Key Points
Higher abundance TET2 mutations are associated with increased response to hypomethylating agents, particularly when ASXL1 is not mutated.
TP53 and PTPN11 mutations are associated with shorter overall survival after hypomethylating agent treatment, but do not predict response.
Abstract
Only a minority of myelodysplastic syndrome (MDS) patients respond to hypomethylating agents (HMAs), but strong predictors of response are unknown. We sequenced 40 recurrently mutated myeloid malignancy genes in tumor DNA from 213 MDS patients collected before treatment with azacitidine (AZA) or decitabine (DEC). Mutations were examined for association with response and overall survival. The overall response rate of 47% was not different between agents. Clonal TET2 mutations predicted response (odds ratio [OR] 1.99, P = .036) when subclones unlikely to be detected by Sanger sequencing (allele fraction <10%) were treated as wild-type (WT). Response rates were highest in the subset of TET2 mutant patients without clonal ASXL1 mutations (OR 3.65, P = .009). Mutations of TP53 (hazard ratio [HR] 2.01, P = .002) and PTPN11 (HR 3.26, P = .006) were associated with shorter overall survival but not drug response. Murine-competitive bone marrow transplantation followed by treatment with AZA demonstrated that Tet2-null cells have an engraftment advantage over Tet2-WT cells. AZA significantly decreased this advantage for Tet2-null cells (P = .002) but not Tet2-WT cells (P = .212). Overall, Tet2 loss appears to sensitize cells to treatment with AZA in vivo, and TET2 mutations can identify patients more likely to respond to HMAs.
Introduction
DNA hypomethylating agents (HMAs) are the only class of drugs approved for the treatment of patients with higher-risk myelodysplastic syndromes (MDS). Azacitidine (AZA) was approved by the Food and Drug Administration (FDA) for MDS in 2004 and was later shown to confer an overall survival benefit compared with supportive care in a randomized phase 3 study.1 Decitabine (DEC), the deoxynucleotide analog of AZA, was approved for the treatment of MDS in 2006 based on its ability to improve blood counts and decrease bone marrow blasts proportions.2 However, only 40% to 50% of patients treated with either AZA or DEC experience hematologic improvement (HI) with these agents, and complete responses (CRs) occur in as few as 10% to 15% of treated patients.3,4 Effective methods for identifying patients who are most likely to respond to treatment with an HMA would be of immediate clinical utility. Clinical features and patient characteristics may help stratify patients according to their response rates, but these models are not sufficiently conclusive to deny eligible patients a trial of AZA or DEC based on their predictions alone.5,6 Better biomarkers of response to HMAs are needed.
Since the FDA approval of AZA and DEC, our understanding of the molecular genetic basis for MDS has expanded dramatically. Recurrent somatic mutations have been identified in more than 40 genes, and many of these mutated genes have been associated with important clinical measures including overall survival.7-9 Because mutated genes underlie the pathogenic mechanisms driving the initiation and progression of MDS, they may represent molecular biomarkers of drug sensitivity or resistance. This is exemplified by the observation that MDS with deletions of the long arm of chromosome 5 (del[5q]) have a striking sensitivity to lenalidomide, whereas MDS patients without this lesion are less likely to have a hematologic response and are much less likely to have a cytogenetic or prolonged response.10 No such cytogenetic correlate has been found for the HMAs, but single-gene mutations involving the pathways targeted by these drugs may be better candidates.
DEC and AZA (which is metabolized into DEC intracellularly) inhibit DNA methyltransferases and decrease the methylation of cytosine residues. Several of the most frequently mutated genes in MDS encode proteins involved in the epigenetic regulation of gene expression such as TET2, DNMT3A, and ASXL1. DNMT3A is a de novo DNA methyltransferase and is a potential target of the HMAs. Somatic mutations of DNMT3A have been shown to decrease its activity, suggesting that pharmacologic targets other than DNMT3A are likely mediators of response to AZA and DEC.11 Loss of function mutations in TET2 impair the ability of this enzyme to oxidize methylcytosine residues and are associated with altered DNA methylation patterns and decreased 5-hydroxymethylcytosine levels in MDS patient samples.12,13 A small study of AZA-treated MDS patients using Sanger sequencing to determine the mutation status of TET2 found that mutations of this gene were associated with a slightly higher rate of response than in wild-type (WT) TET2 patients.14 However, the investigators did not examine these samples for additional mutations that might have modified this result and did not use techniques sensitive enough to identify mutations in small-disease subclones. Subclonal mutations in genes associated with an adverse prognosis, including ASXL1, RUNX1, and NRAS, have been shown to have clinical relevance regardless of their abundance within the dysplastic clone.8,15 These adverse mutations are often associated with disease progression and may mitigate the value of a sensitizing abnormality if they confer resistance to treatment.
We hypothesize that mutations of individual genes may serve as biomarkers of response for MDS patients treated with HMAs. We used massively parallel sequencing to examine 40 recurrently mutated genes in disease samples from 213 MDS patients treated with AZA or DEC. We examined the association of mutational patterns at different mutant allele fractions with response to treatment and overall survival. We used a competitive murine bone marrow transplant model to test the sensitivity of Tet2-null and Tet2 wild-type (Tet2-WT) hematopoietic cells to treatment with AZA.
Materials and methods
Patient samples and response assessment
A total of 213 MDS patients treated with AZA or DEC were included in this study. Samples were obtained from patients treated at the Dana-Farber Cancer Institute (2003-2010, N = 42), the MD Anderson Cancer Center (2003-2010, N = 104), and as part of the DACO-020 (ADOPT) clinical trial of DEC (2005-2006, N = 67). All samples were collected with patient consent under institutional review board–approved protocols in accordance with the Declaration of Helsinki. Response to treatment was assessed using International Working Group (IWG) response criteria revised in 2006. Patients with either a CR, partial response (PR), or HI were considered as “responders” (R, n = 100, 47%), whereas patients described as having “no response,” “stable disease,” “progressive disease,” “death” before response assessment, or “not evaluable” were considered “nonresponders” (NR, n = 113, 53%).
Sample processing, DNA sequencing, and mutation analysis
DNA was extracted from bone marrow mononuclear cells or peripheral blood samples collected before treatment (median 18 days, range 9-119). Whole-genome amplification of DNA for each sample was performed using the REPLI-g kit from QIAGEN. A genotype fingerprint of 22 common single-nucleotide polymorphisms (SNPs) for each sample was generated by MALDI-TOF genotyping (Sequenom). Target regions of 40 genes (supplemental Table 1, available on the Blood Web site) and genotype fingerprint regions were enriched using the Agilent SureSelect hybrid capture system according to the manufacturer’s instructions. Barcoded samples were pooled in equimolar amounts and subjected to 100 nucleotide paired-end sequencing on an Illumina Hi Sequation 2000. Sequence reads were aligned to the human genome (Build 37) using the Burroughs-Wheeler algorithm.16 The Genome Analysis Toolkit was used to clean and locally realign reads before calling missense and insertion/deletion variants using MuTect.17,18 Sample identity was confirmed by matching fingerprint genotype calls. Synonymous variants, noncoding variants more than 6 bases from splice junctions, or variants present in databases of “normal” genomes (dbSNP 132 or NHLBI Exome Sequencing Project) at a population frequency of 1% or more were discarded. Remaining variants were considered candidate somatic mutations.
Competitive murine bone marrow transplants
Age-matched Tet2−/−;Mx-Cre+ and Tet2+/+;Mx-Cre+ donor animals (CD45.2) were treated with pIpC (15 μg/g intraperitoneally [IP]) for 3 nonconsecutive days to induce excision of exon 3 of Tet2.19 Donor bone marrow was harvested 2 weeks post-pIpC and mixed in a 1:2 ratio with bone marrow harvested from 45.1 WT donors (B6.SJL-Ptprca Pepcb/BoyJ; Jackson Labs), and was then transplanted into 45.1 recipients for a total of 1 million cells per recipient. Peripheral blood engraftment was assessed by fluorescence-activated cell sorting at 2 weeks posttransplant, at which point recipient mice were divided into treatment groups (n = 7 per group) and treated with either 5-AZA (2.5 mg/kg IP; Santa Cruz Biotechnology) or vehicle control on the following schedule: 2 weeks on, 2 weeks off. Peripheral blood chimerism and complete blood count were assessed after each round of treatment.
Statistical methods
Categorical variables were compared using the Fisher exact test or χ2 test as appropriate, whereas continuous variables were compared using the Wilcoxon rank-sum test. A Cochran-Mantel-Haenszel test was used to test for differences in response rate by mutational status controlling for treatment. Unadjusted and adjusted logistic regression models were used to predict response to therapy. Models were adjusted for covariates including age (≥70 y vs <70 y), sex, International Prognostic Scoring System (IPSS) risk group (low/intermediate 1 vs intermediate 2/high) and treatment (AZA vs DEC alone vs DEC ± other). The odds ratio (OR) and 95% confidence intervals (CI) were estimated for the risk group (mutated) and compared with the reference group (WT). The Hosmer and Lemeshow goodness-of-fit test was used to assess model fit of logistic regression models. Overall survival was calculated from the time of treatment to the time of death from any cause, or was censored at the date last known alive and was compared using a log-rank test. Unadjusted and adjusted univariate Cox models were also constructed using the same covariates. For the competitive murine experiments, the percent 45.2 chimerism was calculated for each time point. Error bars indicate standard error of the mean (SEM) for each group, and P values for each time point were calculated using a 2-sample Student t test. All P values reported are 2-sided and considered significant at .05. No adjustments were made for multiple hypothesis testing.
Results
Spectrum of mutations
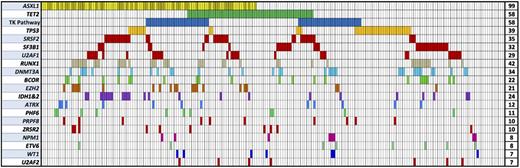
We examined tumor samples collected from 213 patients from 3 different sites before treatment with AZA, DEC, or DEC + another agent. There were no significant differences in pretreatment patient characteristics by treatment site (Table 1) or baseline characteristics as shown in Table 2. Frequently mutated regions of 40 genes previously shown to be somatically mutated in patients with MDS were subject to hybrid capture and massively parallel sequencing (supplemental Table 1). These include the most frequently mutated splicing factors, kinase signaling genes, transcription factors, and epigenetic regulators such as TET2, DNMT3A, ASXL1, and EZH2. With this panel, we identified one or more mutations in 39 genes (Figure 1). In total, 94% of patients had a mutation in at least one recurrently mutated gene. The most frequently mutated genes were ASXL1 (46%), TET2 (27%), RUNX1 (20%), TP53 (18%), and DNMT3A (16%) followed by the splicing factor genes SRSF2 (16%), SF3B1 (15%), and U2AF1 (14%).
Patient characteristics and treatments received by TET2 mutational status
| . | N (%) . | TET-WT . | TET2-mut . | P value* . |
|---|---|---|---|---|
| N | 213 | 155 | 58 | |
| Treatments received | ||||
| AZA alone | 42 (20) | 30 (19) | 12 (21) | .60 |
| DEC alone | 144 (68) | 103 (66) | 41 (71) | |
| DEC + other | 27 (13) | 22 (14) | 5 (9) | |
| Age, ≥70 y | 103 (48) | 72 (46) | 31 (53) | .44 |
| Sex | ||||
| Male | 155 (73) | 118 (76) | 37 (64) | .085 |
| Female | 58 (27) | 37 (24) | 21 (36) | |
| FAB | ||||
| RA | 30 (14) | 19 (12) | 11 (19) | .13 |
| RARS | 24 (11) | 15 (10) | 9 (16) | |
| RAEB | 125 (59) | 97 (63) | 28 (48) | |
| RAEB-t | 6 (3) | 4 (3) | 2 (3) | |
| CMML | 21 (10) | 13 (8) | 8 (14) | |
| Other | 7 (3) | 7 (5) | 0 (0) | |
| IPSS risk group | ||||
| Low | 11 (5) | 5 (3) | 6 (10) | .019 |
| Int-1 | 86 (40) | 56 (36) | 30 (52) | |
| Int-2 | 76 (36) | 61 (39) | 15 (26) | |
| High | 37 (17) | 31 (20) | 6 (10) | |
| Unknown | 3 (1) | 2 (1) | 1 (2) | |
| Cytogenetics | ||||
| Normal or –Y alone | 107 (50) | 68 (44) | 39 (67) | .022 |
| Complex | 51 (24) | 45 (29) | 6 (10) | |
| −7/7q- isolated or +1 | 14 (7) | 12 (8) | 2 (3) | |
| +8 isolated | 11 (5) | 8 (5) | 3 (5) | |
| 20q- isolated | 7 (3) | 6 (4) | 1 (2) | |
| 5q- isolated or +1 | 3 (1) | 3 (2) | 0 (0) | |
| Other | 13 (6) | 9 (6) | 4 (7) | |
| Unknown | 7 (3) | 4 (3) | 3 (5) |
| . | N (%) . | TET-WT . | TET2-mut . | P value* . |
|---|---|---|---|---|
| N | 213 | 155 | 58 | |
| Treatments received | ||||
| AZA alone | 42 (20) | 30 (19) | 12 (21) | .60 |
| DEC alone | 144 (68) | 103 (66) | 41 (71) | |
| DEC + other | 27 (13) | 22 (14) | 5 (9) | |
| Age, ≥70 y | 103 (48) | 72 (46) | 31 (53) | .44 |
| Sex | ||||
| Male | 155 (73) | 118 (76) | 37 (64) | .085 |
| Female | 58 (27) | 37 (24) | 21 (36) | |
| FAB | ||||
| RA | 30 (14) | 19 (12) | 11 (19) | .13 |
| RARS | 24 (11) | 15 (10) | 9 (16) | |
| RAEB | 125 (59) | 97 (63) | 28 (48) | |
| RAEB-t | 6 (3) | 4 (3) | 2 (3) | |
| CMML | 21 (10) | 13 (8) | 8 (14) | |
| Other | 7 (3) | 7 (5) | 0 (0) | |
| IPSS risk group | ||||
| Low | 11 (5) | 5 (3) | 6 (10) | .019 |
| Int-1 | 86 (40) | 56 (36) | 30 (52) | |
| Int-2 | 76 (36) | 61 (39) | 15 (26) | |
| High | 37 (17) | 31 (20) | 6 (10) | |
| Unknown | 3 (1) | 2 (1) | 1 (2) | |
| Cytogenetics | ||||
| Normal or –Y alone | 107 (50) | 68 (44) | 39 (67) | .022 |
| Complex | 51 (24) | 45 (29) | 6 (10) | |
| −7/7q- isolated or +1 | 14 (7) | 12 (8) | 2 (3) | |
| +8 isolated | 11 (5) | 8 (5) | 3 (5) | |
| 20q- isolated | 7 (3) | 6 (4) | 1 (2) | |
| 5q- isolated or +1 | 3 (1) | 3 (2) | 0 (0) | |
| Other | 13 (6) | 9 (6) | 4 (7) | |
| Unknown | 7 (3) | 4 (3) | 3 (5) |
Test includes only known categories, χ2 test used for cytogenetics.
Response vs patient characteristics and treatment
| . | Total . | Nonresponders, n (%) . | Responders, n (%) . | P value* . |
|---|---|---|---|---|
| N | 213 | 113 (53) | 100 (47) | |
| Treatment | ||||
| AZA alone | 42 (20) | 22 (52) | 20 (48) | .96 |
| DEC alone | 144 (68) | 76 (53) | 68 (47) | |
| DEC + Other | 27 (13) | 15 (56) | 12 (44) | |
| Sex | ||||
| Male | 155 (73) | 82 (53) | 73 (47) | .99 |
| Female | 58 (27) | 31 (53) | 27 (47) | |
| Age | ||||
| <70 y | 110 (52) | 64 (58) | 46 (42) | .13 |
| ≥70 y | 103 (48) | 49 (48) | 54 (52) | |
| FAB | ||||
| RA | 30 (14) | 20 (67) | 10 (33) | .008* |
| RARS | 24 (11) | 15 (63) | 9 (38) | |
| RAEB | 125 (59) | 65 (52) | 60 (48) | |
| RAEB-t | 6 (3) | 4 (67) | 2 (33) | |
| CMML | 21 (10) | 4 (19) | 17 (81) | |
| Other | 7 (3) | 5 (71) | 2 (29) | |
| IPSS risk group | ||||
| Low/Int-1 | 97 (46) | 53 (55) | 44 (45) | .78† |
| Int-2/High | 113 (53) | 59 (52) | 54 (48) | |
| Unknown | 3 (1) | 1 (33) | 2 (67) | |
| Cytogenetics | ||||
| Normal | 104 (49) | 49 (47) | 55 (53) | .31 |
| Complex | 51 (24) | 28 (55) | 23 (45) | |
| Other | 51 (24) | 31 (61) | 20 (39) | |
| Unknown | 7 (3) | 5 (71) | 2 (29) |
| . | Total . | Nonresponders, n (%) . | Responders, n (%) . | P value* . |
|---|---|---|---|---|
| N | 213 | 113 (53) | 100 (47) | |
| Treatment | ||||
| AZA alone | 42 (20) | 22 (52) | 20 (48) | .96 |
| DEC alone | 144 (68) | 76 (53) | 68 (47) | |
| DEC + Other | 27 (13) | 15 (56) | 12 (44) | |
| Sex | ||||
| Male | 155 (73) | 82 (53) | 73 (47) | .99 |
| Female | 58 (27) | 31 (53) | 27 (47) | |
| Age | ||||
| <70 y | 110 (52) | 64 (58) | 46 (42) | .13 |
| ≥70 y | 103 (48) | 49 (48) | 54 (52) | |
| FAB | ||||
| RA | 30 (14) | 20 (67) | 10 (33) | .008* |
| RARS | 24 (11) | 15 (63) | 9 (38) | |
| RAEB | 125 (59) | 65 (52) | 60 (48) | |
| RAEB-t | 6 (3) | 4 (67) | 2 (33) | |
| CMML | 21 (10) | 4 (19) | 17 (81) | |
| Other | 7 (3) | 5 (71) | 2 (29) | |
| IPSS risk group | ||||
| Low/Int-1 | 97 (46) | 53 (55) | 44 (45) | .78† |
| Int-2/High | 113 (53) | 59 (52) | 54 (48) | |
| Unknown | 3 (1) | 1 (33) | 2 (67) | |
| Cytogenetics | ||||
| Normal | 104 (49) | 49 (47) | 55 (53) | .31 |
| Complex | 51 (24) | 28 (55) | 23 (45) | |
| Other | 51 (24) | 31 (61) | 20 (39) | |
| Unknown | 7 (3) | 5 (71) | 2 (29) |
Test includes only known categories.
No difference was observed between the 4 individual IPSS categories (P = .24).
Spectrum of mutations in 213 patients in select MDS-associated genes. Each column represents an individual patient sample, and each colored cell represents a mutation of the gene or gene group listed to left of that row. The number of mutations for each row is indicated in the column to the right. Darker bars in the ASXL1 row indicate patients with a p.G642fs mutation. TK Pathway = NRAS, KRAS, CBL, CBLB, JAK2, PTPN11, BRAF, MPL, and KIT.
Spectrum of mutations in 213 patients in select MDS-associated genes. Each column represents an individual patient sample, and each colored cell represents a mutation of the gene or gene group listed to left of that row. The number of mutations for each row is indicated in the column to the right. Darker bars in the ASXL1 row indicate patients with a p.G642fs mutation. TK Pathway = NRAS, KRAS, CBL, CBLB, JAK2, PTPN11, BRAF, MPL, and KIT.
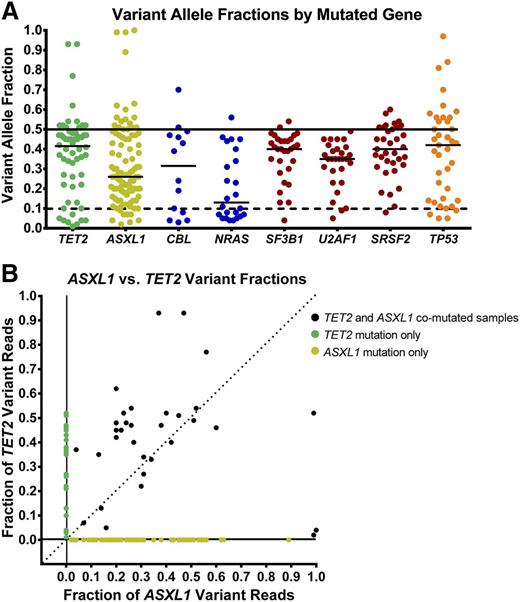
The frequency of mutations identified in these genes was largely similar to those identified in other MDS patient cohorts. Only ASXL1 mutations were more frequent compared with prior studies, many of which examined a greater proportion of lower-risk patients without transfusion dependence; used less sensitive Sanger sequencing of ASXL1; excluded unannotated missense mutations; or excluded insertions in a homopolymeric tract near amino acid 642.7-9 Other previously observed patterns of mutations were identified in this cohort including the paucity of ASXL1 mutations in SF3B1 mutant samples, the near mutual exclusivity of splicing factor mutations, and the lower rate of other gene mutations in patients with TP53 mutations (supplemental Figure 1).20,21 As expected, mutations of TET2, ASXL1, NRAS, EZH2, and SRSF2 were overrepresented in chronic myelomonocytic leukemia (CMML) cases, SF3B1 mutations were predominantly in refractory anemia with ring sideroblasts (RARS) cases, and mutations of TP53, IDH1, and IDH2 were relatively underrepresented in refractory anemia (RA)/RARS patients (supplemental Figure 1). The variant allele fractions (VAFs) of mutations were not uniform and varied greatly for individual genes (Figure 2). For example, splicing factor abnormalities had higher median variant allele fractions, whereas the VAFs for mutations in tyrosine kinase signaling genes were lower, indicative of their frequent presence in disease subclones. Mutations of all genes included some variants present only at low abundance.
Variant allele frequencies in selected genes. (A) Quantitative measure of variant-containing reads estimates the abundance of these mutations (uncorrected for allele copy number). Mutations of TET2 (green), TP53 (orange), and splicing factor genes (red) are often present in the dominant clone, whereas mutations of tyrosine kinase–signaling genes (blue) are often present in smaller clones. Mutations of ASXL1 (yellow) are more widely distributed. (B) Analysis of samples with both TET2 and ASXL1 mutations indicate that ASXL1 mutations are most often codominant with, or smaller than, TET2-mutant clones.
Variant allele frequencies in selected genes. (A) Quantitative measure of variant-containing reads estimates the abundance of these mutations (uncorrected for allele copy number). Mutations of TET2 (green), TP53 (orange), and splicing factor genes (red) are often present in the dominant clone, whereas mutations of tyrosine kinase–signaling genes (blue) are often present in smaller clones. Mutations of ASXL1 (yellow) are more widely distributed. (B) Analysis of samples with both TET2 and ASXL1 mutations indicate that ASXL1 mutations are most often codominant with, or smaller than, TET2-mutant clones.
Clinical findings, variant allele fraction, and response rates
The overall response rate of patients in the study was 47%, with 31% achieving CR according to IWG criteria revised in 2006 (Table 2). There was no significant difference in response by treatment regimen (P = .96) or source of sample (P = .36). IPSS risk groups and cytogenetic abnormalities were not associated with response rate. The only clinical feature significantly associated with response rate was FAB classification (P = .008), driven largely by the high response rate of CMML patients (17/21, 81%). Thirty-five percent of RA/RARS patients achieved a response compared with 47% of refractory anemia with excess blasts (RAEB) patients. No differences were detected in the time to response for each mutation.
In a prior study by Itzykson et al, mutations of TET2 detected by Sanger sequencing were found to predict a nearly twofold greater response rate with AZA.14 In our cohort, TET2 mutant patients showed only a trend toward increased response rates compared with WT (55% vs 44%; OR 1.58 [0.86-2.89], P = .14) and no other mutated gene was associated with a significantly improved overall response rate in univariate analyses (Table 3, supplemental Table 2, and supplemental Figure 2). However, the VAF for mutations of TET2 and several other genes spanned a wide range of values including many likely to be below the detection limit for Sanger sequencing (Figure 2). We hypothesized that mutations capable of sensitizing cells to HMAs are more likely to be associated with a clinical response to treatment when they are present in a major disease clone. For example, even complete elimination of a clone representing <20% of bone marrow cells might not have any effect on the assessment of clinical response. Therefore, we repeated our analysis with mutations present at a VAF of <10% treated as if they were unmutated. For heterozygous mutations, this cutoff represents variants present in <20% of the sample cellularity and is at the estimated limit of sensitivity for Sanger sequencing. In this revised analysis considering VAF, mutations of TET2 were associated with a significantly higher response rate compared with WT (60% vs 43%; OR 1.99 [1.05-3.80], P = .036; adjusted OR 1.98 [1.02-3.85], P = .044), and comparable with that shown by Itzykson et al (Table 3, supplemental Table 2, and supplemental Figure 2).
Association of gene mutations with response rate in logistic regression analysis
| Mutated gene* . | Unadjusted OR (95% CI) . | P value . | Adjusted† OR (95% CI) . | P value . |
|---|---|---|---|---|
| Mutations with VAF ≥10% | ||||
| TET2-mut vs TET2-WT | 1.99 (1.05, 3.80) | .036 | 1.98 (1.02, 3.85) | .044 |
| ASXL-mut vs ASXL1-WT | 0.69 (0.40, 1.20) | .19 | 0.68 (0.38, 1.19) | .17 |
| TET2-mut + ASXL1-WT vs other | 3.65 (1.38, 9.67) | .009 | 3.64 (1.35, 9.79) | .011 |
| TET2-mut + ASXL1-WT vs both WT | 3.40 (1.24, 9.35) | .011 | 3.36 (1.20, 9.38) | .013 |
| TET2-WT + ASXL1-mut vs both WT | 0.77 (0.41, 1.46) | .35 | 0.80 (0.39, 1.46) | .39 |
| TET2-mut + ASXL1-mut vs both WT | 1.11 (0.48, 2.61) | .62 | 1.07 (0.44, 2.61) | .59 |
| CBL-mut vs CBL-WT | 0.27 (0.06, 1.29) | .10 | 0.28 (0.06, 1.40) | .12 |
| Including all mutations | ||||
| TET- mut vs TET2-WT | 1.58 (0.86, 2.89) | .14 | 1.60 (0.85, 3.02) | .15 |
| ASXL1-mut vs ASXL1-WT | 0.77 (0.45, 1.32) | .34 | 0.74 (0.42, 1.30) | .29 |
| TET2-mut + ASXL1-WT vs other | 2.37 (1.00, 5.58) | .049 | 2.40 (0.99, 5.79) | .051 |
| TET2-mut + ASXL1-WT vs both WT | 2.27 (0.91, 5.63) | .055 | 2.27 (0.89, 5.79) | .056 |
| TET2-WT + ASXL1-mut vs both WT | 0.86 (0.45, 1.64) | .16 | 0.84 (0.43, 1.62) | .15 |
| TET2-mut + ASXL1-mut vs both WT | 1.06 (0.47, 2.38) | .67 | 1.04 (0.45, 2.44) | .68 |
| CBL-mut vs CBL-WT | 0.17 (0.04, 0.79) | .023 | 0.18 (0.04, 0.82) | .027 |
| Mutated gene* . | Unadjusted OR (95% CI) . | P value . | Adjusted† OR (95% CI) . | P value . |
|---|---|---|---|---|
| Mutations with VAF ≥10% | ||||
| TET2-mut vs TET2-WT | 1.99 (1.05, 3.80) | .036 | 1.98 (1.02, 3.85) | .044 |
| ASXL-mut vs ASXL1-WT | 0.69 (0.40, 1.20) | .19 | 0.68 (0.38, 1.19) | .17 |
| TET2-mut + ASXL1-WT vs other | 3.65 (1.38, 9.67) | .009 | 3.64 (1.35, 9.79) | .011 |
| TET2-mut + ASXL1-WT vs both WT | 3.40 (1.24, 9.35) | .011 | 3.36 (1.20, 9.38) | .013 |
| TET2-WT + ASXL1-mut vs both WT | 0.77 (0.41, 1.46) | .35 | 0.80 (0.39, 1.46) | .39 |
| TET2-mut + ASXL1-mut vs both WT | 1.11 (0.48, 2.61) | .62 | 1.07 (0.44, 2.61) | .59 |
| CBL-mut vs CBL-WT | 0.27 (0.06, 1.29) | .10 | 0.28 (0.06, 1.40) | .12 |
| Including all mutations | ||||
| TET- mut vs TET2-WT | 1.58 (0.86, 2.89) | .14 | 1.60 (0.85, 3.02) | .15 |
| ASXL1-mut vs ASXL1-WT | 0.77 (0.45, 1.32) | .34 | 0.74 (0.42, 1.30) | .29 |
| TET2-mut + ASXL1-WT vs other | 2.37 (1.00, 5.58) | .049 | 2.40 (0.99, 5.79) | .051 |
| TET2-mut + ASXL1-WT vs both WT | 2.27 (0.91, 5.63) | .055 | 2.27 (0.89, 5.79) | .056 |
| TET2-WT + ASXL1-mut vs both WT | 0.86 (0.45, 1.64) | .16 | 0.84 (0.43, 1.62) | .15 |
| TET2-mut + ASXL1-mut vs both WT | 1.06 (0.47, 2.38) | .67 | 1.04 (0.45, 2.44) | .68 |
| CBL-mut vs CBL-WT | 0.17 (0.04, 0.79) | .023 | 0.18 (0.04, 0.82) | .027 |
Reference group is listed second.
Adjusted for sex, age (<70, ≥70 y), IPSS (Low/Int1 vs Int2/High), and treatment (AZA alone vs DEC alone vs DEC + other); none of the Hosmer and Lemeshow tests indicated a lack of fit for each model.
When all VAFs were considered, only mutations of CBL, which were often of low VAF, were associated with a lower rate of response compared with WT (14% vs 49%) in this analysis (OR 0.17 [0.04-0.79], P = .023; adjusted OR 0.18 [0.04-0.82], P = .027) but was not significant when low VAF mutations were considered WT (20% vs 48%) (OR 0.27 [0.06-1.30], P = .10).
By sequencing multiple genes, we had the opportunity to determine whether mutations in additional genes could modulate the response rates of TET2-mutant patients. We focused on the subset of patients defined by their TET2 and ASXL1 mutation status (at any VAF) because these contained enough patients for a meaningful statistical analysis. Patients with mutated TET2 and unmutated ASXL1 demonstrated an increased overall response rate compared with all others (65% vs 44%; OR 2.37, [1.00-5.58], P = .049) (Table 3). This effect was more pronounced when mutations were required to have a VAF ≥10% (74% vs 44%; OR 3.65, [1.38-9.67], P = .009), representing >10% of patients in this cohort (Table 3).
In vivo model of AZA response in Tet2−/− cells
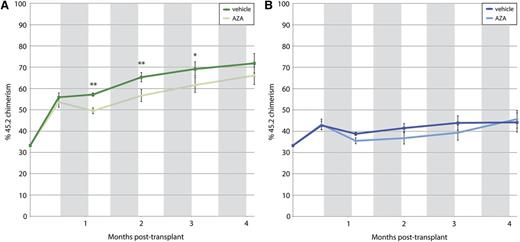
The observed association between TET2 mutations and response to treatment could be mediated directly by TET2 loss-of-function or by indirect or cell-extrinsic effects. To test whether TET2 loss-of-function can sensitize cells to HMAs, we performed a competitive murine bone marrow transplant experiment using hematopoietic cells from Tet2-null and WT littermate donors. As expected, equal numbers of CD45.2+ cells transplanted into CD45.1+ recipients resulted in greater engraftment of Tet2-null cells by 2.5 weeks posttransplant, before treatment with AZA. There was no difference in peripheral blood counts between groups at this time point. Treatment with AZA (2.5 mg/kg M-F × 2 weeks) or vehicle was begun on day 20 posttransplant and repeated starting on days 48, 76, and 104. Regardless of genotype, AZA-treated animals exhibited significant decreases in white blood cell and hematocrit levels and an initial drop in peripheral blood chimerism. For several subsequent cycles, AZA-treated Tet2-null cells maintained a significantly decreased representation in peripheral blood, whereas Tet2-WT cells did not (Figure 3).
Peripheral blood chimerism. Shown over time after competitive bone marrow transplantation with cells from 45.2 Tet2-null mice (A) and 45.2 Tet2-WT mice (B). Gray bars indicate periods of treatment with AZA or vehicle. Tet2-null cells show increased chimerism compared with Tet2-WT cells. Treatment with AZA significantly decreases chimerism in the Tet2-null recipient mice only. *P < .05, **P < .01.
Peripheral blood chimerism. Shown over time after competitive bone marrow transplantation with cells from 45.2 Tet2-null mice (A) and 45.2 Tet2-WT mice (B). Gray bars indicate periods of treatment with AZA or vehicle. Tet2-null cells show increased chimerism compared with Tet2-WT cells. Treatment with AZA significantly decreases chimerism in the Tet2-null recipient mice only. *P < .05, **P < .01.
Associations with overall survival
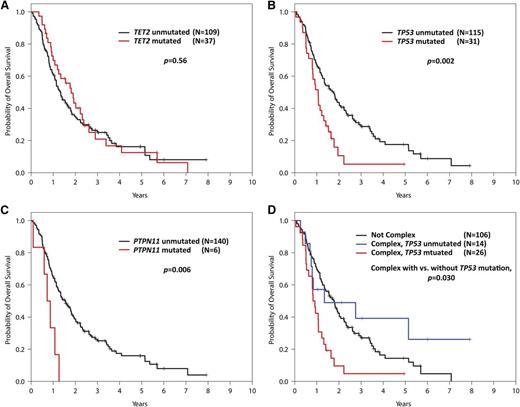
Traditional prognostic models like the IPSS and Revised IPSS (IPSS-R) are based on patient cohorts examined only until they receive disease-modifying therapies such as HMAs or they undergo stem cell transplantation. Response to specific treatments could significantly alter the prognostic impact of adverse disease features or genetic alterations. We explored the relationship between mutation status and overall survival in the subset of patients with available survival data. Of these 146 patients (69%) in our cohort, 119 died during follow-up. The median follow-up for patients remaining alive was 3.8 years (95% CI, 3.1-5.8). Despite its association with response, TET2 mutation status was not associated with overall survival (P = .56), consistent with the finding in Itzykson et al (Figure 4A). Mutations of TP53 were associated with lower overall survival (21% of patients; HR 2.01 [1.29-3.14], P = .002; adjusted HR 1.91 [1.20-3.05], P = .007; Figure 4B) as were the much rarer mutations of PTPN11 (4% of patients, HR 3.26 [1.41-7.58], P = .006; adjusted HR 2.47 [0.98-6.26], P = .056; Figure 4C).
Kaplan-Meier curves for overall survival in the 146 out of 213 study patients with survival data. (A) Survival of patients with and without TET2 mutations. (B) Survival of patients with and without TP53 mutations. (C) Survival of patients with and without PTPN11 mutations. (D) Survival of complex karyotype patients with and without TP53 mutations vs patients without complex karyotypes.
Kaplan-Meier curves for overall survival in the 146 out of 213 study patients with survival data. (A) Survival of patients with and without TET2 mutations. (B) Survival of patients with and without TP53 mutations. (C) Survival of patients with and without PTPN11 mutations. (D) Survival of complex karyotype patients with and without TP53 mutations vs patients without complex karyotypes.
Overall survival in patients with complex karyotypes was strongly associated with TP53 mutation status (Figure 4D). Patients with both a complex karyotype and a TP53 mutation had a median survival of only 0.9 years. In contrast, patients with complex cytogenetics and no TP53 mutation had an overall survival of 1.3 years, which was not different from patients with karyotypes other than complex (median 1.8 years, P = .28). This indicates that the adverse prognostic value ascribed to the complex karyotype is largely driven by its frequent association with TP53 mutations, which could be used to better refine disease risk in this patient population.
Discussion
In our study, the presence of TET2 mutation at >10% allele burden predicted an increased response to HMAs, particularly in the subset that lacked similarly abundant mutations of ASXL1. To achieve this result, we examined tumor samples from 213 MDS patients collected before treatment with HMAs for mutations in 40 genes known to be recurrently mutated in MDS. The patients in our cohort were representative of those studied in clinical trials of AZA and DEC in terms of predicted disease risk and severity of cytopenias. Overall response rates were just under 50% and did not differ by the type of drug patients received. Using sensitive quantitative sequencing techniques, we were able to identify mutations in >90% of patients in patterns similar to those seen in prior multigene studies of MDS.
Our findings are consistent with those of Itzykson et al, who previously reported that 11 of 13 (85%) MDS patients with TET2 mutations detected by Sanger sequencing responded to treatment with AZA compared with a 52% response rate for their overall cohort of 86 patients. In that study, mutations in other genes were not examined, and small subclonal TET2 mutations likely went undetected. Our broader and more sensitive multigene analysis similarly identified TET2 mutations as predictive of response to HMAs in a larger cohort of patients. Surprisingly, consideration of mutations in other genes did not reveal additional predictors of favorable response, and inclusion of low VAF mutations weakened the association between TET2 mutation status and response rate. However, our approach identified the 10% of patients with mutated TET2 and WT ASXL1 as the group most likely to respond to treatment. Potential explanations for this finding include partial resistance to HMAs caused by ASXL1 mutations. In this model, the ASXL1-mutated subclone would be expected to grow in size during disease progression, or relapse, and might confer primary resistance. ASXL1-mutated patients with WT TET2 did have a lower likelihood of response, but this was not statistically significant (OR 0.63 [0.35-1.15], P = .13). Alternatively, we observed that ASXL1 mutations were often subclonal or at a lower VAF than TET2 mutations in comutated patients (Figure 2C). The acquisition of secondary mutations (of which ASXL1 was the most frequent) could indicate more clonally progressive disease that might be inherently more resistant to treatment.
The mechanism by which TET2 mutations might influence response to HMAs is not clear. Altered methylation has been observed in patients with TET2 mutations and in animal models of Tet2 loss. However, measurement of pretreatment DNA methylation by itself has not been found to be predictive of response to HMAs.22 In our murine bone marrow transplant experiment, exposure to AZA preferentially decreased the clonal advantage associated with loss of Tet2 function. This effect may be associated with a greater AZA sensitivity in more actively cycling cells because AZA results in cell division–dependent passive demethylation of DNA. Mice with hematopoietic Tet2 loss are known to have increased myeloid progenitor proliferation.19,23-25
An important finding of our study was that no pattern of mutation was strongly associated with a lack of response to treatment. Responses to HMAs were observed even in patients with mutations that confer a very poor prognosis. Therefore, our data indicate that mutation information alone should not be used as a basis for denying therapy with an HMA if treatment is indicated. Studies examining samples collected at multiple time points are needed to identify mutations predictive of acquired resistance or relapsed disease.26-28
The association between molecular or clinical biomarkers and HMA response may be confounded by the variations in enzymes responsible for the activation and metabolism of AZA and DEC.11,29-31 Patients who demonstrate significant hypomethylation of blood-cell DNA after treatment with AZA or DEC (indicating sufficient exposure to target DNA methyltransferases) may be more likely to have a clinically significant response.22,32 It is possible that the predictive value of cell-intrinsic somatic mutations may be enhanced if controlled for cell-extrinsic variables such as effective dose of HMAs received and activated in cells.
Survival data were collected for more than two-thirds of our cohort. Mutation profiles capable of predicting response to HMAs were not associated with differences in overall survival (Figure 3). However, mutations in 2 genes that were not predictive of response, TP53 and the rarer PTPN11, were each associated with decreased overall survival. The majority of TP53 mutant patients had a complex karyotype, a known adverse risk factor associated with shorter overall survival. More than half of our complex-karyotype patients harbored a TP53 mutation (32/51), and these patients had a very short overall survival (median 0.9 years). However, complex karyotype patients without a detectable TP53 mutation had an overall survival that was no different from the group of patients with noncomplex karyotypes. This indicates that the negative prognostic significance attributed the complex karyotype can be better explained by the TP53 mutation status of these patients and validates the results of recent studies in MDS and AML.7,33 In addition to TP53, mutations in any of 4 additional genes—RUNX1, ASXL1, EZH2, or ETV6—were found to predict shorter overall survival than expected by examining clinical features alone. However, mutations of these 4 genes were not found to be prognostically adverse in this cohort of treated patients (supplemental Figure 3). In contrast, samples for our previous study were collected before the approval of AZA and DEC and therefore came largely from untreated patients. Our results suggest that treatment with HMAs may partially abrogate the adverse prognostic impact of these lesions. If validated, our finding would form a justification for treating patients whose adverse prognosis is driven by mutations in these genes.
The clinical implications of our findings are that response to hypomethylating therapy can be predicted in a subset of patients using molecular genetic features. A more robust predictor might be created by incorporating clinical findings or other biomarkers.5,34,35 Indeed, the Groupe Français des Myélodysplasies has presented a clinically and cytogenetically based prognostic model for AZA-treated patients, although its predictive power is unclear.5,6,36 As with these clinical measures, no mutations identified in our study were reliably strong predictors of primary resistance to treatment in a large number of patients. Therefore, there is no genetic rationale for denying MDS patients the opportunity to be treated with AZA or DEC based on our findings, particularly because there are few alternative therapies approved for this patient population.
In conclusion, means of reliably predicting response to HMAs would be of clinical benefit in the care of patients with MDS. Our study demonstrates that mutation profiles can help in this effort to some extent. Studies examining the mechanism by which these biomarkers might mediate sensitivity or resistance to treatment would be of clinical value and could lead to the discovery of additional therapeutic targets in MDS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by the National Institutes of Health, National Heart, Lung and Blood Institute (R01HL082945), a Department of Defense bone marrow failure research program grant, a Leukemia and Lymphoma Society Scholar Award (B.L.E.), the National Institute of Diabetes and Digestive and Kidney Diseases (K08DK091360), and an ASH Scholar Award (R.B.). In addition, this research was supported in part by the MD Anderson Cancer Center Leukemia Support Grant (CCSG) CA016672, grant MDS P01 CA108631, and the Fundacion Ramon Areces.
Authorship
Contribution: R.B., B.L.E., and D.P.S. designed the study; M.B.-N., R.M.S., G.G.-M., H.K., and D.P.S. collected samples, curated clinical data, and edited the manuscript; R.B., A.P.-L., J.Z., H.W., B.C., and R.C. carried out DNA sequencing; P.S., G.G., J.Z., and R.B. performed DNA sequencing analysis; A.L. performed the murine experiments; R.B., K.S., and D.N. carried out the statistical analysis; and R.B., B.L.E., K.S., and D.N. wrote the manuscript, which was edited by all authors.
Conflict-of-interest disclosure: R.B., B.L.E., G.G.-M., and D.P.S. have consulted for Genoptix and Celgene. R.B., B.L.E., K.S., and D.N. have IP licensed by Genoptix.
Correspondence: Benjamin Ebert, Brigham and Women’s Hospital, 1 Blackfan Circle–Karp CHRB 5.211, Boston, MA 02115; e-mail: bebert@partners.org; and Rafael Bejar, Moores Cancer Center, University of California, San Diego, 3855 Health Sciences Dr MC 0820, La Jolla, CA 92093-0820; e-mail: rabejar@ucsd.edu.