function by in silico prediction (Figure 2B; supplemental Figure 2; supplemental Table 5). One additional patient had a small bi-allelic deletion of the 5′ untranslated region of SAMHD1 (Figure 2C).
Representative examples of somatic SAMHD1 mutations. (A) Electropherogram of SAMHD1 mutations using paired germ-line and tumor DNA. (Upper) Representative results from 2 patients are depicted. (Lower) Whole genome arrays showing (left) copy neutral loss of heterozygosity (cnLOH) or (right) a heterozygous deletion over chromosome 20 involving the SAMHD1 locus. (B) Multiple sequence alignment of SAMHD1 around the mutated residue (arrow) of each patient. Degree of conservation is reflected by background color code (dark blue, highly conserved to white, not conserved). (C) Whole genome array showing a small homozygous deletion at the 5′ end of the SAMHD1 locus in patient CLL 095.
Representative examples of somatic SAMHD1 mutations. (A) Electropherogram of SAMHD1 mutations using paired germ-line and tumor DNA. (Upper) Representative results from 2 patients are depicted. (Lower) Whole genome arrays showing (left) copy neutral loss of heterozygosity (cnLOH) or (right) a heterozygous deletion over chromosome 20 involving the SAMHD1 locus. (B) Multiple sequence alignment of SAMHD1 around the mutated residue (arrow) of each patient. Degree of conservation is reflected by background color code (dark blue, highly conserved to white, not conserved). (C) Whole genome array showing a small homozygous deletion at the 5′ end of the SAMHD1 locus in patient CLL 095.
Summary of SAMHD1 mutations
| Patient number . | Cohort . | Sample ID . | Mutation position . | Mutation type . | Amino acid . | SAMHD1 locus (array) . | VAF (%) . |
|---|---|---|---|---|---|---|---|
| 1 | 1 | CLL077 | g.35 526 336 A>T | Missense | F545L | cnLOH | 99 |
| 2 | 1 | CLL 092 | g.35 580 045 T>A | Missense | M1K | cnLOH | 72 |
| 1 | CLL 092 | g.35 526 886 ->A | Frameshift insertion | C522X | cnLOH | 27 | |
| 3 | 1 | CLL073 | g.35 563 477 A>G | Missense | Y155C | Loss | 94 |
| 4 | 1 | CLL 002 | g.35 533 885 T>C | Missense | L431S | Loss | 92 |
| 5 | 1 | CLL 238 | g.35 532 562 C>A | Missense | D501Y | Normal | 40 |
| 6 | 2 | CLL 242 | g.35 540 906 G>A | Missense | R371H | Normal | 50 |
| 7 | 2 | CLL 069 | g.35 563 507 G>A | Missense | R145Q | cnLOH | 98 |
| 8 | 2 | CLL 228 | g.35 526 256 G>A | Nonsense | W572X | cnLOH | 99 |
| 9 | 2 | CLL354 | g. 35 532 585 A>C | Missense | L493R | Normal | 55 |
| 10 | 2 | CLL 007 | g. 35 559 171 T>C | Missense | H206R | Mosaic cnLOH | 18 |
| 2 | CLL 007 | g.35 559 186 A>T | Missense | I201N | Mosaic cnLOH | 50 | |
| 11 | 3 | CLL 2537 | g.35 540 955 G>A | Missense | E355K | Loss | 99 |
| 12 | 3 | CLL 2353 | g.35 563 508 C>T | Nonsense | R145X | Loss | 87 |
| 13 | 3 | CLL 2247 | g. 35 540 925 T>G | Missense | T365P | Mosaic cnLOH | 66 |
| 14 | 3 | CLL 2198 | g. 35 545 407 T>A | Missense | I300L | Normal | 36 |
| 3 | CLL 2198 | g. 35 563 469 G>A | Missense | P158S | Normal | 45 | |
| 3 | CLL 2198 | g. 35 580 001 C>A | Missense | D16Y | Normal | 48 | |
| 15 | 3 | CLL 2219 | g. 35 547 889 G>A | Missense | L244F | Normal | 39 |
| 3 | CLL 2219 | g. 35 545 437 G>A | Missense | R290C | Normal | 39 | |
| 16 | 3 | CLL 2250 | g. 35 533 826 G>A | Missense | R451C | Normal | 57 |
| 3 | CLL 2250 | g. 35 533 825 C>A | Missense | R451L | Normal | 33 | |
| 17 | 3 | CLL 2462 | g. 35 532 564 A>C | Missense | V500G | cnLOH | 99 |
| Patient number . | Cohort . | Sample ID . | Mutation position . | Mutation type . | Amino acid . | SAMHD1 locus (array) . | VAF (%) . |
|---|---|---|---|---|---|---|---|
| 1 | 1 | CLL077 | g.35 526 336 A>T | Missense | F545L | cnLOH | 99 |
| 2 | 1 | CLL 092 | g.35 580 045 T>A | Missense | M1K | cnLOH | 72 |
| 1 | CLL 092 | g.35 526 886 ->A | Frameshift insertion | C522X | cnLOH | 27 | |
| 3 | 1 | CLL073 | g.35 563 477 A>G | Missense | Y155C | Loss | 94 |
| 4 | 1 | CLL 002 | g.35 533 885 T>C | Missense | L431S | Loss | 92 |
| 5 | 1 | CLL 238 | g.35 532 562 C>A | Missense | D501Y | Normal | 40 |
| 6 | 2 | CLL 242 | g.35 540 906 G>A | Missense | R371H | Normal | 50 |
| 7 | 2 | CLL 069 | g.35 563 507 G>A | Missense | R145Q | cnLOH | 98 |
| 8 | 2 | CLL 228 | g.35 526 256 G>A | Nonsense | W572X | cnLOH | 99 |
| 9 | 2 | CLL354 | g. 35 532 585 A>C | Missense | L493R | Normal | 55 |
| 10 | 2 | CLL 007 | g. 35 559 171 T>C | Missense | H206R | Mosaic cnLOH | 18 |
| 2 | CLL 007 | g.35 559 186 A>T | Missense | I201N | Mosaic cnLOH | 50 | |
| 11 | 3 | CLL 2537 | g.35 540 955 G>A | Missense | E355K | Loss | 99 |
| 12 | 3 | CLL 2353 | g.35 563 508 C>T | Nonsense | R145X | Loss | 87 |
| 13 | 3 | CLL 2247 | g. 35 540 925 T>G | Missense | T365P | Mosaic cnLOH | 66 |
| 14 | 3 | CLL 2198 | g. 35 545 407 T>A | Missense | I300L | Normal | 36 |
| 3 | CLL 2198 | g. 35 563 469 G>A | Missense | P158S | Normal | 45 | |
| 3 | CLL 2198 | g. 35 580 001 C>A | Missense | D16Y | Normal | 48 | |
| 15 | 3 | CLL 2219 | g. 35 547 889 G>A | Missense | L244F | Normal | 39 |
| 3 | CLL 2219 | g. 35 545 437 G>A | Missense | R290C | Normal | 39 | |
| 16 | 3 | CLL 2250 | g. 35 533 826 G>A | Missense | R451C | Normal | 57 |
| 3 | CLL 2250 | g. 35 533 825 C>A | Missense | R451L | Normal | 33 | |
| 17 | 3 | CLL 2462 | g. 35 532 564 A>C | Missense | V500G | cnLOH | 99 |
To investigate the association between SAMHD1 mutations and clinical outcome, we analyzed a further 263 UK clinical trials samples by Sanger sequencing and NGS. These patients were from 2 pretreatment trials (cohort 2, n = 200) and 2 relapsed/refractory trials (cohort 3, n = 63; Materials and methods). We found a mutation frequency of 3% in the pretreatment group and of 11% in the relapsed and refractory group (Table 1). Interestingly, the array analyses of 3 patients showed mosaicism, confirming the acquired nature of the copy number change involving the SAMHD1 locus.
Mutations were distributed across all exons, similar to those identified in AGS (Figure 3A). Three mutations (R145X, R145Q, and I201N) were common to both AGS and CLL patients. When assessed using genome-wide high-resolution arrays, SAMHD1 mutations showed a clear association with chromosome 20 abnormalities involving the SAMHD1 locus (seen in 12 of 18 patients; Table 1; Figure 3B). SAMHD1 mutations were not found to associate with any other described CLL mutations (Figure 3B; supplemental Table 4). As expected for a group of CLL patients requiring treatment, both treatment naïve and relapsed/refractory patients with SAMHD1 mutations showed unmutated immunoglobulin (IgVH) rearrangements in the majority of cases. Only 3 of 11 relapsed/refractory patients had del11q compared with 3 of 7 in the pretreatment group (Figure 3B). Together, these data do not support the notion that the enrichment for SAMHD1 mutations in advanced CLL could be due to a higher prevalence of these confounding poor risk factors in this group.
Schematic representation of SAMHD1 mutations and relationship to other genetic lesions. (A) Schematic representation of SAMHD1 protein. (Upper) Mutations found in current study. (Lower) Known mutations in AGS patients. Mutations common to both diseases are shown in red. (B) Associations between SAMHD1 mutations and other genetic aberrations and clinical outcome. cnLOH, copy neutral loss of heterozygosity; CLL239*, AGS patient; Response, clinical response according to the iwCLL guidelines46 ; PR, partial response; CR, complete response; SD, stable disease; MRD, minimal residual disease; N/A, not available.
Schematic representation of SAMHD1 mutations and relationship to other genetic lesions. (A) Schematic representation of SAMHD1 protein. (Upper) Mutations found in current study. (Lower) Known mutations in AGS patients. Mutations common to both diseases are shown in red. (B) Associations between SAMHD1 mutations and other genetic aberrations and clinical outcome. cnLOH, copy neutral loss of heterozygosity; CLL239*, AGS patient; Response, clinical response according to the iwCLL guidelines46 ; PR, partial response; CR, complete response; SD, stable disease; MRD, minimal residual disease; N/A, not available.
To evaluate the impact of SAMHD1 mutations on clinical outcome, we reviewed primary response data on the clinical trial samples (Figure 3B). For the first-line patients recruited into AdMIRe, the end points of CR and bone marrow MRD were analyzed. The CR rate for the AdMIRe cohort, excluding the SAMHD1-mutated patients, was 137 of 210 patients (65.2%; 95% exact confidence interval [CI]: 58.4-71.7%).34 The proportion of SAMHD1-mutated patients with CR was 0 of 5; this difference in sample proportions was statistically significant (95% CI for difference in proportions: 48.6-81.9%, P = .014).
The MRD negative rate across the AdMIRe cohort, excluding the SAMHD1-mutated patients, was 108 of 188 patients (57.4%; 95% exact CI: 50.0-64.6%) The proportion of SAMHD1-mutated patients who were MRD negative was 0 of 5; this difference in sample proportions was statistically significant (95% CI for difference in proportions: 40.1-74.8%, P = .035).
Patients with SAMHD1 mutations treated in CLL 201 and CLL 202 clinical trials showed outcomes as expected for this cohort of relapsed/refractory patients (Figure 3B). The end point of overall survival was analyzed. The standard log-rank test for comparing the Kaplan-Meier survival curves (supplemental Figure 3) for WT SAMHD1 and mutated SAMHD1 samples gave P = .666, which suggests that the 2 curves are not significantly different. However, as there were only 6 samples with SAMHD1 mutations in our overall survival analysis, a larger cohort will be needed to draw more definite conclusions.
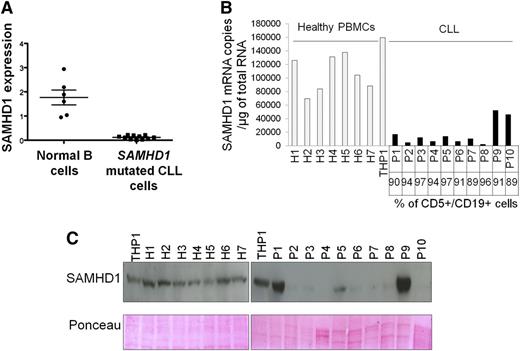
SAMHD1 is expressed in normal B cells18,37 (Figure 4A). To interrogate the impact of the SAMHD1 variants, we compared SAMHD1 mRNA expression in highly purified samples from 11 SAMHD1 mutated CLL patients with that of normal B cells. Using quantitative RT-PCR, we found statistically significant lower expression levels of mRNA in SAMHD1-mutated CLL cells (Figure 4A). Where possible, we sequenced cDNA extracted from SAMHD1-mutated CLL cells by Sanger sequencing using primers designed to cross intron-exon boundaries. This confirmed that, at least in these patients, SAMHD1 transcripts in CLL cells consisted exclusively of mutant SAMHD1 (supplemental Figure 4A). Western blots performed on paired samples also confirmed reduced protein expression compared with WT SAMHD1 CLL (supplemental Figure 4B).
Expression profiles of SAMHD1 in primary CLL cells. (A) cDNA pools from B cells (CD19+, CD5−) sorted from peripheral blood of healthy donors and SAMHD1-mutated CLL cells (CD19+, CD5+) were subjected to quantitative PCR analysis using ABL as a control gene. (B) Measurement of SAMHD1 mRNA by quantitative RT-PCR. The percentage of leukemia cells (CD5+/CD19+) is indicated. Statistical comparison of SAMHD1 mRNA levels between each CLL sample and the mean of the healthy PBMCs was performed. The black bars indicate P < .01. (C) Measurement of SAMHD1 protein levels by western blot in 10 CLL samples and 7 healthy PBMCs. Ponceau dye staining was used to control protein load.
Expression profiles of SAMHD1 in primary CLL cells. (A) cDNA pools from B cells (CD19+, CD5−) sorted from peripheral blood of healthy donors and SAMHD1-mutated CLL cells (CD19+, CD5+) were subjected to quantitative PCR analysis using ABL as a control gene. (B) Measurement of SAMHD1 mRNA by quantitative RT-PCR. The percentage of leukemia cells (CD5+/CD19+) is indicated. Statistical comparison of SAMHD1 mRNA levels between each CLL sample and the mean of the healthy PBMCs was performed. The black bars indicate P < .01. (C) Measurement of SAMHD1 protein levels by western blot in 10 CLL samples and 7 healthy PBMCs. Ponceau dye staining was used to control protein load.
Furthermore, we assessed SAMHD1 mRNA (Figure 4B) and protein levels (Figure 4C) in 10 untreated CD19+, CD5+ samples from consecutive CLL patients compared with healthy control PBMCs and observed a marked reduction of SAMHD1 mRNA and protein expression in 8 of 10 cases. One case (P9) showed significant mRNA and protein expression, and another case (P10) had no protein expression despite presence of mRNA. Together, these observations suggest that SAMHD1 mRNA and protein expression in CLL is heterogeneous and that in addition to mutations within the SAMHD1 coding region itself, other control mechanisms are involved in regulating its expression in CLL.
Next, we extended our analysis to other malignancies. SAMHD1 mutations have been identified in colorectal,38 lung, and urinary tract cancer.38 However, WGS analysis of 1006 different tumor cell lines showed that SAMHD1 mutations in solid tumors are rare40 (supplemental Table 6), and of only 16 cell lines with mutations, 5 were of hemopoietic/lymphoid and 11 of solid tumor origin. SAMHD1 is expressed in all normal human tissues17 (supplemental Figure 5). Assessment of SAMHD1 protein (supplemental Figure 6A) and mRNA (supplemental Figure 6B) expression in different tumor cell lines revealed low level expression in all cell lines tested with the exception of the human acute monocytic leukemia cell line, THP1, known to express normal levels of WT SAMHD1.9 Interestingly, alternative SAMHD1 transcripts (supplemental Figure 6C) were identified in all the cell lines tested except in THP1 cells. Additionally, primary breast cancer specimens from 34 individuals demonstrated reduced to absent protein expression of SAMHD1 in 50% of cases (supplemental Figure 6D).
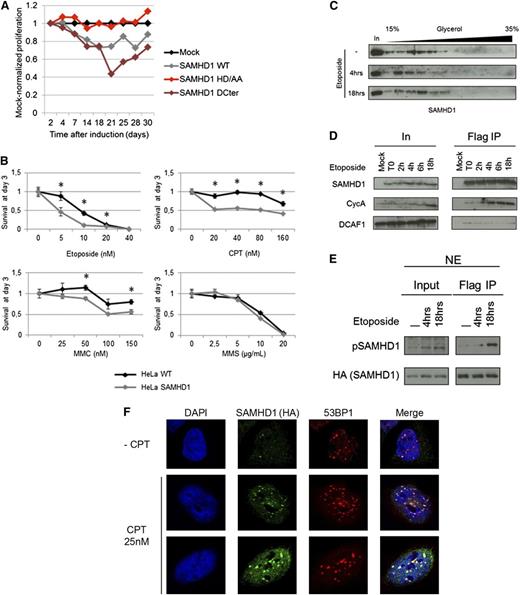
There is evidence that an imbalance in the dNTP pool produces a hypermutator phenotype and that decreased levels of dNTPs interfere with proper DNA replication and repair.22,23 Therefore, we investigated the potential role of SAMHD1 in cellular proliferation and DNA damage response. HeLa cells were transduced with lentiviral vector expressing, under the control of an inducible promoter, either WT SAMHD1, a dNTP triphosphohydrolase-inactive HD mutant,9,13 or a C-terminal deletion mutant that has been characterized as a constitutively active form33 (supplemental Figure 7). Expression of WT SAMHD1, but not HD mutant, resulted in slow proliferation compared with cells transduced with empty vector (Figure 5A). Interestingly, the antiproliferative activity of SAMHD1 was amplified when the constitutively active mutant (SAMHD1ΔCter) was expressed, suggesting a key role for SAMHD1 enzymatic activity. Next, HeLa cells expressing WT SAMHD1 were treated with 3 DNA damaging agents, camptothecin, mitomycin C, and etoposide (known to induce DNA double-strand breaks [DSBs]), and with a methylating agent, methyl methanesulfonate. The proportion of living cells was measured after 3 days and compared with baseline (Figure 5B). In this system, WT SAMHD1 increased cell death after treatment with DNA DSB agents. However, no effect on cell survival was seen with the methylating agent, suggesting that SAMHD1 is involved specifically in DNA DSB repair. To explore whether SAMHD1 acquires new function following DNA damage by engaging in specific protein interactions, we analyzed its distribution in nuclear extracts prepared from mock- or etoposide-treated HeLa SAMHD1 cells using a glycerol gradient sedimentation assay that allows size-dependent separation of protein complexes. We found that SAMHD1 resides in a high-molecular-weight complex (HMW) in untreated cells and converts to a low-molecular-weight complex after 4-hour treatment with etoposide (Figure 5C). Conversion from the HMW to the low-molecular weight complex is transient, because SAMHD1 was recovered in the HMW complex 18 hours after etoposide treatment.
SAMHD1 effects on cell proliferation and cell viability in response to DNA damage agents. (A) HeLa cells stably expressing an inducible empty vector (Mock) or a vector encoding for SAMHD1 wild-type (WT) SAMHD1 mutated in the HD domain (HD/AA) or truncated at amino acid 575 (ΔCter) were induced with doxycycline. Results are expressed as the proliferation ratio of SAMHD1-transduced cells divided by mock-transduced cell. A representative experiment out of 3 is shown. (B) HeLa and HeLa cells stably expressing SAMHD1 were treated with increasing concentrations of DNA damaging agents, and the percentage of living cells was measured 3 days after treatment. Each graph shows the mean of 3 independent experiments. *A t-test P < .05. (C) HeLa cells stably expressing SAMHD1 were mock treated or treated with 5 nM of etoposide for 4 or 18 hours. Nuclear extracts were prepared and separated on a glycerol gradient. The localization of SAMHD1 along the gradient was assessed by western blot. (D) HeLa (Mock) and HeLa cells stably expressing Flag-HA–tagged SAMHD1 were treated with 5 nM etoposide for the indicated time. Flag IPs were performed and SAMHD1 interactions with CycA and DCAF1 were assessed by western blot. (E) Flag IPs were performed using nuclear extract of HeLa cells stably expressing Flag-HA–tagged SAMHD1 after treatment with 5 nM etoposide. SAMHD1 and SAMHD1 phosphorylated on threonine 592 (pSAMHD1) levels were determined by western blot. (F) SAMHD1 colocalizes with 53BP1 at the site of DSBs. HeLa cells expressing SAMHD1-HA were mock-treated or treated with 25 nM CPT. SAMHD1 and 53BP1 intranuclear localizations were determined by immunofluorescence using specific antibodies and confocal microscopy.
SAMHD1 effects on cell proliferation and cell viability in response to DNA damage agents. (A) HeLa cells stably expressing an inducible empty vector (Mock) or a vector encoding for SAMHD1 wild-type (WT) SAMHD1 mutated in the HD domain (HD/AA) or truncated at amino acid 575 (ΔCter) were induced with doxycycline. Results are expressed as the proliferation ratio of SAMHD1-transduced cells divided by mock-transduced cell. A representative experiment out of 3 is shown. (B) HeLa and HeLa cells stably expressing SAMHD1 were treated with increasing concentrations of DNA damaging agents, and the percentage of living cells was measured 3 days after treatment. Each graph shows the mean of 3 independent experiments. *A t-test P < .05. (C) HeLa cells stably expressing SAMHD1 were mock treated or treated with 5 nM of etoposide for 4 or 18 hours. Nuclear extracts were prepared and separated on a glycerol gradient. The localization of SAMHD1 along the gradient was assessed by western blot. (D) HeLa (Mock) and HeLa cells stably expressing Flag-HA–tagged SAMHD1 were treated with 5 nM etoposide for the indicated time. Flag IPs were performed and SAMHD1 interactions with CycA and DCAF1 were assessed by western blot. (E) Flag IPs were performed using nuclear extract of HeLa cells stably expressing Flag-HA–tagged SAMHD1 after treatment with 5 nM etoposide. SAMHD1 and SAMHD1 phosphorylated on threonine 592 (pSAMHD1) levels were determined by western blot. (F) SAMHD1 colocalizes with 53BP1 at the site of DSBs. HeLa cells expressing SAMHD1-HA were mock-treated or treated with 25 nM CPT. SAMHD1 and 53BP1 intranuclear localizations were determined by immunofluorescence using specific antibodies and confocal microscopy.
We and others have recently uncovered the phosphorylation of SAMHD1 at threonine 592 (Thr592) by CyclinA2/CDK1 as a key regulatory mechanism of its antiviral activity.33,41 Interestingly, we found that treatment of cells with etoposide increased the pool of SAMHD1 interacting with CyclinA (Figure 5D) and induced SAMHD1 phosphorylation at the Thr592 (Figure 5E). To further explore the involvement of SAMHD1 in DNA damage response, we determined whether SAMHD1 is recruited to the site of DNA damage. HeLa cells expressing SAMHD1 were mock-treated or treated with camptothecin and stained for both 53BP1 and SAMHD1. 53BP1 foci were seen in untreated cells presumably reflecting DSBs that resulted from replication stress. An increase in number and size of 53BP1 foci was observed on camptothecin treatment. Interestingly, SAMHD1 colocalized with 53BP1 foci in both untreated and in CPT-treated cells (Figure 5F). These results suggest that SAMHD1 is recruited to the site of DNA damage. Taken together, our results show that SAMHD1 is involved in the response to DNA DSBs and engages in specific protein interactions on DNA damage.
Discussion
In this article, we present a detailed molecular description of patients with SAMHD1-mutated CLL recruited into clinical trials and begin to explore the role of SAMHD1 in CLL pathogenesis. We focused initially on the molecular characterization of an AGS patient in whom a homozygous germ-line mutation in SAMHD1 had been identified previously.7 Remarkably, this patient developed CLL at only 24 years of age; only 0.2% of patients diagnosed with CLL are 20 to 34 years of age, and the median age at presentation is 72 years.42 To our knowledge, there have been no other reports of CLL in AGS patients, although only a small number of patients beyond the age of 20 years are known to us. When we used targeted NGS, WES, and a genome-wide SNP array, approaches to test CLL-derived B cells from this patient, we confirmed the SAMHD1 mutation status and identified a large region of homozygosity consistent with inheritance from consanguineous parents carrying shared haplotypes. Importantly, we did not identify any acquired CLL-related sequence changes2,4,5,30,31 or structural variants1,43 in the leukemia cells. These data support the notion that a homozygous SAMHD1 germ-line mutation was sufficient to cause early-onset CLL in this particular patient. Alternatively, other currently unknown acquired or germ-line driver mutations could be involved in the CLL pathogenesis.
The AGS CLL patient findings together with previous longitudinal CLL WGS results in which an acquired homozygous SAMHD1 mutation was identified led us to perform detailed molecular studies of SAMHD1 using well-characterized samples from patients recruited into clinical trials. Significantly, our sequencing data revealed that SAMHD1 mutations occur at a fourfold higher frequency in the relapse/refractory cohort, and the majority are likely deleterious as shown by in silico analyses. Furthermore, the genome-wide SNP array data showed that the mutations in this group associate with cnLOH or monoallelic losses encompassing SAMHD1. WGS and WES analyses have been used previously to document recurrent mutations in CLL2,,-5 and most recently, SAMHD1 mutations have been noted at a statistically significant frequency of 4%.5 In contrast, extensive WES studies of solid tumor cell lines indicate that genomic mutations of SAMHD1 in solid tumors are rare events: only 16 mutations were identified in 1006 samples, and this did not reach statistical significance (E. Papaemmanuil, personal communication, January 2013).
Recently, Edelmann et al presented the results of an array-based study of untreated CLL patients recruited into the German CLL8 multicenter trial for first-line treatment.43 In our study, only cohort 2 represented an equivalent pretreatment cohort. For this patient group, the copy number aberration (CNA) findings in our study were consistent with those of the CLL8 study with no CNAs involving SAMHD1 being detected. For cnLOH, no regions involving SAMHD1 either in the 144 paired or 209 unpaired analyzed cases were reported in the CLL8 study, whereas we report 3 cases (1.5%) in our cohort 2. All 3 of our cnLOH cases are >10 Mb in size (∼14, ∼29, and ∼33 Mb), but would have been excluded by Edelmann et al43 because they did not coincide with a proven tumor-specific cnLOH or CNA. Additional reasons that may explain why cnLOH regions involving SAMHD1 were not reported in the CLL8 samples include sample size (direct comparison was only possible for 144 paired CLL8 cases) and differences in the array platform and analysis thresholds used for making cnLOH calls. For example, it is possible that mosaicism, a feature observed in 1 of the 3 cases in our cohort 2, may not have been called in the CLL8 study due to a reduced number of nonhomozygous intervening SNP calls. Similarly, background noise may have an effect.
To follow-up on our genomic studies, we examined SAMHD1 mRNA and protein expression in CLL and solid tumors. SAMHD1 is widely expressed across most normal tissues.18,37 Interestingly, although SAMHD1 mutations were expressed and clearly associated with markedly reduced mRNA and protein levels, reduced expression of SAMHD1 was also seen in most, but not all, WT SAMHD1 CLL. One case showed significant mRNA and protein expression, and another case had no protein expression despite presence of mRNA. Similarly, SAMHD1 mutations were only rarely found in solid tumors, but reduced expression was seen across many cancer cell lines and primary breast cancer. Together, these data suggest (1) that SAMHD1 mutations might lead to increased degradation of mutant mRNA and/or protein and (2) that other mechanisms apart from mutations must also be responsible for SAMHD1 regulation. Interestingly, methylation of the SAMHD1 promoter at CpG island resulting in SAMHD1 transcriptional repression has recently been reported in CD4+ T cell lines such as Jurkat and SupT1.44 This further strengthens the idea that a low level of SAMHD1 expression might involve different regulatory mechanisms.
As a prelude to more detailed functional studies, we presented a series of experiments that provide the first evidence for the potential involvement of SAMHD1 in the regulation of cell proliferation and response to DNA damage-inducing agents. The regulation of cell proliferation by SAMHD1 requires its HD domain (Figure 5A). However, our experiments do not determine whether the THP activity and/or the more recently reported nuclease activity of SAMHD114 are involved. Indeed, the SAMHD1 HD domain is required for both activities.14
We showed that SAMHD1 engages in specific interactions on DNA damage, and its expression regulates cell survival in response to DNA damage-inducing agents. Importantly, we found that SAMHD1 colocalizes with 53BP1 in DNA repair foci after induction of DSB. Together, these data suggest that SAMHD1 plays a role in the cellular response to DNA damage. The precise molecular mechanisms underlying these observations are unknown at present. Further studies, including its effect on cellular dNTP availability and genetic experiments of SAMHD1 knockout models, will be critical to elucidate further the role of SAMHD1 in DNA damage response.
From a clinical perspective, it is possible that SAMHD1 mutations in CLL lead to chemotherapy resistance. This is supported by several lines of evidence. (1) Loss of SAMHD1 expression results in enhanced resistance to DSB-inducing agent (Figure 5B). (2) SAMHD1 mutations are enriched in the relapsed/refractory cohort without apparent increase of poor risk confounding factors such as del11q or unmutated IgVH. (3) The small number of patients with SAMHD1 mutations in our cohort had a poorer response to first-line chemo-immunotherapy. Alternatively, SAMHD1 mutations might be associated with disease aggressiveness, defined by a short time to first treatment or transformation to Richter’s syndrome. However, in our small number of SAMHD1-mutated relapsed/refractory patients, there was no difference in overall survival compared with WT SAMHD1 patients, suggesting that mutations may not be linked to CLL aggressiveness. Clearly, analyses of larger patient cohorts are required to address this question fully, including also patients with monoclonal B-cell lymphocytosis or Richter’s syndrome.
There are important implications of these findings. First, our data illustrate that low-frequency mutations can be candidate drivers of disease and that large collaborative efforts are required to gain insight into the biological and clinical consequences of these low-frequency mutations. Second, mutations in SAMHD1 are specific to the hematopoietic system and can be founder mutations that occur as early as in the germ-line DNA. This supports their role in the early stages of leukemogenesis. Third, the evidence we present indicates that SAMHD1 mutations lead to exclusive and reduced expression of mutant mRNA. This poses considerable challenges to the development of drugs targeting the defect. Provided protein function is preserved, 1 possible strategy might be to increase SAMHD1 expression by preventing SAMHD1 degradation.45
If our results are confirmed by larger studies, the main implication will be to avoid the use of DNA damaging agents in patients with mutations in SAMHD1. In this regard, understanding how SAMHD1 determines the fate of DNA-damaged cell is an important challenge.
In conclusion, we presented recurrent mutations in SAMHD1 as newly identified molecular events in CLL and demonstrated that the protein SAMHD1 likely engages in specific interactions in response to DNA damage. More work is needed to fully elucidate its role both in regulation of DNA damage response and in leukemogenesis and/or disease progression. Only the analysis of large cohorts of well-characterized clinical trial samples will reveal whether SAMHD1 mutations emerge as clinically significant response predictors of chemotherapy resistance.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Molecular Virology laboratory for critical reading of the manuscript and Sabine Laurent-Chabalier for technical assistance.
This work was supported by grants from the European Research Council (250333), Sidaction (Fonds de dotation PIERRE BERGE), Agence Nationale de Recherche sur le Sida, and European FP7 “HIT HIDDEN HIV” contract 305762 (to M.B.). T.L. was supported by Ministère de l'Enseignement surpérieur et de la Recherche fellowships. This work was supported by the Oxford Partnership Comprehensive Biomedical Research Centre with funding from the Department of Health’s National Institute for Health Research Biomedical Research Centres funding scheme. A.S., A.T.T., J.C.T., and S.J.L.K. are supported also by the Health Innovation Challenge Fund (HICF-1009-026), a parallel funding partnership between the Wellcome Trust and the Department of Health. S.J.L.K. is also supported by the Wellcome Trust Core Award Grant (090532/Z/09/Z). Y.J.C. acknowledges the Manchester Biomedical Research Centre and the Greater Manchester Comprehensive Local Research Network. The research leading to these results has received funding from the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant 241779.
The views expressed in this publication are those of the authors and not necessarily those of the Department of Health or the Wellcome Trust.
Authorship
Contribution: R.C., T.L., M.B., and A.S conceived and designed the experiments and prepared the manuscript; R.C., T.L., A.B., P.R., A.T.T., H.D., J.M., J.B., M.R., Y.-L.L., P.P., and A.T. performed experiments; S.A., P.H., F.S., A.P., and J.G.J. provided samples; R.C., T.L., M.B., A.S., G.W.C., M.T., D.R.C., S.J.H., M.T.R., S.J.L.K., J.C.T, J.R., D.B., and Y.J.C., analyzed data; and A.S. and M.B. oversaw all of the work.
Conflict-of-interest disclosure: M.T.R. and D.B are employees of Illumina Inc., a public company that develops and markets systems for genetic analysis. The remaining authors declare no competing financial interests.
Correspondence: Anna Schuh, Oxford NIHR Biomedical Research Centre, Molecular Diagnostic Laboratory, Level 4 John Radcliffe Site, University of Oxford, Oxford, UK; e-mail: anna.schuh@ndcls.ox.ac.uk; or Monsef Benkirane, Institut de Genetique Humaine, CNRS UPR1142, Laboratoire de Virologie Moleculaire, Montpellier, France; e-mail: monsef.benkirane@igh.cnrs.fr.
References
Author notes
R.C., T.L., M.B., and A.S. contributed equally to this work.