Key Points
Absence of STAT3 serine phosphorylation restricts activated K-Ras–driven myeloproliferative disease in a mouse model.
A mitochondrial function of STAT3 supports K-Ras–driven, factor-independent growth of myeloid progenitors in vitro.
Abstract
Juvenile myelomonocytic leukemia, acute myeloid leukemia (AML), and other myeloproliferative neoplasms (MPNs) are genetically heterogeneous but frequently display activating mutations in Ras GTPases and activation of signal transducer and activator of transcription 3 (STAT3). Altered STAT3 activity is observed in up to 50% of AML correlating with poor prognosis. Activated STAT proteins, classically associated with tyrosine phosphorylation, support tumor development as transcription factors, but alternative STAT functions independent of tyrosine phosphorylation have been documented, including roles for serine-phosphorylated STAT3 in mitochondria supporting transformation by oncogenic Ras. We examined requirements for STAT3 in experimental murine K-Ras–dependent hematopoietic neoplasia. We show that STAT3 is phosphorylated on S727 but not Y705 in diseased animals. Moreover, a mouse with a point mutation abrogating STAT3 S727 phosphorylation displayed delayed onset and decreased disease severity with significantly extended survival. Activated K-Ras required STAT3 for cytokine-independent growth of myeloid progenitors in vitro, and mitochondrially restricted STAT3 and STAT3-Y705F, both transcriptionally inert mutants, supported factor-independent growth. STAT3 was dispensable for growth of normal or K-Ras–mutant myeloid progenitors in response to cytokines. However, abrogation of STAT3-S727 phosphorylation impaired factor-independent malignant growth. These data document that serine-phosphorylated mitochondrial STAT3 supports neoplastic hematopoietic cell growth induced by K-Ras.
Introduction
Juvenile myelomonocytic leukemia (JMML) and acute myeloid leukemia (AML), which can develop from myeloproliferative neoplasms (MPNs), are characterized by aberrant expansion of myeloid cells that leads to morbidity due to infection, hemorrhage, anemia, or organ infiltration. Oncogenic mutations contribute to JMML and AML; mutant Ras GTPases occur in 35% of JMML and 15% to 44% of AML cases.1,2 Mutation frequency probably underestimates involvement of activated Ras pathways because mutations in upstream regulators (eg, NF-1, FLT3, or c-Kit) occur in up to 40% of myeloid leukemia,3-5 activating Ras–mitogen-activated protein kinase (MAPK) signaling. Signal transducer and activator of transcription (STAT) proteins have been implicated in MPN and AML (typically STAT3 and STAT5). STAT3 was initially characterized as a latent factor, activated by phosphorylation on tyrosine (Y705) and serine (S727) in response to cytokines. Y705-phosphorylated STAT3 accumulates in the nucleus, initiating transcription of target genes. STAT3 target genes govern proliferation, migration, differentiation, angiogenesis, and survival,6 giving STAT3 important nuclear roles in human cancers.7,8 Up to 50% of AML patients display enhanced STAT3 tyrosine phosphorylation, correlating with poor prognosis.9-13 The paradigm for the role of STAT3 in AML posits constitutive tyrosine phosphorylation due to cytokines (eg, interleukin-6 [IL-6]14 ) or mutations in tyrosine kinases (eg, FLT315 or less frequently Janus kinase 2 [JAK2]16 ). However, oncogenic Ras mutations do not result in STAT3 tyrosine phosphorylation17 and yet drive hematopoietic malignancies. Hence, STAT3 may have distinct functions in the context of activated Ras.
Noncanonical tyrosine phosphorylation-independent functions of STAT3 have been identified. These noncanonical functions fall into 2 categories. Non-Y705–phosphorylated STAT3 may contribute to transcription in cooperation with other transcription factors.18,19 Second, STAT3 accumulates in mitochondria, also without tyrosine phosphorylation.17,20 In mitochondria, STAT3 augments electron transport chain activity, adenosine triphosphate production, Warburg-like effects, and tumorigenic growth in Ras-transformed cells. These mitochondrial functions depend on phosphorylation of STAT3 at S727 but not on Y705.17
Much research on STAT3 phosphorylation has focused on pY705 following cytokine stimulation or tyrosine kinase oncogene activation, but not on STAT3 phosphorylation status (Y705 or S727) during Ras transformation in vivo. We examined the requirement for STAT3 in a mouse model of Ras-driven hematopoietic malignancies, in which the K-RasG12D oncogene is activated following deletion of a Lox-STOP-Lox cassette by Cre recombinase controlled by the Mx promoter, which is activated in hematopoietic progenitors among other cells.21-25 We report that STAT3 is phosphorylated on S727 but not Y705 in hematopoietic progenitors expressing oncogenic K-Ras. Moreover, a point mutation blocking S727 phosphorylation significantly attenuated K-Ras–induced disease. Mutant forms of STAT3 unable to support transcription (Y705F, which cannot become tyrosine phosphorylated, and mitochondrially restricted STAT3 unable to accumulate in nuclei) mediated growth factor–independent proliferation of bone marrow progenitors driven by activated K-Ras, in absence of endogenous STAT3. These results imply that STAT3 mitochondrial functions support K-Ras–driven hematologic malignancy in absence of STAT3 nuclear transcriptional functions.
Material and methods
Antibodies and cytokines
Antibodies used: DX-5-allophycocyanin (APC)-Cy7, IL-7Rα-APC-Cy7 (A7R34), TER119-APC-Cy7, CD11b-APC-Cy7 (M1/70), B220-APC-Cy7 (RA3-6B2), CD4-APC-Cy7 (H129.19), CD8α-APC-Cy7 (53-6.7), c-Kit-fluorescein isothiocyanate (FITC) (2B8), Sca-1-phycoerythrin (PE)-Cy7 (D7), CD34-APC (RAM34), FCγR-Alexa 700 (2.4G2) (BioLegend); STAT3 (pY705)-Alexa 488 (BD Pharmingen); STAT3 (pS727), STAT3 (pY705) (Cell Signaling Technology); pSTAT5 (Zymed); phosphorylated extracellular signal-regulated kinase (pERK) (Santa Cruz Biotechnology). Cytokines (IL-3, IL-6, stem cell factor [SCF], FLT-3L, and granulocyte macrophage–colony-stimulating factor [GM-CSF]) were from Peprotech.
Cell culture
293T cells were grown in Dulbecco modified Eagle medium (DMEM) under standard conditions. Bone marrow was cultured in DMEM with 10% fetal calf serum (FCS), gentamycin, 100 µM β-mercaptoethanol, IL-3 (10 ng/mL), IL-6 (10 ng/mL), SCF (50 ng/mL), and FLT-3L (50 ng/mL). Recombinant retroviruses were generated in HEK293T cells.17
Mice
K-RasG12DLSL mice23 were interbred with Mx-1-Cre,26 STAT3-LoxP,27 and/or STAT3-S727A28 mice. Resulting strains were injected intraperitoneally with 100 µg of polyinosinic-polycytidilic acid (poly(I:C); Sigma-Aldrich). Mice were analyzed for MPNs every 2 weeks after injection by inspection of coat condition, posture, palpable splenomegaly, and peripheral blood counts using a Mindray BC 3200 hematology analyzer or a Hemavet 950FS (Drew Scientific) on blood collected from the retro-orbital sinus or submandibular vein. Animals were maintained in a specific pathogen-free vivarium, with neomycin sulfate (2 mg/mL) added to drinking water to control intestinal flora.29 Mice at the time of injection with poly(I:C) were 5 to 6 weeks of age. All mice were on the C57BL6 background. All work with experimental animals was in accordance with protocols approved by the NYU Langone Medical Center (NYULMC) Institutional Animal Care and Use Committee.
Bone marrow transplant assays
Wild-type recipient mice (C57BL6/J) were purchased from Taconic. Bone marrow cells harvested from mice of appropriate genotypes were injected IV in the retro-orbital sinus of 7-week-old lethally irradiated (2 × 550 rad) recipients. Hematopoietic reconstitution was allowed to proceed for 4 weeks prior to injection of poly(I:C), and mice were subsequently monitored for early signs of MPN development by peripheral leukocyte counts. Animals were killed at 2 intervals for analysis by flow cytometry.
Colony formation unit assays
Colony formation assays were performed on freshly isolated or retrovirally transduced marrow. Mice were injected with poly(I:C) and rested 2 weeks. Bone marrow was harvested and lineage depleted using EasySep-biotin (StemCell Technologies). Lineage-negative marrow was plated at 103 cells per well in methylcellulose lacking growth factor and cytokines (M3231; StemCell Technologies) or supplemented with 50 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3 and 3 units/mL erythropoietin (EPO) (M3434; StemCell Technologies). Colonies were counted after 10 days.
For retroviral transduction, marrow was cultured in DMEM, 10% FCS, gentamycin, 100 µM β-mercaptoethanol, IL-3 (10 ng/mL), IL-6 (10 ng/mL), SCF (50 ng/mL), and FLT3L (50 ng/mL) for 24 hours prior to transduction with pMIG-based constructs.20 Transduced marrow was cultured for a further 48 hours and sorted for lineage-negative, green fluorescent protein (GFP)–positive (transduced) populations. Cells (103) were cultured in methylcellulose lacking growth factors and cytokines (M3231; StemCell Technologies) or supplemented with 50 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3, and 3 units/mL EPO (M3434; StemCell Technologies). Colonies were counted after 10 days.
Subcellular fractionation and immunoblotting
Mitochondria were isolated using differential centrifugation. Cells (108) harvested by centrifugation were washed with cold phosphate-buffered saline (PBS) and resuspended in 10 volumes of buffer A (220 mM sorbitol, 70 mM sucrose, 50 mM 4-morpholinepropanesulfonic acid [MOPS], pH 7.4, 1 mM EDTA, plus protease inhibitors). Cells were homogenized and centrifuged at 800g for 10 minutes twice. Crude mitochondrial fractions were isolated from supernatants at 10 000g for 10 minutes, washed in buffer A, and resuspended in 250 mM sucrose, 1 mM EDTA, 10 mM Tris-HCl, pH 7.4, plus protease inhibitors (buffer B). Mitochondria were purified on a sucrose-Percoll gradient (80%, 52%, and 26%) at 60 000g for 45 minutes. Mitochondria were collected from the interface between the 26% and 52% layers and washed twice in buffer B at 7800g for 10 minutes. Organelle-free cytosol fractions were prepared from supernatants by centrifugation at 100 000g for 2 hours. Mitochondria were lysed in 150 mM NaCl, 50 mM Tris (pH 7.4), 1 mM EDTA, 0.5% Triton X-100, protease inhibitors, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 mM dithiothreitol and resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Blots were developed with 1:15 000 dilution of fluor-conjugated secondary antibody (Li-Cor).
Intracellular phospho-flow cytometry
Bone marrow cells were treated with Fc block and stained with biotin-conjugated antibodies against committed lineage epitopes (DX-5, IL-7Rα, TER119, Mac-1, CD19, CD4, and CD8α), streptavidin-brilliant violet 421, c-Kit–APC, and appropriate phospho-specific antibodies, and analyzed by flow cytometry, as described.30 In brief, erythrocyte-depleted marrow was fixed in 2% paraformaldehyde for 10 minutes at 37°C and incubated in 90% chilled methanol overnight, incubated in PBS with 4% calf serum for 1 hour, treated with Fc block, and stained with relevant antibodies. Fluorescence-activated cell sorter (FACS) data were analyzed using Cyflogic (CyFlo Ltd) or FlowJo software (TreeStar).
Results
Experimental design to assess the role of STAT3 serine phosphorylation in Ras-dependent disease
To assess the role of STAT3 in Ras-dependent MPNs, we bred Mx-1–Cre:K-RasG12DLSL:STAT3-LoxP/SA mice (referred to as Mx:K-Ras:F/SA). These mice contain a single conditional allele of K-RasG12DLSL,23 a single conditional allele of wild-type STAT3,27 and a single mutant allele of STAT3-S727A, which cannot be serine phosphorylated.28 Phosphorylation of Y705 is independent of S727.31 In absence of Cre expression, these mice express a single wild-type allele of K-Ras and a wild-type and S727A allele of STAT3. Following Mx-1–Cre induction by poly(I:C) injection, hematopoietic cells express K-RasG12D following deletion of the LSL cassette22,32 and only mutant STAT3-S727A, due to deletion of the conditional wild-type STAT3 allele.29 Experimental mice were compared with control Mx-Cre mice lacking either K-RasG12DLSL or STAT3-S727A alleles and therefore either do not express oncogenic Ras or express wild-type STAT3. To avoid possible gene dosage effects, all STAT3-expressing mice were Mx-1-Cre:STAT3-LoxP/+ or SA (with or without K-RasG12DLSL) and therefore expressed a single allele of STAT3 following poly(I:C) injection.
K-RasG12D drives STAT3 S727 but not Y705 phosphorylation in vivo
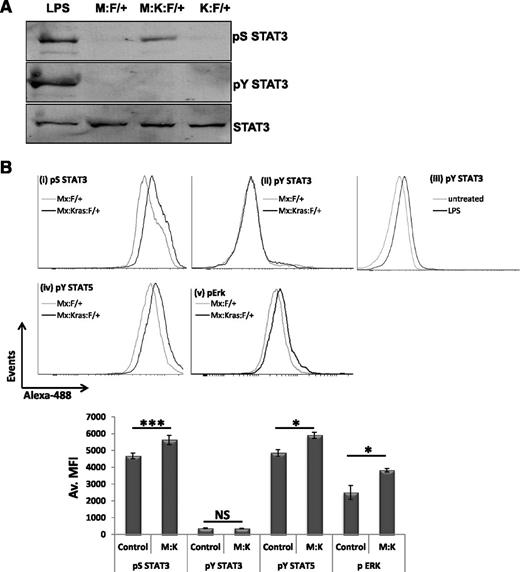
Poly(I:C)-injected Mx-1–Cre:K-RasG12DLSL:STAT3-LoxP/+ mice (referred to as Mx:K-Ras:F/+) are expected to develop MPNs with shared characteristics of JMML, including expansion of myeloid progenitors in the marrow and periphery, splenomegaly, and extramedullary hematopoiesis.22,32 Induction of Mx-Cre effectively deleted the LSL cassette in hematopoietic cells (allowing oncogenic K-Ras expression) and the conditional STAT3 allele, leaving intact only a STAT3-S727A allele (supplemental Figure 1, see supplemental Data available at the Blood Web site). Phosphorylation of STAT3 on S727 and Y705 was determined by immunoblotting of peripheral blood leukocytes. No STAT3 phosphorylation was observed in Mx:F/+ or K-Ras:F/+ animals, indicating that no STAT3 phosphorylation resulted from injection of poly(I:C) itself in absence of oncogenic K-Ras expression (Figure 1A, lanes 2, 4). However, activation of K-RasG12D in Mx:K-Ras:F/+ mice (Figure 1A, lane 3) led to S727 phosphorylation, similar to that observed in peripheral blood of lipopolysaccharide (LPS)-challenged mice (Figure 1A, lane 1). Although STAT3 Y705 phosphorylation was readily detected in LPS-challenged mice (Figure 1A, lane 1), this posttranslational modification was undetectable following activation of oncogenic K-RasG12D (Figure 1A, lane 3).
STAT3 is phosphorylated on S727, not Y705, in K-RasG12D-driven MPNs in vivo. STAT3-Flox mice of the following genotypes, Mx:STAT3F/+ (M:F/+), Mx:K-RasG12DLSL:F/+ (M:K:F/+), or K-RasG12DLSL:STAT3F/+ (K:F/+), were injected with a single 100-µg dose of poly(I:C). Mice were allowed to recover for 2 weeks before peripheral blood was collected, red blood cells lysed, and total leukocytes assessed for STAT3 phosphorylation on S727 (pS STAT3) or Y705 (pY STAT3) by immunoblotting (A). Antibody against total STAT3 served as a loading control. As a positive control for STAT3 phosphorylation, wild-type mice were injected with 30 µg of LPS 4 hours prior to harvesting peripheral blood leukocytes (LPS). (B) After 2 weeks of recovery following poly(I:C) injection, bone marrow was harvested from Mx:F/+ (Control) or Mx:K-Ras:F/+ mice (M:K) and expression of (i) pS727 STAT3, (ii) pY705 STAT3, (iv) pSTAT5, and (v) pERK was determined in lineage-negative, c-Kit+ cell populations by flow cytometery using phospho-specific antibodies. As a positive control for STAT3 Y705 phosphorylation (iii), mice were injected with 30 µg of LPS 4 hours prior to harvesting bone marrow. Pooled data from 3 animals for each genotype are plotted in the histogram showing the MFI ± standard deviation. Statistical significance was calculated using the Student t test. *P < .05, ***P < .001. MFI, mean fluorescence intensity.
STAT3 is phosphorylated on S727, not Y705, in K-RasG12D-driven MPNs in vivo. STAT3-Flox mice of the following genotypes, Mx:STAT3F/+ (M:F/+), Mx:K-RasG12DLSL:F/+ (M:K:F/+), or K-RasG12DLSL:STAT3F/+ (K:F/+), were injected with a single 100-µg dose of poly(I:C). Mice were allowed to recover for 2 weeks before peripheral blood was collected, red blood cells lysed, and total leukocytes assessed for STAT3 phosphorylation on S727 (pS STAT3) or Y705 (pY STAT3) by immunoblotting (A). Antibody against total STAT3 served as a loading control. As a positive control for STAT3 phosphorylation, wild-type mice were injected with 30 µg of LPS 4 hours prior to harvesting peripheral blood leukocytes (LPS). (B) After 2 weeks of recovery following poly(I:C) injection, bone marrow was harvested from Mx:F/+ (Control) or Mx:K-Ras:F/+ mice (M:K) and expression of (i) pS727 STAT3, (ii) pY705 STAT3, (iv) pSTAT5, and (v) pERK was determined in lineage-negative, c-Kit+ cell populations by flow cytometery using phospho-specific antibodies. As a positive control for STAT3 Y705 phosphorylation (iii), mice were injected with 30 µg of LPS 4 hours prior to harvesting bone marrow. Pooled data from 3 animals for each genotype are plotted in the histogram showing the MFI ± standard deviation. Statistical significance was calculated using the Student t test. *P < .05, ***P < .001. MFI, mean fluorescence intensity.
Peripheral blood is a mixture of mostly mature cells, whereas the cell of origin in this disease model is confined largely to bone marrow but accumulates in spleen or liver during disease.24 To ensure we were addressing STAT3 phosphorylation in cell populations responsible for disease onset, we assessed STAT3 phosphorylation in Lin−–c-Kit+ progenitors. pS727-STAT3 was detected in response to activated K-Ras in Mx:K-Ras:F/+ animals (Figure 1Bi), as was phospho-Erk (Figure 1Bv). However, no pY705-STAT3 was detected, although it was readily detected in progenitors from LPS-stimulated mice (compare panels Figure 1Bii and Figure 1Biii). Interestingly, pY-STAT5 was detected in K-Ras–activated progenitors from Mx:K-Ras:F/+ mice, consistent with previous data that K-RasG12D confers heightened and prolonged pY-STAT5.30 These data suggest that pY705-STAT3 is unlikely to be involved in K-Ras–driven hematopoietic progenitor expansion, whereas pS727 STAT3 and activated STAT5 may be functionally relevant. We have previously shown that pS727 in Ras-transformed cells requires activated MAPK kinase (MEK).33 Consistent with this notion, incubation of bone marrow from K-Ras–activated mice with a MEK inhibitor (PD0325901, 1 µM, 2 hours) caused loss of detectable pS727 (data not shown), suggesting that pS727-STAT3 is downstream of the activated Ras-MAPK pathway.
Loss of STAT3 S727 phosphorylation attenuated K-Ras–induced malignancy
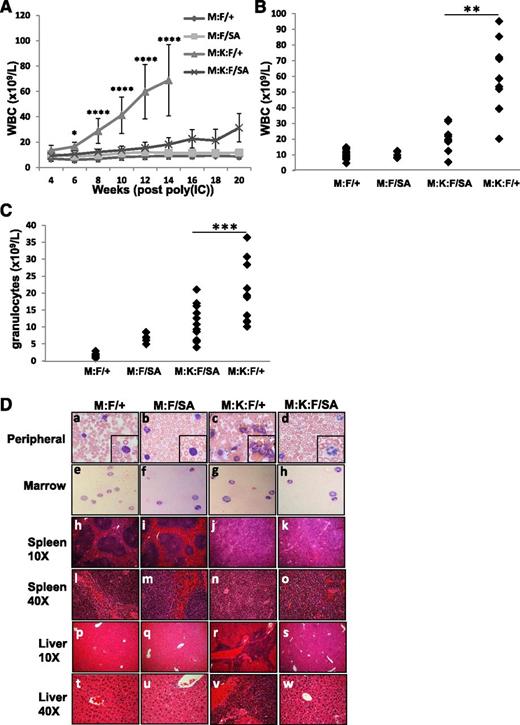
We examined disease incidence and progression in Mx:K-Ras mice crossed to a knock-in strain expressing STAT3-S727A, which cannot be serine phosphorylated (Mx:K-Ras:F/SA). STAT3-S727A (SA) animals showed no overt immune phenotype, as previously reported.28 Mx:F/+, Mx:F/SA, Mx:K-Ras:F/+, and Mx:K-Ras:F/SA mice were injected with poly(I:C) and monitored for MPNs. As expected, Mx:F/+ and Mx:F/SA animals, which did not express K-RasG12D, showed no signs of MPNs. All Mx:K-Ras:F/+ animals rapidly developed disease, typified by expansion of peripheral leukocytes (mean 79.3 × 109 L−1 at 14 weeks, Figure 2A-B). Most notable was massive expansion of peripheral granulocyte populations (mean 18.4 × 109 L−1, Figure 2C). However, in Mx:K-Ras:F/SA mice, which expressed K-RasG12D in the context of mutant STAT3 that could not be serine phosphorylated, these laboratory findings were greatly attenuated. Loss of STAT3-S727 phosphorylation significantly reduced expansion of peripheral leukocytes (mean 18.9 × 109L−1 at 14 weeks) and granulocytes (mean 10.4 × 109 L−1) (Figure 2A-C and supplemental Figure 2). At each time point and at end point, statistically significant increases in peripheral leukocytes and granulocytes were observed for animals expressing activated K-RasG12D and wild-type STAT3, compared with Mx:F/+ wild-type mice (supplemental Figure 2). However, more modest increases were observed only at later time points for animals expressing STAT3-S727A and K-RasG12D. The increases in peripheral leukocytes eventually observed in Mx:K-Ras:F/SA mice did not arise from cells that failed to delete the floxed wild-type STAT3 allele (not shown). Differences observed between animals with wild-type or mutant STAT3 were statistically significant (supplemental Figure 2). Mx:F/SA animals, which expressed STAT3-S727A but not K-RasG12D, showed modestly elevated peripheral granulocyte counts compared with control (7 × 109 L−1 and 1.4 × 109 L−1, respectively), but these animals were otherwise phenotypically normal (Figure 2C). Elevated granulocyte populations have been reported for animals lacking hematopoietic STAT329 or its downstream target SOCS3,34 likely due to an impaired ability to downregulate JAK1 activity in response to endogenous cytokines. Expression of a single STAT3-S727A allele may partially recapitulate the STAT3-null phenotype, but elevation of peripheral granulocytes has not been reported previously in these mice.35
Blocking STAT3 S727 phosphorylation ameliorates K-RasG12D–driven MPNs. Control animals (M:F/+) lacking K-RasG12D, Mx:STAT3F/SA (M:F/SA), Mx:K-RasG12DLSL:STAT3F/+ (M:K:F/+), or MxK-RasG12DLSL:STAT3F/SA (M:K:F/SA) mice were injected with poly(I:C) and peripheral white blood cells enumerated every 2 weeks (A). Each point represents the mean of cohorts of 5 animals, with error bars representing 1 standard deviation from the mean. *P < .05 and ****P < .0001 for comparisons between the M:K:F/+ and M:K:F/SA cohorts (calculated by 2-way ANOVA with the Tukey multiple comparison test). On the day animals were sacrificed due to morbidity (week 14 for Mx:K-Ras:F/+) or at the end of the experiment in the case of all other cohorts (week 20), peripheral white blood cells (B) or granulocytes (C) were counted and plotted. Each point represents cell numbers from an independent animal. **P < .01 and ***P < .001 for comparisons between the M:K:F/+ and M:K:F/SA cohorts (1-way ANOVA with the Tukey multiple comparison test). (D) Animals were sacrificed and tissues collected for histological analysis (20 weeks post poly(I:C) injection for Mx:F/+, Mx:F/SA, Mx:K-Ras:F/SA; 14 weeks for Mx:K-Ras:F/+). The peripheral blood (subpanels a-d) and bone marrow smears (subpanels e-h) were prepared with Wright-Giemsa stain and imaged with a 60× objective (inset: 2× digital enlargement). Spleen (subpanels h-o) and liver sections (subpanels p-w) were stained with hematoxylin and eosin and imaged with a 10× or 40× objective, as indicated. ANOVA, analysis of variance.
Blocking STAT3 S727 phosphorylation ameliorates K-RasG12D–driven MPNs. Control animals (M:F/+) lacking K-RasG12D, Mx:STAT3F/SA (M:F/SA), Mx:K-RasG12DLSL:STAT3F/+ (M:K:F/+), or MxK-RasG12DLSL:STAT3F/SA (M:K:F/SA) mice were injected with poly(I:C) and peripheral white blood cells enumerated every 2 weeks (A). Each point represents the mean of cohorts of 5 animals, with error bars representing 1 standard deviation from the mean. *P < .05 and ****P < .0001 for comparisons between the M:K:F/+ and M:K:F/SA cohorts (calculated by 2-way ANOVA with the Tukey multiple comparison test). On the day animals were sacrificed due to morbidity (week 14 for Mx:K-Ras:F/+) or at the end of the experiment in the case of all other cohorts (week 20), peripheral white blood cells (B) or granulocytes (C) were counted and plotted. Each point represents cell numbers from an independent animal. **P < .01 and ***P < .001 for comparisons between the M:K:F/+ and M:K:F/SA cohorts (1-way ANOVA with the Tukey multiple comparison test). (D) Animals were sacrificed and tissues collected for histological analysis (20 weeks post poly(I:C) injection for Mx:F/+, Mx:F/SA, Mx:K-Ras:F/SA; 14 weeks for Mx:K-Ras:F/+). The peripheral blood (subpanels a-d) and bone marrow smears (subpanels e-h) were prepared with Wright-Giemsa stain and imaged with a 60× objective (inset: 2× digital enlargement). Spleen (subpanels h-o) and liver sections (subpanels p-w) were stained with hematoxylin and eosin and imaged with a 10× or 40× objective, as indicated. ANOVA, analysis of variance.
Histological analysis of Mx:F/+, Mx:F/SA, Mx:K-Ras:F/+, and Mx:K-Ras:F/SA mice showed that Mx:K-Ras:F/+ animals displayed characteristics of MPNs that were not observed in the control cohorts or Mx:F/SA cohorts and were less pronounced in Mx:K-Ras:F/SA mice (Figure 2D). Peripheral blood of poly(I:C)-injected Mx:K-Ras:F/+ animals was dominated by expansion of myeloid cells (Figure 2Dc), and spleens and livers displayed morphologic signs of extramedullary hematopoiesis (Figure 2Dj,r), clearly visible at high magnification (Figure 2Dn,v). Of note, although Mx:K-Ras:F/SA animals also displayed signs of disorganized splenic architecture (Figure 2Dk), these phenotypes developed to a reduced extent compared with the Mx:K-Ras:F/+ cohort (Figure 2Dn-o). Similarly, peripheral blood from Mx:K-Ras:F/SA animals (Figure 2Dd) did not display myeloid cell expansion to the extent observed in blood from Mx:K-Ras:F/+ animals (Figure 2Dc). Moreover, morphologic evidence of extramedullary hematopoiesis, which was readily detected in liver of Mx:K-Ras:F/+ mice (Figure 2Dr,v), was reduced or absent in livers from Mx:K-Ras:F/SA animals (Figure 2Ds,w). Together, these data show that a point mutation in STAT3 at S727 capable of blocking phosphorylation diminishes the severity of K-RasG12D–driven disease, consistent with a critical role for S727 phosphorylation.
Loss of STAT3 S727 phosphorylation increases survival of animals with K-Ras–induced malignancy
Mx:K-Ras:F/+ mice developed signs of morbidity (hunched posture, labored breathing, and palpable splenomegaly). Necropsy revealed significantly enlarged spleens (mean 581 mg compared with 89 mg in controls, Table 1). Mx:K-Ras:F/SA mice were outwardly normal at similar time points, with no palpable splenomegaly; however, necropsy revealed enlarged spleens relative to normal animals (mean 294 mg, Table 1), approximately half the size observed in Mx:K-Ras:F/+ mice. Liver weights did not vary by genotype (Table 1).
Loss of STAT3 S727 phosphorylation reduces splenomegaly
| Genotype . | Average spleen weight, mg ± SD . | Average liver weight, mg ± SD . |
|---|---|---|
| Mx:F/+ | 89 ± 12 | 1296 ± 187 |
| Mx:F/SA | 91 ± 15 | 1110 ± 131 |
| Mx:K-Ras:F/+ | 581 ± 157 | 1283 ± 153 |
| Mx:K-Ras:F/SA | 294 ± 73 | 1228 ± 203 |
| Genotype . | Average spleen weight, mg ± SD . | Average liver weight, mg ± SD . |
|---|---|---|
| Mx:F/+ | 89 ± 12 | 1296 ± 187 |
| Mx:F/SA | 91 ± 15 | 1110 ± 131 |
| Mx:K-Ras:F/+ | 581 ± 157 | 1283 ± 153 |
| Mx:K-Ras:F/SA | 294 ± 73 | 1228 ± 203 |
Spleen and liver of animals of the indicated genotypes were weighed at autopsy. Each cohort contained a minimum of 5 animals; significant differences between the indicated groups were calculated using the Student t test. Mx:F/+ and Mx:F/SA animals were analyzed at 250 days post poly(I:C) injection; Mx:K-RasF/+ mice were analyzed at 70 to 100 days; Mx:K-Ras:F/SA at 100 to 150 days.
SD, standard deviation.
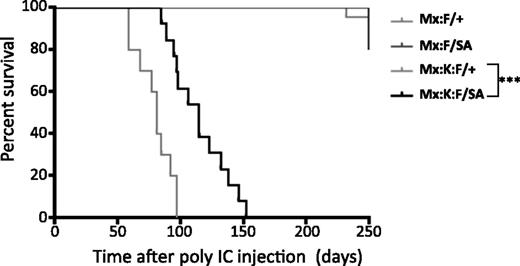
Survival analysis following poly(I:C) injection showed that the STAT3-S727A allele significantly increased lifespan compared with Mx:K-Ras:F/+ animals (Figure 3). Mice expressing K-RasG12D and wild-type STAT3 (Mx:K-Ras:F/+) displayed median survival of 81 days. In contrast, Mx:K-Ras:F/SA mice displayed significantly increased median survival (115 days, P < .001, log-rank test). Exact cause of death was not determined for either genotype, although morbidity correlated with increased peripheral leukocytosis (severe for Mx:K-Ras:F/+ animals; milder for Mx:K-Ras:F/SA mice) and splenomegaly (Table 1). Together with the laboratory hematological parameters, these survival statistics confirmed the importance of STAT3 serine phosphorylation for development of MPNs and morbidity.
Blocking STAT3 S727 phosphorylation significantly increases the life of K-RasG12D animals. Kaplan-Meier survival curves of animals of the indicated genotypes. Control animals (Mx:F/+) and Mx:STAT3F/SA (Mx:F/SA) animals survived until the experiment was terminated at over 300 days. Mx:K-Ras:F/+ (Mx:K:F/+) mice died rapidly from MPNs with a median survival of 83 days. Mx:K-Ras:F/SA (Mx:K:F/SA) animals survived significantly longer (***P < .001 log-rank test) with a median survival of 115 days. Cohorts included at least 10 animals, except Mx:F/SA (5 animals).
Blocking STAT3 S727 phosphorylation significantly increases the life of K-RasG12D animals. Kaplan-Meier survival curves of animals of the indicated genotypes. Control animals (Mx:F/+) and Mx:STAT3F/SA (Mx:F/SA) animals survived until the experiment was terminated at over 300 days. Mx:K-Ras:F/+ (Mx:K:F/+) mice died rapidly from MPNs with a median survival of 83 days. Mx:K-Ras:F/SA (Mx:K:F/SA) animals survived significantly longer (***P < .001 log-rank test) with a median survival of 115 days. Cohorts included at least 10 animals, except Mx:F/SA (5 animals).
Expansion of myeloid progenitor populations in the spleen is ameliorated in absence of STAT3-S727 phosphorylation
Hematopoietic activation of K-Ras leads to rapid expansion of peripheral leukocytes, due at least in part to increased hematopoietic stem cell progenitor populations in bone marrow and expansion of myeloid cells in peripheral organs.22,24,32 We examined the role of STAT3-S727 phosphorylation in progenitor expansion in marrow and spleen during disease progression. To confirm that any observed changes were due to cell autonomous effects of K-RasG12D and STAT3 mutations within the hematopoietic compartment rather than potentially confounding effects of Mx-Cre expression in peripheral tissues, we examined animals following bone marrow transplant. Irradiated isogenic wild-type recipients were reconstituted with marrow from Mx:K-Ras:F/+ or Mx:K-Ras:F/SA mice, or from Mx:F/+ mice as controls. Recipients were injected with poly(I:C) after successful engraftment and monitored for early disease as indicated by a doubling of peripheral leukocyte counts. The frequency of Lin−–c-Kit+ (MP), Lin−–Sca+–c-Kit+ (LSK), and various myeloid progenitor cell populations was examined in bone marrow and spleen by flow cytometry. A modest increase in granulocyte-macrophage progenitor (GMP) cells was observed in K-RasG12D animals with mild disease, which became more pronounced in mice reconstituted with Mx:K-Ras:F/+ marrow but not in mice reconstituted with Mx:K-Ras:F/SA marrow as the disease progressed (Figure 4A,C). There was also a modest decrease in the frequency of LSK cells in mice reconstituted with Mx:K-Ras:F/+ marrow, consistent with expectations,30 accompanied by a decrease in megakaryocyte-erythroid progenitor (MEP) cell frequencies. These laboratory findings were modestly but consistently more severe in mice reconstituted with Mx:K-Ras:F/+ relative to Mx:K-Ras:F/SA marrow. Myeloid progenitor populations also increased in diseased spleen, with common myeloid progenitor (CMP), GMP, and MEP frequencies trending higher as disease progressed (Figure 4B,D). Of note, increases in CMP and particularly GMP numbers and frequencies were consistently reduced in the absence of pSTAT3-S727, suggesting mitigation of disease. However, increased splenic mature myeloid and granulocytic CD11b+ and Gr1+/CD11b+ cell frequencies did not differ significantly due to STAT3 status (supplemental Figure 3).
Activated K-Ras-induced increase in splenic hematopoietic progenitor cells requires STAT3 S727. Animals reconstituted with bone marrow of the indicated genotypes were injected with poly(I:C) and allowed to develop early (cohort 1, 1.5-fold increase in peripheral white blood cell counts) and later signs of MPNs (cohort 2, more than twofold increase). Bone marrow (A,C) and splenic leukocytes (B,D) were harvested and the proportions and total numbers of MPs, LSK, CMPs (Lin−, cKit+, FCγR−, CD34+), GMPs (Lin−, cKit+, FCγR+, CD34+), and MEPs (Lin−, cKit+, FCγR−, CD34lo) were assessed by flow cytometry. Representative flow cytometry dot plots are shown in panels A and B. Proportion and total numbers of cells per organ are shown for the early (1) and later cohorts (2) in panels C and D. Number of animals per cohort were: 2 Mx:F/+ (cohort 1); 2 Mx:F/+ (cohort 2); 6 Mx:K-Ras:F/+ (cohort 1), 4 Mx:K-Ras:F/+ (cohort 2); 5 Mx:K-Ras:F/SA (cohort 1); 4 Mx:K-Ras:F/SA (cohort 2). Statistical significance was calculated using the Student t test. *P < .05.
Activated K-Ras-induced increase in splenic hematopoietic progenitor cells requires STAT3 S727. Animals reconstituted with bone marrow of the indicated genotypes were injected with poly(I:C) and allowed to develop early (cohort 1, 1.5-fold increase in peripheral white blood cell counts) and later signs of MPNs (cohort 2, more than twofold increase). Bone marrow (A,C) and splenic leukocytes (B,D) were harvested and the proportions and total numbers of MPs, LSK, CMPs (Lin−, cKit+, FCγR−, CD34+), GMPs (Lin−, cKit+, FCγR+, CD34+), and MEPs (Lin−, cKit+, FCγR−, CD34lo) were assessed by flow cytometry. Representative flow cytometry dot plots are shown in panels A and B. Proportion and total numbers of cells per organ are shown for the early (1) and later cohorts (2) in panels C and D. Number of animals per cohort were: 2 Mx:F/+ (cohort 1); 2 Mx:F/+ (cohort 2); 6 Mx:K-Ras:F/+ (cohort 1), 4 Mx:K-Ras:F/+ (cohort 2); 5 Mx:K-Ras:F/SA (cohort 1); 4 Mx:K-Ras:F/SA (cohort 2). Statistical significance was calculated using the Student t test. *P < .05.
Mitochondrial STAT3 mediates K-RasG12D–driven growth of bone marrow progenitors
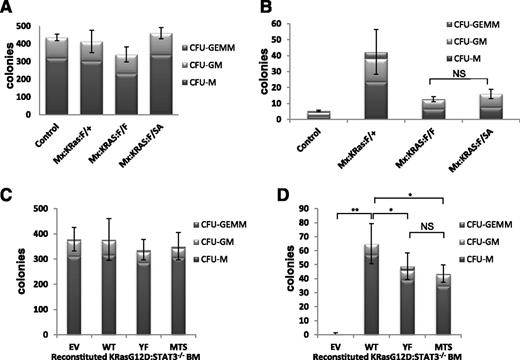
The dependence of MPN symptoms on STAT3-S727 phosphorylation in absence of detectable Y705 phosphorylation prompted us to explore STAT3 function in progenitor cell proliferation. To characterize the requirements for STAT3 that support K-RasG12D-driven hematopoietic disease, we assayed in vitro progenitor proliferation in the presence and absence of growth factors because oncogenic Ras confers factor-independent myeloid cell growth. We plated lineage-negative bone marrow from poly(I:C)-injected Mx:F/+, Mx:K-Ras:F/+, Mx:K-Ras:F/F, or Mx:K-Ras:F/SA mice in methylcellulose, unsupplemented or supplemented with SCF, IL-3, IL-6, EPO, and GM-CSF. Presence of myeloid colonies was scored quantitatively after 10 days, based on typical colony morphologies. Each genotype supported formation of myeloid colonies in approximately equivalent numbers and lineages in growth factor-supplemented medium (Figure 5A), indicating the presence of approximately equal numbers of progenitors responding similarly to growth factor stimulation. When plated in absence of growth factors, marrow from Mx:K-Ras:F/+ mice readily formed myeloid colonies, indicative of malignancy as previously observed22 ; in contrast and as expected, marrow from control animals without expression of oncogenic K-RasG12D failed to grow without growth factor supplementation (Figure 5B). Strikingly, progenitors from poly(I:C)-injected Mx:K-Ras:F/F mice that deleted STAT3 but expressed oncogenic K-RasG12D formed markedly fewer colonies than K-RasG12D–expressing progenitors that were wild type for STAT3 (Figure 5B), documenting a requirement for STAT3 expression in support of K-Ras–driven proliferation without growth factors. We next considered whether this STAT3 requirement reflected canonical transcriptional functions only augmented by pS727, or a noncanonical activity highly dependent on serine phosphorylation. Progenitors from bone marrow of poly(I:C)-injected Mx:K-Ras:F/SA mice expressing STAT3, which cannot be S727 phosphorylated, were also impaired for growth without added growth factors, in a manner similar to STAT3-null cells (Figure 5B). These results indicate that the STAT3 requirement for K-RasG12D–dependent growth factor-independent proliferation required S727, likely due to its ability to be phosphorylated in response to activated K-Ras.
Nontranscriptional, mitochondrial STAT3 supports factor-independent colony formation. Control, Mx:K-Ras:F/+, Mx:K-Ras:F/F, or Mx:K-Ras:F/SA mice were injected with poly(I:C) and allowed to recover for 1 week. Bone marrow was harvested and lineage depleted. Cells (103) were plated in methylcellulose supplemented with 50 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3, 3 units/mL EPO, and 20 ng/mL GM-CSF (A) or without growth factors (B). Colonies were counted and identified after 10 days in culture. (C-D) Mx:K-Ras:F/F mice were injected with poly(I:C) and allowed to recover for 1 week. Bone marrow was harvested and transduced with pMIG retroviruses expressing WT, YF, MTS, or EV. Lineage-negative, GFP-positive cells were sorted and 103 cells per plate were cultured in methylcellulose supplemented with 50 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3, 3 units/mL EPO, and 20 ng/mL GM-CSF (C) or without growth factors (D). Colony numbers and cell types were enumerated after 10 days. Histograms represent the mean of triplicate experiments and error bars are 1 standard deviation from the mean of the total number of colonies. *P < .05 and **P < .001 of differences between groups calculated by the 1-way ANOVA and the Tukey multiple comparison test. EV, empty vector control; MTS, mitochondrially restricted STAT3 mutant; NS, not significant; WT, wild-type STAT3; YF, STAT3 Y705F mutant.
Nontranscriptional, mitochondrial STAT3 supports factor-independent colony formation. Control, Mx:K-Ras:F/+, Mx:K-Ras:F/F, or Mx:K-Ras:F/SA mice were injected with poly(I:C) and allowed to recover for 1 week. Bone marrow was harvested and lineage depleted. Cells (103) were plated in methylcellulose supplemented with 50 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3, 3 units/mL EPO, and 20 ng/mL GM-CSF (A) or without growth factors (B). Colonies were counted and identified after 10 days in culture. (C-D) Mx:K-Ras:F/F mice were injected with poly(I:C) and allowed to recover for 1 week. Bone marrow was harvested and transduced with pMIG retroviruses expressing WT, YF, MTS, or EV. Lineage-negative, GFP-positive cells were sorted and 103 cells per plate were cultured in methylcellulose supplemented with 50 ng/mL SCF, 10 ng/mL IL-6, 10 ng/mL IL-3, 3 units/mL EPO, and 20 ng/mL GM-CSF (C) or without growth factors (D). Colony numbers and cell types were enumerated after 10 days. Histograms represent the mean of triplicate experiments and error bars are 1 standard deviation from the mean of the total number of colonies. *P < .05 and **P < .001 of differences between groups calculated by the 1-way ANOVA and the Tukey multiple comparison test. EV, empty vector control; MTS, mitochondrially restricted STAT3 mutant; NS, not significant; WT, wild-type STAT3; YF, STAT3 Y705F mutant.
STAT3-S727 phosphorylation in absence of Y705 phosphorylation is indicative of a mitochondrial rather than nuclear activity.17,20 STAT3 accumulates in mitochondria regardless of S727 phosphorylation status, including in cells of the marrow (supplemental Figure 4), but remains inactive in absence of S727 phosphorylation.36 To directly distinguish whether the STAT3 requirement in K-Ras–induced progenitor proliferation was nuclear or mitochondrial, we transduced STAT3-null progenitors with GFP-tagged retroviruses encoding wild-type STAT3, STAT3-Y705F mutant (YF) which cannot be tyrosine phosphorylated, or a mitochondrially restricted STAT3 mutant (MTS-S3) anchored in mitochondria and unable to accumulate in nuclei or support transcription.17,20 Bone marrow from poly(I:C)-injected Mx:K-Ras:F/F mice lacking endogenous STAT3 was transduced with control retrovirus or retroviruses expressing wild-type or mutant forms of STAT3. GFP+ bone marrow cells, indicative of retroviral transduction, were FACS sorted into Lin+ and Lin− populations. Lin−-GFP+ progenitor cells were plated in methylcellulose in the presence or absence of added growth factors, and colony formation was enumerated after 10 days. The more abundant Lin+-GFP+ control population was analyzed by immunoblotting for recombinant STAT3 expression (supplemental Figure 5). Equal numbers of colonies formed regardless of STAT3 status when cultured in presence of growth factors (Figure 5C), reinforcing the notion that STAT3 is dispensable for proliferative responses to growth factors. In contrast, cells transduced with vectors lacking STAT3 failed to thrive when cultured in absence of growth factors (Figure 5D EV). As expected, bone marrow transduced with a wild-type STAT3-expressing retrovirus formed colonies, even when cultured without growth factors, at an efficiency comparable to colony formation by cells expressing endogenous STAT3, indicative of successful reconstitution (compare Figure 5D WT with Figure 5B Mx:K-Ras:F/+). Strikingly, STAT3-null progenitors transduced with STAT3-YF or STAT3-MTS also supported colony growth when cultured in absence of growth factors (Figure 5D). This result suggests that the primary requirement for STAT3 in Ras-driven progenitor proliferation depends on its mitochondrial role, in contrast to STAT3 being dispensable for growth factor-driven proliferation. A role for mitochondrial STAT3 is even more striking when the disparity in expression of MTS-STAT3 relative to wild type or YF STAT3 is considered (supplemental Figure 5). MTS-STAT3 expression at steady state was significantly reduced compared with the more ubiquitously expressed isoforms, possibly due to the limited number of mitochondria present in myeloid progenitors. Nonetheless, this protein accumulation was sufficient to support factor-independent colony growth of K-RasG12D–expressing progenitors. This result demonstrates that a noncanonical mitochondrial function of STAT3, independent of tyrosine phosphorylation and nuclear accumulation, is instrumental for K-Ras–driven but not growth factor-driven myeloid cell proliferation.
Discussion
In this study, we defined a function for STAT3 that participates in initiation and progression of K-RasG12D–driven MPNs, distinct from normal hematopoiesis. Specifically, STAT3-S727 phosphorylation facilitated Ras-dependent MPNs, leading to more rapid onset and greater severity of disease. We found that STAT3 is necessary for K-RasG12D–driven growth factor-independent proliferation of myeloid cells in vitro, dependent on S727. Experimental evidence demonstrated that mitochondrial expression of STAT3 is sufficient to mediate K-Ras–dependent myeloid proliferation in vitro. Using a mouse model in which each experimental allele was expressed from its endogenous locus, we showed that oncogenic K-Ras expression led to STAT3 serine but not tyrosine phosphorylation. Mutation of a single serine phosphorylation site in STAT3 attenuated disease in vivo and factor-independent cell proliferation in vitro. These effects of STAT3 were hematopoietic cell-autonomous because expression of the STAT3-S727A mutant allele exclusively within the hematopoietic compartment following BMT was sufficient to attenuate or delay disease.
Oncogenic function of STAT3 in response to activated Ras is transcription independent
Although STAT3 was first characterized as a transcription factor, recent evidence has revealed expanded functions, including roles in modulating cancer metabolism. In this study, we provide evidence that the mitochondrial but not nuclear activity of STAT3 supports K-RasG12D–driven MPN. Metabolic regulation by STAT3 is mediated at least in part from within mitochondria, dependent on serine 727 phosphorylation.36 Tyrosine phosphorylation of STAT3 is necessary for the transcription of most of its target genes; in contrast, S727 phosphorylation contributes to, but is not required for, STAT3 transcriptional functions.28,35 We did not observe tyrosine phosphorylation of STAT3 in peripheral blood or hematopoietic progenitors expressing oncogenic K-Ras, consistent with the lack of protein tyrosine kinase dependence of this oncoprotein. Mutation of STAT3 Y705 did not prevent oncogene-driven proliferation of myeloid progenitors in vitro. Together, these data support the notion that STAT3 Y705 phosphorylation is dispensable for K-RasG12D–induced MPN, consistent with a role for STAT3 outside the nucleus. Moreover, artificial restriction of STAT3 to the mitochondrial matrix in absence of cytoplasmic and nuclear accumulation was sufficient for K-RasG12D–dependent, cytokine-independent progenitor cell proliferation in vitro. This mitochondrially restricted STAT3 is incapable of entering cell nuclei, activating gene transcription, or supporting cell transformation by the tyrosine kinase oncogene Src.17 Extrapolating from the in vitro cell growth requirements suggests that a mitochondrial role of STAT3 is the critical function that contributes to K-RasG12D–driven myeloid malignancy, similar to its role in other Ras-driven transformations.17
Mitochondrial activities of STAT3 are complex. The best described role of mitochondrial STAT3 is its ability to augment electron transport chain activity,17,20 although how this is achieved by a relatively small pool of mitochondrial STAT337 remains unclear. STAT3 also impedes opening the mitochondrial transition pore, thereby delaying cell death in response to toxic insults38 and possibly to the apoptotic effects of oncoprotein expression. STAT3 regulates mitochondrial function during normal hematopoiesis, at least in part by limiting reactive oxygen species (ROS).39 Interestingly, mitochondrial STAT3 appeared dispensable for progenitor growth in vitro in the presence of growth factors (Figure 5), suggesting that its role in MPNs may be oncogene specific, distinct from normal hematopoiesis. It remains to be determined whether regulation of oxidative phosphorylation, mitochondrial transition pore opening, or some undescribed mitochondrial function contributes to K-Ras–induced MPN and factor-independent proliferation.
STAT3 is an important signaling molecule during cytokine stimulation. The use of the STAT3-S727A mice in this study enabled the nuclear activities of STAT3 to proceed (albeit at a reduced capacity28 ) while significantly impairing STAT3 mitochondrial activity. Complete loss of STAT3 from the hematopoietic compartment leads to colitis due to impaired innate immune regulation,29,40 likely due at least in part to failure to induce the negative regulator SOCS334,41 in response to cytokines such as IL10.37 Colitis was not observed in STAT3-S727A animals, particularly on the C57BL6 background.28,35 Importantly, we did not observe any overt phenotype in animals expressing a single copy of STAT3-S727A in absence of oncogenic Ras (Mx:F/SA mice) other than a modest increase in peripheral granulocytes, consistent with this mutant STAT3 allele being sufficient to maintain essential cytokine signaling. We observed that STAT5, unlike STAT3, was tyrosine-phosphorylated in proliferating malignant cells. STAT5 is activated by the myeloid cytokines IL3, IL5, and GM-CSF42 and has been implicated in myeloid leukemia.43,44 Increased sensitivity of STAT5 to cytokine-stimulated tyrosine phosphorylation occurs in the K-RasG12D mouse model.30 We speculate that tyrosine-phosphorylated STAT5 drives expression of essential STAT target genes necessary for proliferation in malignant cells. This hypothesis is consistent with our data that STAT3 is dispensable for myeloid progenitor proliferation in the presence of cytokines in vitro. Transcriptional functions of STAT3 may therefore be uninvolved or superfluous, due to the presence of activated STAT5.
Serine-phosphorylated STAT3 is a target of oncogenic Ras signaling
Blocking STAT3-S727 phosphorylation without affecting its tyrosine phosphorylation–dependent nuclear functions that transduce cytokine signals could provide a chemotherapeutic approach to MPNs. Loss of S727 phosphorylation might be expected to reduce malignant burden in patients with activated K-Ras, while retaining critical nuclear activities of tyrosine-phosphorylated STAT3. STAT3 S727 is imbedded in an MAPK consensus motif45 and is a substrate for enzymes in the MAPK family.31 ERK1 phosphorylates STAT3 in response to growth factor stimulation,46 a pathway activated by Ras signaling.47 STAT3 and ERK2 interact48 and STAT3 is serine phosphorylated in cells when ERK2 is active. Indeed, H-, K-, and N-Ras transformation of fibroblasts leads to STAT3-S727 phosphorylation in an MEK-dependent manner,33 and a phosphomimetic version of STAT3 renders Ras-transformed fibroblasts partially resistant to MEK inhibition, suggesting that STAT3-S727 phosphorylation is a critical transformation target of oncogenic Ras.33 Constitutive ERK phosphorylation is observed in 74% to 83% of myeloid leukemias, but not in normal marrow.42,43 Inhibition of ERK phosphorylation causes growth arrest and can induce apoptosis in tumor blasts but not normal bone marrow.49,50 Moreover, MEK inhibition abrogates MPNs in K-RasG12D mice.51 Given the efficacy of MEK inhibition in experimental K-Ras–driven MPNs,51,52 targeting STAT3 S727 phosphorylation, possibly in conjunction with inhibition of STAT5 or JAK activity,53 could provide a viable therapeutic approach.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Liang Hu and Yeray Arteaga for expert technical assistance, members of the Levy laboratory for helpful discussions, Carl Weidinger for advice and help with experimental procedures, and Lawrence Gardner for critical reading of the manuscript and helpful discussions.
The authors also thank the NYU Langone Medical Center flow cytometry and histology cores, supported in part by grant 5P30CA016087-32 from the National Cancer Institute at the National Institutes of Health (NIH), for their help in preparing and analyzing samples. D.J.G. acknowledges support from a Special Fellow award from the Leukemia & Lymphoma Society, the National Health and Medical Research Council of Australia (GNT1024929 and GNT1063914), and the Victorian Government’s Operational Infrastructure Support Program. C.L. is supported by a Leukemia & Lymphoma Society Career Development Program Fellowship. I.A. is supported by the NIH (National Cancer Institute grants RO1CA133379, RO1CA105129, RO1CA149655, and National Institute of General Medical Sciences grant RO1GM088847), the Leukemia & Lymphoma Society (TRP program grants), The V Foundation for Cancer Research, the Irma T. Hirschl Trust, and the St. Baldrick’s Foundation for Cancer Research. D.E.L. acknowledges funding from the NIH (National Institute of Allergy and Infectious Diseases grants R01AI28900 and U54AI057158) and the Dr Louis A. Schneider Foundation Trust.
Authorship
Contribution: D.J.G., I.J.M., and C.L. designed and performed experiments, analyzed and interpreted the data, and wrote the manuscript; I.A. interpreted data; and D.E.L. designed experiments, analyzed and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for D.J.G. is Monash Institute of Medical Research, Clayton, Victoria, Australia.
The current affiliation for C.L. is Institut Gustave Roussy and Institut National de la Santé et de la Recherche Médicale U1009, Villejuif, France; and Université Paris-Sud 11, Orsay, France.
Correspondence: David. E. Levy, NYU School of Medicine, 550 First Ave, New York, NY 10016; e-mail: david.levy@nyumc.org; or Daniel J. Gough, MIMR-PHI Institute of Medical Research, 27-31, Wright St, Clayton, Victoria 3068, Australia; e-mail: daniel.gough@mimr-phi.org.
References
Author notes
D.J.G. and I.J.M. made equal contributions.