Key Points
miR-217 enhances the GC reaction by dampening genotoxic-induced Bcl-6 degradation in GC B cells.
miR-217 is an oncogene and its overexpression provides a model of miRNA-induced mature B-cell lymphomagenesis.
Abstract
microRNAs are a class of regulators of gene expression that have been shown critical for a great number of biological processes; however, little is known of their role in germinal center (GC) B cells. Although the GC reaction is crucial to ensure a competent immune response, GC B cells are also the origin of most human lymphomas, presumably due to bystander effects of the immunoglobulin gene remodeling that takes place at these sites. Here we report that miR-217 is specifically upregulated in GC B cells. Gain- and loss-of-function mouse models reveal that miR-217 is a positive modulator of the GC response that increases the generation of class-switched antibodies and the frequency of somatic hypermutation. We find that miR-217 down-regulates the expression of a DNA damage response and repair gene network and in turn stabilizes Bcl-6 expression in GC B cells. Importantly, miR-217 overexpression also promotes mature B-cell lymphomagenesis; this is physiologically relevant as we find that miR-217 is overexpressed in aggressive human B-cell lymphomas. Therefore, miR-217 provides a novel molecular link between the normal GC response and B-cell transformation.
Introduction
microRNAs (miRNAs) are negative regulators of gene expression that influence virtually all biological processes. A high proportion of human miRNAs are located in cancer-associated genomic regions,1 and deregulated miRNA expression is a hallmark of most cancer types, including lymphomas.2-4 Indeed, miRNA profiling is increasingly recognized as a valuable tool for the classification and even prognosis of lymphoma. However, the role of specific miRNAs in the development of B-cell lymphoma has rarely been addressed in vivo;5 specifically, functional evidence for the involvement of miRNAs in the development of mature B-cell lymphomas is very scarce. This void is particularly important because the vast majority of human B-cell lymphomas originate from mature B cells.
The predominance of mature B-cell lymphomas is most likely due to the unique molecular events taking place in B cells during the immune response. B cells generate a hugely diverse repertoire of antibodies, which enable specific immune responses against virtually any pathogen the organism may encounter. Antibody diversity is achieved in 2 stages. The first takes place during B-cell differentiation through V(D)J recombination. The second stage is unique to mature B cells that have encountered antigen and takes place in germinal centers (GCs), allowing the generation of higher affinity memory B cells and plasma cells.
The GC reaction involves the clonal expansion of antigen-specific B lymphocytes and the generation of B-cell subclones with related antigen specificities, from which those expressing immunoglobulins with improved affinity for the antigen are positively selected.6 Molecularly, this is triggered by the so-called somatic hypermutation (SHM), which introduces point mutations on the variable region of the Ig molecule—responsible for antigen recognition. In addition, GCs are the home of the class switch recombination (CSR) reaction, a region-specific recombination reaction between 2 switch regions of the immunoglobulin heavy chain (IgH) locus that generates antibodies with different isotypes. SHM and CSR are both initiated by activation-induced deaminase (AID), which deaminates cytosines on the immunoglobulin locus.7 This initial lesion on DNA is subsequently processed by DNA repair and recombination factors to allow the fixation of a mutation, in the case of SHM, or the generation of a DNA double-strand break and a recombination reaction, in the case of CSR.8
The fact that inactivation of AID causes an immunodeficiency in humans9 highlights the importance of the GC reaction in the immune response. However, the generation of mutations and double-strand breaks associated with the GC reaction entails a risk for the genome integrity of mature B lymphocytes, resulting in a permissive environment for lymphomagenesis.10-12 Indeed, the most prominent hallmark of mature B-cell lymphomas is the presence of recurrent chromosomal translocations that involve the immunoglobulin locus and an oncogene.13,14 These illegitimate junctions are a direct consequence of AID activity in the GC reaction, because AID deficiency prevents the generation of c-myc-IgH translocations associated with Burkitt lymphoma (BL)15-17 and reduces the incidence of mature B-cell lymphoma in different mouse models.15,18 In addition, Bcl-6, a transcriptional repressor indispensable for the GC reaction, seems to provide an environment tolerant to the DNA damage associated with AID-dependent events, which contributes to its oncogenic activity.19 Remarkably, close to 80% of all human B-cell lymphomas originate from B cells that are GC experienced—generally known as mature B-cell lymphomas13—the most aggressive of which are BL and diffuse large B-cell lymphomas (DLBCLs).
Tight regulation of the GC reaction is thus critical with regard both to the efficacy of the immune response and to lymphoma development. miRNAs promote subtle changes in gene expression through imperfect pairing with their target mRNAs.20,21 Very often a single miRNA can bind hundreds of different target mRNAs, thus acting as a regulator of a gene network rather than individual genes.20,22 miRNAs have previously been implicated in the regulation of the immune response and tumorigenic processes and mice whose GC B cells are depleted of miRNAs mount inefficient GC responses.23 However, there is little direct evidence for the role of individual miRNAs in this context, and to date, only miR-155 has been clearly associated with the GC response.24-26 Here we show that miR-217 is upregulated after in vitro and in vivo stimulation of mature B cells. Using gain and loss of function in vivo models, we found that miR-217 is a positive regulator of the GC reaction and an oncogene in GC B cells.
Methods
miR-217 detection by miRNA microarray hybridization and quantitative reverse transcriptase-polymerase chain reaction
RNA was extracted with Trizol (Invitrogen). Mouse miRNA microarray analysis was performed with paired samples of nonstimulated and stimulated B cells.27 For quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) of the miR-217 precursor, total RNA was converted to cDNA using random primers (Roche), reversed transcribed with SuperScript II (Invitrogen), and quantified by SYBR Green assay (Applied Biosystems). Primers are detailed in supplemental Methods available on the Blood Web site. Mature miR-217 was quantified from total RNA with miR-217 miRCURY LNA primers (Exiqon). U6 and sno-142 (Exiqon) were used as normalization controls.
T cell-dependent immunizations
Groups of 7 to 9 littermate mice were immunized by footpad injection with 50 μg of 4-hydroxy-3-nitrophenylacetyl (NP) hapten conjugated to chicken γ globulin (NP-CGG; Biosearch Technologies) in complete Freund’s adjuvant. Mice were euthanized 14 days after immunization for analysis of primary response or were reimmunized with 50 μg of NP-CGG in incomplete Freund’s adjuvant.
Somatic mutation analysis
For analysis of mutations at the Sμ and JH4 regions, GC B cells were purified by cell sorting of pooled Peyer’s patches (4-6 animals per genotype). DNA was extracted, and the Sμ and JH4 regions were PCR-amplified using specific oligonucleotides (supplemental Methods). Amplification products were purified and sequenced by next-generation sequencing (NGS)28 or cloned and sequenced by conventional Sanger sequencing.
RNAseq analysis and miRNA target prediction
GC (CD19+Fas+GL7+) B cells from pooled Peyer’s patches (4-6 animals per genotype) from control and miR-217TG mice were isolated by cell sorting. Total RNA was extracted with Trizol (Invitrogen) and sequenced by RNAseq. Differential expression analysis was done using the edgeR package from Bioconductor,29 and genes with P ≤ .05 were considered for analysis. Pathway analysis of the significantly up- and downregulated genes in miR-217TG vs control GC B cells was performed by ingenuity pathway analysis software (Ingenuity Systems).
Significantly downregulated genes in miR-217TG GC B cells were analyzed for the presence of 835 different miRNAs target sites based on predicted and experimentally validated miRNA-mRNA interactions. Scores for every predictive algorithm were normalized, and the compound score of predictive and experimental databases was calculated.30 See supplemental Methods for more details.
miR-217 expression in Ramos cells, immunobloting, and cell cycle analysis
Ramos cells were transduced with miR-217 pmiRNA or control vectors27 and selected by green fluorescent protein (GFP) expression by flow-activated cell sorting (FACS). Cells were treated for 6 hours with etoposide (Sigma-Aldrich) in the presence or absence of caffeine (Sigma-Aldrich), lysed in radioimmunoprecipitation assay buffer, and immunobloted with anti-Bcl-6 (N-3; Santa Cruz Biotechnology) or α-tubulin (Sigma-Aldrich) antibodies or fixed overnight with 70% ethanol and stained with 2.5 μg/mL 4′,6 diamidino-2-phenylindole for flow cytometry cell cycle analysis.
Human samples and animal procedures
The use of human samples was approved by the ethics committee of the Instituto de Salud Carlos III. The study was conducted in accordance with the Declaration of Helsinki.
All animal procedures conformed to European Union Directive 2010/63EU and Recommendation 2007/526/EC regarding the protection of animals used for experimental and other scientific purposes, enforced in Spanish law under Real Decreto 1201/2005.
Results
miR-217 expression is upregulated in activated B cells
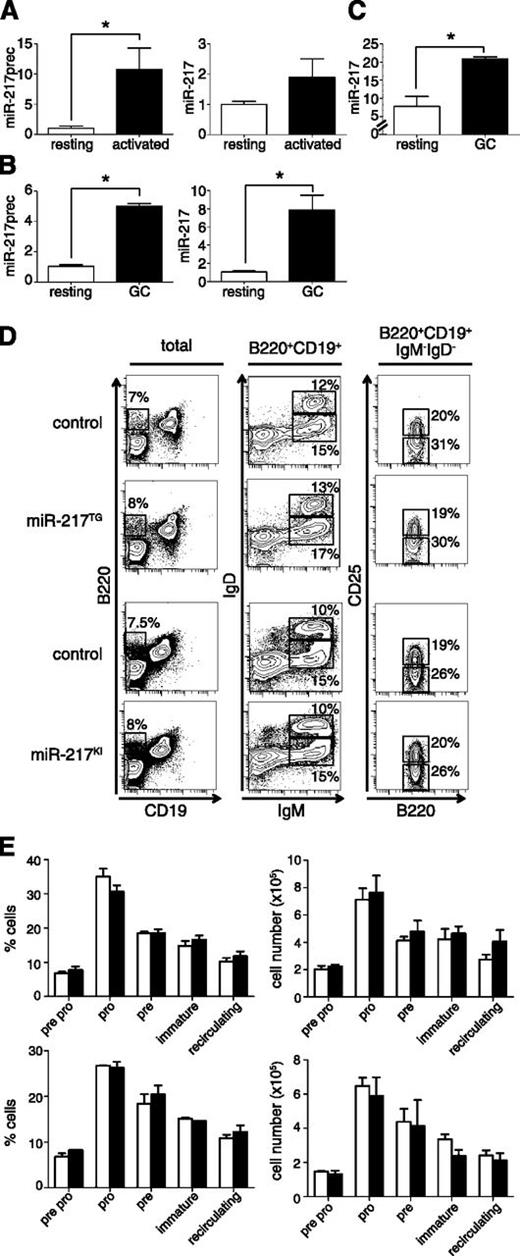
To identify miRNAs that play a role during B-cell activation, we profiled miRNA expression in mature B cells during in vitro activation with lipopolysaccharide (LPS) + interleukin (IL)4, which promotes AID expression, SHM in the switch regions, and CSR. Microarray analysis showed that one of the few miRNAs whose expression is upregulated during this process is miR-21727 (supplemental Figure 1A). Induction of precursor and mature miR-217 during B-cell activation in vitro was confirmed by qRT-PCR (Figure 1A). In addition, we generated GCs in vivo by immunizing mice with sheep red blood cells and conducted qRT-PCR on purified resting and GC B cells. miR-217 precursor and mature miR-217 expression increased by five- and eightfold, respectively, in GC cells (Figure 1B). Similarly, we found that miR-217 expression is increased in GC human B cells compared with resting B cells (Figure 1C). Together, these results show that miR-217 expression is induced during B-cell activation both in mouse and humans.
miR-217 expression is upregulated in activated B cells. (A) qRT-PCR analysis of the expression of the (left) miR-217 precursor and (right) mature miR-217 in B cells after 1 (open bars) and 3 days (filled bars) of activation with LPS + IL4. Data from 3 (miR-217 precursor) and 2 (mature miR-217) independent experiments are shown (P = .05 for miR-217 precursor and P = .27 for miR-217). Each experiment was performed with 2 individual mice. (B) qRT-PCR of the (left) miR-217 precursor and (right) mature miR-217 in resting (open bars) or GC (filled bars) B cells isolated from spleens of wild-type C57BL/6 mice 10 days after immunization with sheep red blood cells. Data are means ± standard deviation from 2 independent experiments with 10 total immunized mice (P = .0002 for miR-217 precursor and P = .01 for miR-217). Two-tailed Student t test: P values: *P < .05. (C) Quantification of miR-217 expression in human resting (CD19+CD27−IgD−) and GC (CD19+CD10+) samples as measured by miRNA array hybridization (data were obtained from GSE29493, P = .003, 2-tailed Student t test). (D) Representative fluorescence-activated cell sorter (FACS) analysis of bone marrow from miR-217TG, miR-217KI and littermate control mice. Plots show the expression of (left) B220/CD19 in total live lymphocytes, (center) IgD/IgM gated in B220+CD19+ cells, and (right) CD25/B220 gated in B220+CD19+IgM−IgD− cells. The proportion of cells is indicated in each gated in B220+ cells. (E) Quantification of the proportions and absolute cell numbers of developing bone marrow B-cell subsets in control (open bars), (upper) miR-217TG (filled bars), and (lower) miR-217KI mice (filled bars). The proportions of different B-cell subsets were quantified within the B220+ population.
miR-217 expression is upregulated in activated B cells. (A) qRT-PCR analysis of the expression of the (left) miR-217 precursor and (right) mature miR-217 in B cells after 1 (open bars) and 3 days (filled bars) of activation with LPS + IL4. Data from 3 (miR-217 precursor) and 2 (mature miR-217) independent experiments are shown (P = .05 for miR-217 precursor and P = .27 for miR-217). Each experiment was performed with 2 individual mice. (B) qRT-PCR of the (left) miR-217 precursor and (right) mature miR-217 in resting (open bars) or GC (filled bars) B cells isolated from spleens of wild-type C57BL/6 mice 10 days after immunization with sheep red blood cells. Data are means ± standard deviation from 2 independent experiments with 10 total immunized mice (P = .0002 for miR-217 precursor and P = .01 for miR-217). Two-tailed Student t test: P values: *P < .05. (C) Quantification of miR-217 expression in human resting (CD19+CD27−IgD−) and GC (CD19+CD10+) samples as measured by miRNA array hybridization (data were obtained from GSE29493, P = .003, 2-tailed Student t test). (D) Representative fluorescence-activated cell sorter (FACS) analysis of bone marrow from miR-217TG, miR-217KI and littermate control mice. Plots show the expression of (left) B220/CD19 in total live lymphocytes, (center) IgD/IgM gated in B220+CD19+ cells, and (right) CD25/B220 gated in B220+CD19+IgM−IgD− cells. The proportion of cells is indicated in each gated in B220+ cells. (E) Quantification of the proportions and absolute cell numbers of developing bone marrow B-cell subsets in control (open bars), (upper) miR-217TG (filled bars), and (lower) miR-217KI mice (filled bars). The proportions of different B-cell subsets were quantified within the B220+ population.
Generation of B cell-specific miR-217 mouse models
To investigate the role of miR-217 during B-cell activation, we generated 2 independent mouse models of B cell-specific miR-217 overexpression. In the first, 4 independent transgenic mouse lines were generated with a construct encoding the miR-217 precursor and a GFP reporter gene under the control of regulatory elements of the mouse κ light chain (Igκ) gene (miR-217TG). In the second strategy, the miR-217 precursor/GFP construct was preceded by a transcriptional stop cassette flanked by LoxP sites and inserted within the endogenous Rosa26 locus (ROSA26miR217ki/+ mice) (supplemental Figure 1B; supplemental Methods); specific expression of miR-217 in B cells was achieved by crossing ROSA26miR217ki/+ mice with CD19−Creki/+ mice (hereafter, miR-217KI mice). ROSA26miR217+/+CD19−Creki/+ mice were used as controls. B cells from miR-217TG and miR-217KI mice showed full GFP and miR-217 expression in mature splenic B cells (supplemental Figure 1C-D). GFP expression was not detected in non-B-cell lineages, such as T, myeloid, or epithelial cells (data not shown). The proportions and absolute numbers of bone marrow and peripheral B cells in the miR-217TG and miR-217KI models were similar to those in wild-type littermate controls (Figure 1D-E), indicating that the miR-217TG and miR-217KI mouse models allow B cell-specific miR-217 overexpression without perturbing B-cell differentiation.
miR-217 expression enhances the GC reaction
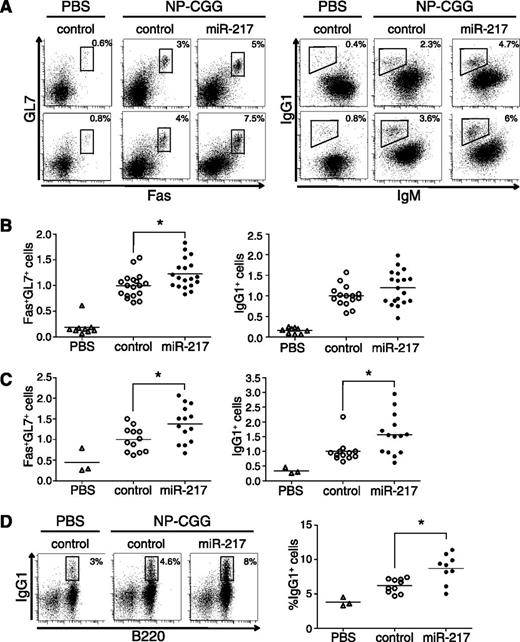
To address the role of miR-217 expression during B-cell activation in GCs, we analyzed the B-cell response to a T cell-dependent antigen in miR-217TG mice. We first immunized miR-217TG mice and wild-type littermate controls with NP-CGG. As an immunization control, mice were injected with phosphate-buffered saline (PBS). The lymph nodes of NP-CGG-immunized wild-type mice contained GC B cells (Fas+GL7+) and B cells that had undergone CSR (IgG1+); but in miR-217TG mice, the response to NP-CGG immunization was greater, with 22% more GC B cells (P = .01) and 20% more IgG1+ B cells (P = .08; Figure 2A-B). A similarly enhanced GC response to NP-CGG immunization was found in miR-217KI mice (supplemental Figure 2A). We next analyzed the secondary B-cell immune response in control and miR-217TG mice. The enhancement of the GC reaction in miR-217TG mice was notably greater in secondary immunization assays, with 40% more Fas+GL7+ B cells and 60% more IgG1+-switched B cells than controls (P = .019 and .018, respectively; Figure 2A,C). In addition, we found that the proportion of CD138+ plasma cells is larger in miR-217TG mice than in controls (P = .02; supplemental Figure 2B). To analyze the influence of miR-217 overexpression on the long-lived memory B-cell compartment, we quantified IgG1+ B cells in spleens from control and miR-217KI mice 1 year after immunization. The proportion of memory B cells was 40% higher in miR-217KI mice than in controls (P = .003; Figure 2D).
miR-217 expression in B cells enhances the GC response. (A) Representative FACS analysis of B220+ lymphocytes in lymph nodes after primary (upper) or secondary (lower) NP-CGG immunization of miR-217TG mice or littermate controls. (B) miR-217TG mice (closed circles) or littermate controls (open circles) were immunized by subcutaneous injection of NP-CGG in complete Freund’s adjuvant, and 14 days after immunization, lymph nodes were analyzed for the presence of GC B cells (Fas+GL7+) and switched B cells (IgG1+). Mice injected with PBS (open triangles) were used as nonimmunized controls. Each symbol represents an individual mouse. Data were normalized to the mean value of control mice in each experiment; results are shown for a total of 4 experiments performed with 4 independent miR-217TG mouse lines (P = .01 for GC B cells and P = .08 for switched B cells). (C) Fourteen days after a primary immunization as in B, mice were reimmunized by subcutaneous injection of NP-CGG and analyzed 7 days later. Symbol code as in B. Data from 2 independent experiments was normalized to the mean response of control mice in each experiment (P = .019 for GC and P = .018 for switched B cells). (D) Spleen memory IgG1+ B cells analyzed by flow cytometry 1 year after NP-CGG immunization of miR-217KI mice. The FACS plots show the frequency of cells in the gate referred to proportion within B220+ cells. Symbol code as in B (P = .003).
miR-217 expression in B cells enhances the GC response. (A) Representative FACS analysis of B220+ lymphocytes in lymph nodes after primary (upper) or secondary (lower) NP-CGG immunization of miR-217TG mice or littermate controls. (B) miR-217TG mice (closed circles) or littermate controls (open circles) were immunized by subcutaneous injection of NP-CGG in complete Freund’s adjuvant, and 14 days after immunization, lymph nodes were analyzed for the presence of GC B cells (Fas+GL7+) and switched B cells (IgG1+). Mice injected with PBS (open triangles) were used as nonimmunized controls. Each symbol represents an individual mouse. Data were normalized to the mean value of control mice in each experiment; results are shown for a total of 4 experiments performed with 4 independent miR-217TG mouse lines (P = .01 for GC B cells and P = .08 for switched B cells). (C) Fourteen days after a primary immunization as in B, mice were reimmunized by subcutaneous injection of NP-CGG and analyzed 7 days later. Symbol code as in B. Data from 2 independent experiments was normalized to the mean response of control mice in each experiment (P = .019 for GC and P = .018 for switched B cells). (D) Spleen memory IgG1+ B cells analyzed by flow cytometry 1 year after NP-CGG immunization of miR-217KI mice. The FACS plots show the frequency of cells in the gate referred to proportion within B220+ cells. Symbol code as in B (P = .003).
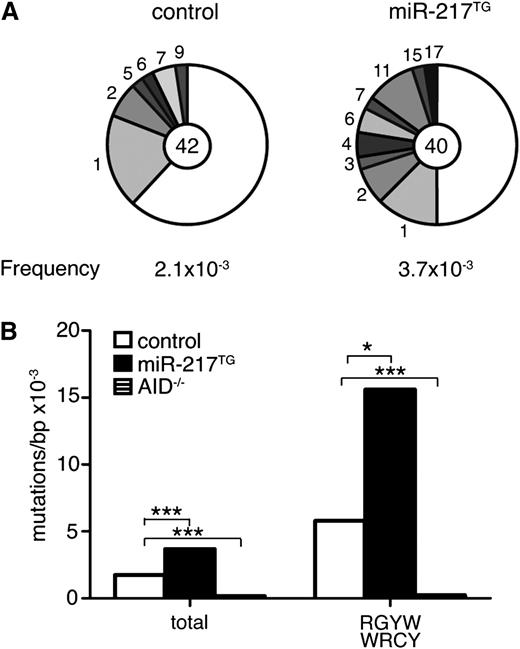
To further characterize the role of miR-217 in the GC reaction, we examined the extent of SHM in miR-217TG mice in vivo. We isolated Fas+GL7+ GC B cells from Peyer’s patches of control and miR-217TG mice and quantified the mutation frequency in an intronic DNA region downstream of the JH4 segment of the IgH locus, a region that accumulates mutations but cannot be subject to affinity maturation events. Conventional Sanger sequencing of the JH4 intronic region showed that miR-217TG B cells have a higher mutation load than wild-type controls (control, 2.1 × 10−3/bp and miR-217TG, 3.7 × 10−3/bp, P = .028; Figure 3A). Likewise, GC B cells from miR-217KI mice accumulated more mutations downstream of JH4 than did wild-type controls (supplemental Figure 2C). Similar results were obtained when we analyzed SHM frequency at the JH4 intronic region by NGS, which allows large numbers of mutations to be analyzed at very high depth.28 No mutations were detected in B cells isolated from AID−/− mice, used as a negative control. We detected total mutation frequencies of 1.7 × 10−3/bp in control animals and 3.7 × 10−3/bp in miR-217TG mice (P = 1.9 × 10−9), a 2.2-fold difference (Figure 3B). This difference was even higher (2.7-fold) when the analysis was limited to cytosines located in AID mutational hotspots RGYW/WRCY (where R = A/G, Y = C/T, and W = A/T) (5.8 × 10−3/bp in controls vs 15.6 × 10−3/bp in miR-217TG; Figure 3B), confirming that miR-217 overexpression in B cells increases the load of SHM in GCs. These results indicate that overexpression of miR-217 in B cells increases the efficiency of GC formation, CSR and SHM, and the generation of terminally differentiated B cells in vivo.
miR-217 expression in B cells enhances SHM. (A-B) Quantification of mutations in the JH4 intronic sequence of GC (Fas+GL7+) B cells isolated from pooled Peyer’s patches of 6 control and 6 miR-217TG mice: (A) Sanger sequencing. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated at the periphery. Mutation frequencies are indicated below each chart, and the total numbers of independent sequences analyzed are indicated in the chart centers. Statistical significance was determined by a 2-tailed Student t test (P = .028). (B) NGS quantification of the total JH4 mutation frequency and of the mutation frequency at cytosines within the RGYW/WRCY AID hotspots in JH4 sequences in GC B cells from control (open bars) and miR-217TG mice (filled bars). The mutation frequency in the JH4 sequence of AID−/− splenic B cells is shown in hatched bars. A total of 300 000 kb/genotype was sequenced. Statistical significance of the mean mutation frequency at each position was determined by a paired Student t test (P = 1.9 × 10−9 for total mutation frequency and P = .002 for mutation frequency at WRCY hotspots).
miR-217 expression in B cells enhances SHM. (A-B) Quantification of mutations in the JH4 intronic sequence of GC (Fas+GL7+) B cells isolated from pooled Peyer’s patches of 6 control and 6 miR-217TG mice: (A) Sanger sequencing. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated at the periphery. Mutation frequencies are indicated below each chart, and the total numbers of independent sequences analyzed are indicated in the chart centers. Statistical significance was determined by a 2-tailed Student t test (P = .028). (B) NGS quantification of the total JH4 mutation frequency and of the mutation frequency at cytosines within the RGYW/WRCY AID hotspots in JH4 sequences in GC B cells from control (open bars) and miR-217TG mice (filled bars). The mutation frequency in the JH4 sequence of AID−/− splenic B cells is shown in hatched bars. A total of 300 000 kb/genotype was sequenced. Statistical significance of the mean mutation frequency at each position was determined by a paired Student t test (P = 1.9 × 10−9 for total mutation frequency and P = .002 for mutation frequency at WRCY hotspots).
Inhibition of endogenous miR-217 impairs the GC reaction
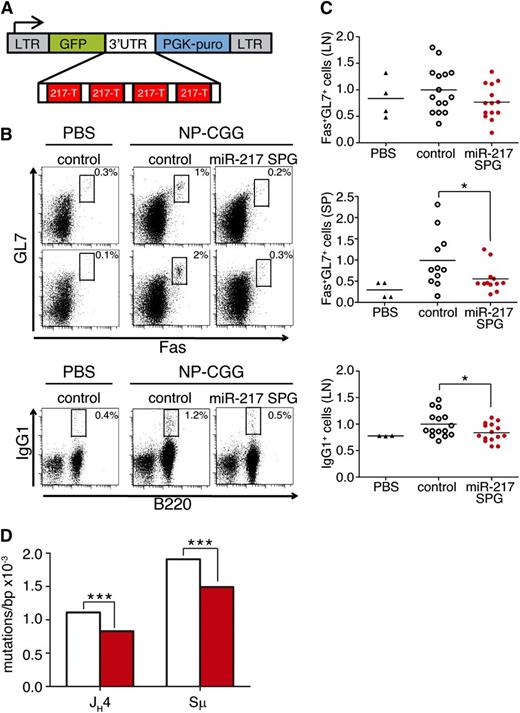
To address whether endogenous miR-217 expression plays a physiological role in GCs, we generated bone marrow mouse chimeras in which endogenous miR-217 expression was inhibited by the expression of a miR-217 sponge (miR-217SPG) construct (Figure 4A), where sequences complementary to miR-217 were cloned in tandem within the 3′untranslated region of a reporter gene and thus function as competitive inhibitors for the binding of the miRNA to its endogenous binding sites.31,32 Bone marrow cells from wild-type mice were transduced with miR-217SPG or control retroviruses and transferred into lethally irradiated CD45.1 congenic recipient mice and immunized 4 weeks later with NP-CGG. Chimeric mice injected with PBS were used as immunization negative controls. Spleen and lymph nodes from immunized miR-217SPG mice contained fewer GC and IgG1+ switched B cells than immunized control chimeric mice, and indeed, the proportions of these cell subsets in immunized miR-217SPG mice were as low as in PBS-injected, nonimmunized animals (Figure 4B-C). We next analyzed SHM in Peyer’s patch GC B cells from miR-217SPG and control chimeric mice by NGS. We found that miR-217SPG GC B cells had a 25% lower mean mutation frequency than control B cells at the JH4 intronic region (P = 3.7 × 10−5; Figure 4D). Very similar results were obtained when we analyzed the mutation load in a fragment of the μ switch region (Sμ) of IgH (P = 4 × 10−4), where mutations accumulate concomitantly with CSR (Figure 4D). These results thus show that inhibition of endogenous miR-217 attenuates the GC reaction and decreases the frequency of somatic mutations associated with both the SHM and the CSR reactions.
Inhibition of endogenous miR-217 expression in B cells impairs the GC response. (A) miR-217-Sponge (miR-217SPG) retroviral construct. Four miR-217 complementary sites, separated by 4-nt spacers, were placed downstream of GFP in the MGP vector. (B-C) GC response of mouse chimeras reconstituted with control or miR-217SPG retrovirally transduced bone marrow precursors analyzed 14 days after NP-CGG immunization. (B) Representative FACS analysis of B220+ GC cells (Fas+GL7+) in (upper) lymph nodes and (lower) spleen and of switched (IgG1+) lymph node cells after NP-CGG immunization. Plots are gated on CD45.1+GFP+ cells. (C) Quantification of GC and switched B cells in mouse chimeras reconstituted with control (open circles) or miR-217 SPG (in red) retrovirally transduced bone marrow precursors. Nonimmunized mice injected with PBS (triangles) were included as immunization controls. Each symbol represents an individual mouse. Data are normalized to the mean response of control mice of 2 independent experiments. Statistical significance was determined by a 2-tailed Student t test vs control immunized mice (P = .12 for LN GCs, P = .05 for SP GCs, and P = .04 for switched B-cell generation). (D) NGS quantification of mutations in (left) JH4 and (right) Sµ sequences in GFP+CD45.1+ GC (Fas+GL7+) B cells isolated from pooled Peyer’s patches of 8 control (open bars) and 8 miR-217SPG (red bars) mouse chimeras. Data are from 2 (JH4) and 1 (Sµ) independent experiments. At least 190 000 kb was sequenced per genotype. Statistical significance of the mean total mutation frequency at each position was determined by a paired Student t test (P = 3.7 × 10−5 for JH4 SHM and P = 4 × 10−4 for Sµ SHM).
Inhibition of endogenous miR-217 expression in B cells impairs the GC response. (A) miR-217-Sponge (miR-217SPG) retroviral construct. Four miR-217 complementary sites, separated by 4-nt spacers, were placed downstream of GFP in the MGP vector. (B-C) GC response of mouse chimeras reconstituted with control or miR-217SPG retrovirally transduced bone marrow precursors analyzed 14 days after NP-CGG immunization. (B) Representative FACS analysis of B220+ GC cells (Fas+GL7+) in (upper) lymph nodes and (lower) spleen and of switched (IgG1+) lymph node cells after NP-CGG immunization. Plots are gated on CD45.1+GFP+ cells. (C) Quantification of GC and switched B cells in mouse chimeras reconstituted with control (open circles) or miR-217 SPG (in red) retrovirally transduced bone marrow precursors. Nonimmunized mice injected with PBS (triangles) were included as immunization controls. Each symbol represents an individual mouse. Data are normalized to the mean response of control mice of 2 independent experiments. Statistical significance was determined by a 2-tailed Student t test vs control immunized mice (P = .12 for LN GCs, P = .05 for SP GCs, and P = .04 for switched B-cell generation). (D) NGS quantification of mutations in (left) JH4 and (right) Sµ sequences in GFP+CD45.1+ GC (Fas+GL7+) B cells isolated from pooled Peyer’s patches of 8 control (open bars) and 8 miR-217SPG (red bars) mouse chimeras. Data are from 2 (JH4) and 1 (Sµ) independent experiments. At least 190 000 kb was sequenced per genotype. Statistical significance of the mean total mutation frequency at each position was determined by a paired Student t test (P = 3.7 × 10−5 for JH4 SHM and P = 4 × 10−4 for Sµ SHM).
miR-217 regulates a DNA damage response and repair gene network and stabilizes Bcl-6 expression in GC B cells
To identify the genes regulated by miR-217 in activated B cells, we performed a genome-wide comparison of the gene expression profiles of miR-217TG and wild-type GC B cells by RNAseq. Comparison of GC B cells from Peyer’s patches of miR-217TG and wild-type mice identified 1740 differentially expressed genes with P ≤ .05 (supplemental Table 1), 1236 of which were downregulated. This set of downregulated genes was significantly enriched in predicted miR-217 targets (P = 9.75 × 10−7; Figure 5A), suggesting that a high proportion of the downregulated genes identified in miR-217TG GC B cells are direct miR-217 targets. To identify the gene networks altered by miR-217 expression in GC B cells, we performed a bioinformatics analysis on the genes differentially expressed in miR-217TG vs control GC (Fas+GL7+) B cells. Ingenuity pathway analysis revealed that miR-217 regulates a DNA damage response (DDR) and repair gene network in GC B cells (P = 10−31; Figure 5B). Interestingly, this gene network contained 2 main hubs, RNA pol II and replication protein A, which are directly linked to AID activity.33-37 The miR-217-regulated gene network included a number of genes linked to the DDR (Nbs1 and Rad50 components of the Mre11-Rad50-Nbs1 [MRN] complex),38 DNA repair and CSR (Wrn, Lig4, and Xrcc2),11,39,40 and genes of the cohesin complex (SMC2, SMC3, PDS5B, and SA2) involved in chromatid cohesion, as well as in DNA replication, transcription, recombination, and repair41,42 ; all of them were downregulated by miR-217.
miR-217 regulates a DNA damage response and repair gene network and dampens Bcl-6 degradation in GC B cells. (A-B) RNAseq analysis of genome-wide gene expression in control and miR-217TG GC B cells isolated from Peyer’s patches. Pools of 4 to 6 mice per genotype were used in each experiment. Data from 2 independent experiments are shown. (A) Enrichment in miRNA prediction targets within the 1236 mRNAs whose expression was down-regulated in miR-217TG vs control GC B cells. The GEO accession number for the RNAseq data is GSE47877. (B) Ingenuity pathway analysis of the genes differentially expressed in control vs miR-217TG GC B cells, showing enrichment for a DDR and repair gene network among the genes regulated by miR-217 (P = 10−31). Downregulated genes are shown in green; upregulated genes are shown in red. (C) Quantification of Nbs1, Xrcc2, Lig4, and Pds5b expression in Ramos cells transduced with control (open bars) or miR-217 (filled bars) retrovirus analyzed by qRT-PCR. Two-tailed Student t test with P values: *P < .05. (D) Cell cycle analysis of etoposide-treated control- and miR-217-transduced Ramos cells. Representative cell cycle plots of untreated (−Etop) or etoposide-treated (6 hours, 10 μM) (+Etop) Ramos cells. Fraction of cells in S cycle ± standard deviation is shown. The lower graph depicts the change in the proportion of cells at each phase of cell cycle in response to etoposide treatment (open bars, control-transduced cells; black bars, miR-217-transduced cells). Bars represent means ± standard deviations from 3 independent experiments (P = .004 for S phase cells, 2-tailed Student t test). (E) (Upper) Immunoblot of Bcl-6 in Ramos cells transduced with control or miR-217 retrovirus treated for 6 hours with the indicated doses of etoposide. (Lower) Immunoblot of Bcl-6 in miR-217-transduced cells left untreated (−Etoposide) or treated with 10 μM Etposide for 6 hours (+Etoposide) in the presence of increases doses of caffeine (0 mM [−], 2 mM [+], and 5 mM [++]. (F) Analysis of Bcl-6 expression in GC cells by flow cytometry. (Upper) Representative dot plots of Fas, GL7, and Bcl-6 staining of Peyer’s patch B cells. Quantification of Bcl-6 expression levels in B220+Fas+GL7+ GC B cells from Peyer’s patches of littermate controls (white bar) and miR-217KI mice (black bar) (MFI, mean fluorescence intensity; n = 3 mice/genotype, P = .0006, 2-tailed Student t test). (G) Bcl-6 expression in spleens of control and miR-217TG mice analyzed by immunohistochemical staining. A representative picture is shown. The proportion of Bcl-6+ stained in follicle areas was quantified in 205 and 201 spleen follicles of control and 10 miR-217TG mice, respectively. Statistical significance was determined by 2-tailed Student t test (P = .0008).
miR-217 regulates a DNA damage response and repair gene network and dampens Bcl-6 degradation in GC B cells. (A-B) RNAseq analysis of genome-wide gene expression in control and miR-217TG GC B cells isolated from Peyer’s patches. Pools of 4 to 6 mice per genotype were used in each experiment. Data from 2 independent experiments are shown. (A) Enrichment in miRNA prediction targets within the 1236 mRNAs whose expression was down-regulated in miR-217TG vs control GC B cells. The GEO accession number for the RNAseq data is GSE47877. (B) Ingenuity pathway analysis of the genes differentially expressed in control vs miR-217TG GC B cells, showing enrichment for a DDR and repair gene network among the genes regulated by miR-217 (P = 10−31). Downregulated genes are shown in green; upregulated genes are shown in red. (C) Quantification of Nbs1, Xrcc2, Lig4, and Pds5b expression in Ramos cells transduced with control (open bars) or miR-217 (filled bars) retrovirus analyzed by qRT-PCR. Two-tailed Student t test with P values: *P < .05. (D) Cell cycle analysis of etoposide-treated control- and miR-217-transduced Ramos cells. Representative cell cycle plots of untreated (−Etop) or etoposide-treated (6 hours, 10 μM) (+Etop) Ramos cells. Fraction of cells in S cycle ± standard deviation is shown. The lower graph depicts the change in the proportion of cells at each phase of cell cycle in response to etoposide treatment (open bars, control-transduced cells; black bars, miR-217-transduced cells). Bars represent means ± standard deviations from 3 independent experiments (P = .004 for S phase cells, 2-tailed Student t test). (E) (Upper) Immunoblot of Bcl-6 in Ramos cells transduced with control or miR-217 retrovirus treated for 6 hours with the indicated doses of etoposide. (Lower) Immunoblot of Bcl-6 in miR-217-transduced cells left untreated (−Etoposide) or treated with 10 μM Etposide for 6 hours (+Etoposide) in the presence of increases doses of caffeine (0 mM [−], 2 mM [+], and 5 mM [++]. (F) Analysis of Bcl-6 expression in GC cells by flow cytometry. (Upper) Representative dot plots of Fas, GL7, and Bcl-6 staining of Peyer’s patch B cells. Quantification of Bcl-6 expression levels in B220+Fas+GL7+ GC B cells from Peyer’s patches of littermate controls (white bar) and miR-217KI mice (black bar) (MFI, mean fluorescence intensity; n = 3 mice/genotype, P = .0006, 2-tailed Student t test). (G) Bcl-6 expression in spleens of control and miR-217TG mice analyzed by immunohistochemical staining. A representative picture is shown. The proportion of Bcl-6+ stained in follicle areas was quantified in 205 and 201 spleen follicles of control and 10 miR-217TG mice, respectively. Statistical significance was determined by 2-tailed Student t test (P = .0008).
Bcl-6 expression is regulated by the DDR through a signaling pathway that promotes Bcl-6 degradation.43 As we found that miR-217 downregulates the expression of a number of genes involved in DDR and repair in GC B cells, we hypothesized that miR-217 could stabilize Bcl-6 expression in these cells. To test this hypothesis, we transduced Ramos cells, a human BL GC B-cell line, with miR-217-GFP and control retroviral vectors and analyzed Bcl-6 expression on genotoxic stress induction. We first verified that genes linked to the DDR and downregulated by miR-217 expression in mouse GC B cells (Nbs1, Xrcc2, Lig4, and Pds5b) were also downregulated by miR-217 in human Ramos cells (Figure 5C). To determine if miR-217 impacts on the response to genotoxic stress, we analyzed the effect of etoposide treatment on the cell cycle of control- and miR-217-transduced Ramos cells. Expectedly, we found that etoposide-induced DDR promoted an increase in the fraction of cells in the S phase of the cell cycle, consistent with a replicative delay (Figure 5D). However, this S phase increase was milder in miR-217-transduced cells (Figure 5D), which suggested that miR-217 may contribute to bypass the DDR response induced by etoposide. Immunoblot analysis showed that etoposide treatment of control Ramos cells rapidly induced Bcl-6 degradation (Figure 5E), in agreement with previous results.43 However, Ramos cells overexpressing miR-217 were partially protected against genotoxic stress-induced Bcl-6 degradation and showed increased Bcl-6 expression levels (Figure 5E). Caffeine treatment protected miR-217 Ramos cells from etoposide-induced Bcl-6 degradation, indicating that an ataxia telangiectasia mutated/ataxia telangiectasia and Rad3-related–dependent pathway is involved in this process (Figure 5E, lower panels). Analysis of Bcl-6 protein levels in GC B cells from control and miR-217KI mice further confirmed that miR-217 expression promotes the accumulation of Bcl-6 (Figure 5F). Finally, quantification of Bcl-6 expression in spleens from miR-217TG and control mice revealed that miR-217TG mice have larger Bcl-6+ follicle areas in spleens (P = .0008; Figure 5G). Overall, these data indicate that miR-217 regulates the GC reaction by modulating the expression levels of a gene network involved in DDR and repair and by dampening genotoxic stress-induced Bcl-6 degradation in GC B cells.
Deregulated miR-217 expression promotes mature B-cell lymphomas
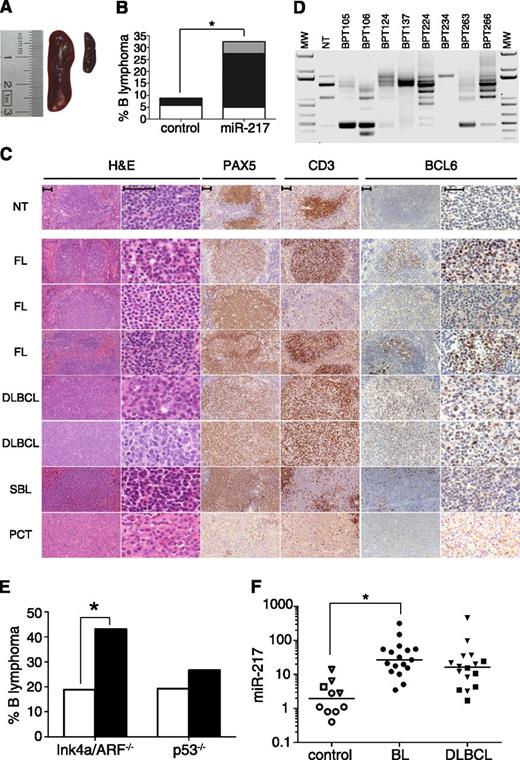
GC B cells are highly prone to oncogenic transformation. Our results show that miR-217 is a positive regulator of the GC reaction, presumably by downregulating the expression of DNA repair genes and by stabilizing Bcl-6 expression, which could enhance the susceptibility of these cells to oncogenic events. To test whether miR-217 overexpression in B cells affects the development of mature B-cell lymphomas, we monitored the incidence of B-cell lymphoma in a group of 34 control and 40 miR-217KI mice. Mice were euthanized and analyzed at an end point of 90 weeks or earlier if they showed signs of disease. miR-217KI mice showed early signs of disease more frequently than control mice did (14 miR-217KI and 7 control mice had to be euthanized before 90 weeks; supplemental Figure 3A). miR-217KI mice had larger spleens than control animals, with a fraction showing clear splenomegaly (20% vs 0% spleens larger than 20 mm; Fisher’s exact test, P = .0058; Figure 6A; Table 1). Histopathological evaluation of the spleens showed that a large proportion (33%) of miR-217KI mice developed B-cell lymphoma (P = .028 vs control mice; Figure 6B). Flow cytometry analysis of lymphomas from miR-217KI mice showed frequent alterations in the expression of the B-cell surface molecules B220, IgD, IgM, and Igκ (supplemental Figure 3B; Table 1). We found that lymphomas in miR-217KI mice had features of mature GC or post-GC B-cell origin.44 Most (70%) were classified as follicular lymphomas (FL), although we also observed other mature B-cell lymphomas, such as DLBCL and B-cell plasmacytomas (PCT) (Figure 6B-C; Table 1). To further characterize the origin of these lymphomas, we performed molecular analysis of their V(D)J rearrangements by PCR amplification and sequencing. We found that a fraction of the lymphomas yielded unique amplification bands (Figure 6D), which were confirmed to correspond with single rearrangements (Table 2), thus providing proof of their clonal origin. In addition, all of the rearrangements subject to this analysis contained mutations (SHM+) in their V genes (Table 2) and an additional fraction showed Bcl-6 expression (Figure 6C; Table 1), confirming their GC/post-GC origin.
miR-217 expression promotes mature B-cell lymphomas. (A-D) Thirty-four control and 40 miR-217KI mice were monitored over 90 weeks for the appearance of B-cell lymphoma. (A) Representative pictures of (right) a nontumoral control spleen and (left) a miR-217KI spleen with splenomegaly. (B) Quantification of B-cell lymphoma incidence in control mice and miR-217KI mice. The proportions of FL (black), DLBCL (white), or other lymphomas (gray) are shown. Statistical significance was determined by 2-tailed Student t test (P = .028). (C) Representative hematoxilin and eosin (H&E) and immunohistochemical stainings for Pax5, CD3, and Bcl-6 in spleens from miR-217KI mice with B-cell lymphomas. Scale bar is 50 μm. NT, nontumoral; SBL, small B-cell lymphoma. (D) PCR analysis of V(D)J rearrangements in tumor samples from miR-217KI mice. DNA was isolated from total spleen and amplified using a V-degenerate primer in combination with an antisense primer downstream of JH4. Sequencing results are shown in Table 2. Mouse IDs are shown. MW, molecular weight marker. (E) Quantification of B-cell lymphoma incidence in control (open bars) and miR-217KI (filled bars) Ink4a/Arf−/− and p53−/− mice. Statistical significance was determined by 2-tailed Student t test (P = .03 in miR217KI Ink4a/Arf−/− vs control Ink4a/Arf−/− and P = .33 in miR217KI p53−/− vs control p53−/−). (F) miR-217 relative expression in human control samples (open circles, peripheral blood CD19+ B cells; open squares, tonsils; open triangles, lymph nodes), in BL samples (closed circles) and in DLBCL samples (closed triangles, GC B GCB-DLBCL; closed squares, activated B cell, ABC-DLBCL) as determined by qRT-PCR. Statistical significance was determined by 2-tailed Student t test (P = .05 for BL).
miR-217 expression promotes mature B-cell lymphomas. (A-D) Thirty-four control and 40 miR-217KI mice were monitored over 90 weeks for the appearance of B-cell lymphoma. (A) Representative pictures of (right) a nontumoral control spleen and (left) a miR-217KI spleen with splenomegaly. (B) Quantification of B-cell lymphoma incidence in control mice and miR-217KI mice. The proportions of FL (black), DLBCL (white), or other lymphomas (gray) are shown. Statistical significance was determined by 2-tailed Student t test (P = .028). (C) Representative hematoxilin and eosin (H&E) and immunohistochemical stainings for Pax5, CD3, and Bcl-6 in spleens from miR-217KI mice with B-cell lymphomas. Scale bar is 50 μm. NT, nontumoral; SBL, small B-cell lymphoma. (D) PCR analysis of V(D)J rearrangements in tumor samples from miR-217KI mice. DNA was isolated from total spleen and amplified using a V-degenerate primer in combination with an antisense primer downstream of JH4. Sequencing results are shown in Table 2. Mouse IDs are shown. MW, molecular weight marker. (E) Quantification of B-cell lymphoma incidence in control (open bars) and miR-217KI (filled bars) Ink4a/Arf−/− and p53−/− mice. Statistical significance was determined by 2-tailed Student t test (P = .03 in miR217KI Ink4a/Arf−/− vs control Ink4a/Arf−/− and P = .33 in miR217KI p53−/− vs control p53−/−). (F) miR-217 relative expression in human control samples (open circles, peripheral blood CD19+ B cells; open squares, tonsils; open triangles, lymph nodes), in BL samples (closed circles) and in DLBCL samples (closed triangles, GC B GCB-DLBCL; closed squares, activated B cell, ABC-DLBCL) as determined by qRT-PCR. Statistical significance was determined by 2-tailed Student t test (P = .05 for BL).
Characterization of B-cell lymphomas in miR-217KI mice
| Mouse ID* . | B-cell lymphoma† . | Grade‡ . | Splenomegaly§ . | Phenotype¶ . |
|---|---|---|---|---|
| BPT048 | FL | Mature B cell, B220+ CD19+ sIgM+ sIgκ+ Fas+GL7+, Pax5low BCL6+; numerous infiltrating T cells | ||
| BPT100 | FL | Yes | Mature B cells, Pax5+ BCL6− and N/A | |
| BPT105 | FL | Mature B cell, B220+ CD19+ sIgM− sIgκ+, Pax5+ BCL6+ | ||
| BPT106 | FL | Mature B cell, B220+ CD19+ sIgM− sIgκ+, Pax5low BCL6+ | ||
| BPT109 | PCT | Aggressive | Mature B cell, Pax5- BCL6- and N/A | |
| BPT124 | FL | Aggressive | Mature B cell, B220low CD19+ sIgMhigh sIgκ+, Pax5low BCL6- | |
| BPT137 | FL | Mature B cell, B220+ CD19+ sIgMlow sIgκ+, Pax5+ BCL6- | ||
| BPT148 | DLBCL | Aggressive | Yes | Mature B cell, B220+ CD19+ sIgMlow sIgκ+, Pax5low BCL6+; numerous infiltrating T cells |
| BPT224 | DLBCL | Aggressive | Yes | Mature B cell, B220+ CD19+ sIgMlow sIgκlow, Pax5low BCL6+; numerous infiltrating T cells |
| BPT226 | FL | Aggressive | Yes | Mature B cell, B220+ CD19+ sIgMlow sIgκlow, Pax5low BCL6+; numerous infiltrating T cells |
| BPT234 | SBL | Mature B cell, B220+ CD19+ sIgMhigh sIgκ+, Pax5+ BCL6- | ||
| BPT263 | FL | Yes | Mature B cell, B220high CD19+ sIgM+ sIgκ+, Pax5+ BCL6+ | |
| BPT266 | FL | Aggressive | Mature B cell, B220+ CD19+ sIgMlow sIgκ+, Pax5low BCL6− |
| Mouse ID* . | B-cell lymphoma† . | Grade‡ . | Splenomegaly§ . | Phenotype¶ . |
|---|---|---|---|---|
| BPT048 | FL | Mature B cell, B220+ CD19+ sIgM+ sIgκ+ Fas+GL7+, Pax5low BCL6+; numerous infiltrating T cells | ||
| BPT100 | FL | Yes | Mature B cells, Pax5+ BCL6− and N/A | |
| BPT105 | FL | Mature B cell, B220+ CD19+ sIgM− sIgκ+, Pax5+ BCL6+ | ||
| BPT106 | FL | Mature B cell, B220+ CD19+ sIgM− sIgκ+, Pax5low BCL6+ | ||
| BPT109 | PCT | Aggressive | Mature B cell, Pax5- BCL6- and N/A | |
| BPT124 | FL | Aggressive | Mature B cell, B220low CD19+ sIgMhigh sIgκ+, Pax5low BCL6- | |
| BPT137 | FL | Mature B cell, B220+ CD19+ sIgMlow sIgκ+, Pax5+ BCL6- | ||
| BPT148 | DLBCL | Aggressive | Yes | Mature B cell, B220+ CD19+ sIgMlow sIgκ+, Pax5low BCL6+; numerous infiltrating T cells |
| BPT224 | DLBCL | Aggressive | Yes | Mature B cell, B220+ CD19+ sIgMlow sIgκlow, Pax5low BCL6+; numerous infiltrating T cells |
| BPT226 | FL | Aggressive | Yes | Mature B cell, B220+ CD19+ sIgMlow sIgκlow, Pax5low BCL6+; numerous infiltrating T cells |
| BPT234 | SBL | Mature B cell, B220+ CD19+ sIgMhigh sIgκ+, Pax5+ BCL6- | ||
| BPT263 | FL | Yes | Mature B cell, B220high CD19+ sIgM+ sIgκ+, Pax5+ BCL6+ | |
| BPT266 | FL | Aggressive | Mature B cell, B220+ CD19+ sIgMlow sIgκ+, Pax5low BCL6− |
Mouse identification number.
SBL, small B-cell lymphoma.
Lymphoma classification grade.
Splenomegaly: spleens larger than 20 mm length.
Phenotype of tumoral cells determined by morphological features, flow cytometry, and immunohistochemica stainings. N/A, not analyzed.
Analysis of VDJ gene rearrangements in clonal B-cell lymphomas of miR-217KI mice
| Mouse ID* . | V gene† . | D gene† . | J gene† . | V-D-J junction . | SHM‡ . |
|---|---|---|---|---|---|
| BPT105 | IGHV1-66*01 | IGHD2-5*01 | IGHJ4*01 | GGGGGCTATAGTCAAATGAGGGGG | + |
| BPT124 | IGHV1S16*01 | IGHD1-2*01 | IGHJ2*01 | GAGGGGATTACTACGGCTACC | + |
| BPT137 | IGHV1S127*01 | IGHD4-1*02 | IGHJ2*01 | GACTGGGACGTCGG | + |
| BPT234 | IGHV1-47*01 | IGHD2-4*01 | IGHJ1*01 | ATGATTACGACCACC | + |
| Mouse ID* . | V gene† . | D gene† . | J gene† . | V-D-J junction . | SHM‡ . |
|---|---|---|---|---|---|
| BPT105 | IGHV1-66*01 | IGHD2-5*01 | IGHJ4*01 | GGGGGCTATAGTCAAATGAGGGGG | + |
| BPT124 | IGHV1S16*01 | IGHD1-2*01 | IGHJ2*01 | GAGGGGATTACTACGGCTACC | + |
| BPT137 | IGHV1S127*01 | IGHD4-1*02 | IGHJ2*01 | GACTGGGACGTCGG | + |
| BPT234 | IGHV1-47*01 | IGHD2-4*01 | IGHJ1*01 | ATGATTACGACCACC | + |
IGBLAST, immunoglobulin BLAST; IMGT international ImMunoGeneTics information system; NCBI, National Center for Biotechnology Information.
Mouse identification number.
Assignment of V, D, and J genes by NCBI/IGBLAST according to the IMGT database.
SHM.
To examine the contribution of tumor suppressor pathways to limiting the B-cell lymphomagenesis induced by miR-217, we analyzed the incidence of B-cell lymphoma in miR-217KI mice in the Ink4a/Arf−/− and p53−/− backgrounds. Ink4a/Arf−/− and p53−/− mice often developed histiocytic sarcomas (40%) and T-cell lymphomas (70%), respectively (data not shown).45-47 In addition, we found that roughly 20% of both control Ink4a/Arf−/− and p53−/− tumor suppressor-deficient mice generated B-cell lymphomas. This incidence was not significantly altered by miR-217 overexpression in the p53−/− background, but in the Ink4a/Arf−/− background, miR-217 overexpression increased B-cell lymphoma incidence to 43% (Figure 6E), although the mean survival of miR-217KI Ink4a/Arf−/− mice was not significantly altered (supplemental Figure 3A). B-cell lymphomas in miR-217KI Ink4a/Arf−/− mice showed histopathological features of mature GC or post-GC B-cell lymphomas (supplemental Figure 3). These results suggest that the Ink4a/Arf, but not the p53, tumor suppressor pathway acts as a surveillance mechanism against the lymphomagenic events induced by miR-217 in mature B cells.
To determine if miR-217 expression levels are altered in mature B-cell human lymphomas, we conducted a qRT-PCR analysis of miR-217 expression in a collection of BL and DLBCL samples. Expression of miR-217 was higher in BL and DLBCL than in control tonsil, lymph node, and peripheral blood B-cell samples (BL vs control, P = .05; Figure 6F). In agreement with these findings, analysis of published data of copy number variation in a cohort of DLBCL cases revealed that the genomic region that contains the miR-217 chromosomal location (MCR 1694) is amplified in a fraction of the cases48 (GSE11318). Together, these data indicate that miR-217 gain of function is associated with human mature B-cell lymphomas.
Discussion
The results presented in this study identify miR-217 as a positive regulator of the GC reaction and as an oncogene that promotes mature B-cell lymphomagenesis. miR-217 is specifically upregulated as a result of B-cell stimulation in the context of the GC reaction during the immune response, and our gain and loss of function approaches directly establish the functional relevance of miR-217 in vivo. miR-217 overexpression boosted the number of GC B cells and promoted the SHM and CSR reactions, and conversely, inhibition of endogenous miR-217 limited these events. Interestingly, miR-217 gain of function did not promote any measurable alterations in B-cell differentiation, suggesting that the function of miR-217 in the B-cell lineage is restricted to the context of GCs and antibody diversification.
miRNAs modulate the expression of gene networks in a cell context-specific manner.20,22 Here, RNAseq analysis showed that miR-217 regulates the expression of a gene network involved in DDR and repair, including Rad50, Nbs1, Wrn, Lig4, and XRCC2, as well as a set of genes of the cohesin complex, all of which are down-regulated by miR-217. GC B cells are intrinsically prone to genome instability: (1) they are programmed to undergo AID-mediated gene remodeling; (2) the intense proliferation of GC B cells subjects them to replicative stress6 ; and (3) the GC reaction depends on Bcl-6, a master transcriptional repressor that dampens the DDR in GC B cells and whose deregulation generates B-cell lymphomas.18,49 Our data show that miR-217 downregulates a network of genes that sense and repair genotoxic events on DNA, which in turn can increase the tolerance of GC B cells to DNA damage, very much like Bcl-6. Notably, we found that miR-217 protects Bcl-6 from genotoxic stress-induced degradation,43 suggesting that both molecules are part of the same network that renders GC cells permissive to genomic instability and prone to malignant transformation.
Consistent with this idea, we found that miR-217 overexpression promotes B-cell lymphomagenesis. Interestingly, mice that overexpress miR-217 resemble Bcl-6-overexpressing Iμ-HABCL6 mice in that both show increased GC formation and develop long latency mature B-cell lymphomas.49 We also found that Ink4a/Arf but not p53 loss sensitizes B cells to miR-217-promoted lymphomagenesis. Although it is possible that a protective role of p53 is masked by the early appearance of T-cell lymphomas in p53−/− mice, our data suggest that Ink4a rather than the Arf-p53 oncogenic stress pathway plays the predominant role in protecting GC B cells against the transforming activity of miR-217. This result is in agreement with the finding that Ink4a protein is frequently lost through gene methylation or deletion in aggressive B-cell lymphomas, whereas ARF silencing is a rarer event.50-53
Deregulation of miRNA expression in human B-cell lymphomas has been extensively documented.1-4,48 In some instances, the functional relevance of miRNA deregulation has been tested in genetically modified mouse models. This is the case of Eμ-miR-155 transgenic mice, which developed acute lymphoblastic leukemia/high grade lymphoma54,55 or miR15/miR16 deficiency, which promoted chronic lymphocytic leukemia,56 among other examples.57,58 Evidence for miRNAs involved in GC or post-GC lymphomagenesis was thus far restricted to the combined action of the miR-17-92 miRNA cluster.59 Here we find that miR-217 overexpression in B cells promotes lymphomas of GC or post-GC origin, most likely by impinging on the regulation of the DDR and Bcl-6. Accordingly, we found increased levels of miR-217 in BL and DLBCL, 2 of the most aggressive lymphomas arising from GCs. Our results identify miR-217 as a novel molecular link between the GC response and B-cell transformation and provide an in vivo model of mature B-cell lymphomagenesis.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all the members of the B Cell Biology Laboratory for helpful discussions, D. S. Rao and D. Baltimore for kindly providing us with the MGP vector, G. Roncador for the anti-Bcl-6 antibody, L. Belver, O. Fernandez-Capetillo, J. Mendez, and M. Serrano for critical reading of the manuscript, O. Dominguez, D. Pisano, F. Sanchez-Cabo, J. M. Ligos for technical advice, and S. Bartlett for English editorial support.
N.B.-I. is a fellow of the research training program funded by the Ministerio de Economía y Competitividad, A.R.R. is supported by the Spanish National Cardiovascular Research Centre, V.G.d.Y. is a Ramón y Cajal Investigator (RYC-2009-04503), and R.N.-C. is supported by the Juan de la Cierva research program. This work was funded by grants from the Ministerio de Economía y Competitividad (SAF2010-21394) and the European Research Council Starting Grant program (BCLYM-207844). A.P.-M. and R.N.-C. were funded by grants from Ministerio de Ciencia e Innovación (BIO2010-17527) and the Government of Madrid (P2010/BMD-2305).
Authorship
Contribution: V.G.d.Y., N.B.-I., P.P.-D., and S.M.M. performed experiments; N.M., L.D.L., and M.A.P. collected and prepared human samples; D.F.R. constructed the original backbone for miR-217 transgene cloning; R.N.-C. and A.P.-M. performed bioinformatics analyses; M.C. did histopathological evaluation; V.G.d.Y., N.B.-I., and A.R.R. analyzed the data; V.G.d.Y. and A.R.R. designed experiments and wrote the manuscript; and A.R.R. supervised the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for P.P.-D. is Department of Pathology, New York University (NYU) Cancer Institute, New York University School of Medicine, New York, NY.
The current affiliation for M.C. is Roche Diagnostics GmbH, Penzberg, Germany.
Correspondence: Almudena R. Ramiro, B Cell Biology Laboratory, Centro Nacional de Investigaciones Cardiovasculares, Melchor Fernández Almagro 3, Madrid 28029, Spain; e-mail: aramiro@cnic.es; or Virginia G. de Yébenes, B Cell Biology Laboratory, Centro Nacional de Investigaciones Cardiovasculares, Melchor Fernández Almagro 3, Madrid 28029, Spain; vgarciay@cnic.es.

![Figure 5. miR-217 regulates a DNA damage response and repair gene network and dampens Bcl-6 degradation in GC B cells. (A-B) RNAseq analysis of genome-wide gene expression in control and miR-217TG GC B cells isolated from Peyer’s patches. Pools of 4 to 6 mice per genotype were used in each experiment. Data from 2 independent experiments are shown. (A) Enrichment in miRNA prediction targets within the 1236 mRNAs whose expression was down-regulated in miR-217TG vs control GC B cells. The GEO accession number for the RNAseq data is GSE47877. (B) Ingenuity pathway analysis of the genes differentially expressed in control vs miR-217TG GC B cells, showing enrichment for a DDR and repair gene network among the genes regulated by miR-217 (P = 10−31). Downregulated genes are shown in green; upregulated genes are shown in red. (C) Quantification of Nbs1, Xrcc2, Lig4, and Pds5b expression in Ramos cells transduced with control (open bars) or miR-217 (filled bars) retrovirus analyzed by qRT-PCR. Two-tailed Student t test with P values: *P < .05. (D) Cell cycle analysis of etoposide-treated control- and miR-217-transduced Ramos cells. Representative cell cycle plots of untreated (−Etop) or etoposide-treated (6 hours, 10 μM) (+Etop) Ramos cells. Fraction of cells in S cycle ± standard deviation is shown. The lower graph depicts the change in the proportion of cells at each phase of cell cycle in response to etoposide treatment (open bars, control-transduced cells; black bars, miR-217-transduced cells). Bars represent means ± standard deviations from 3 independent experiments (P = .004 for S phase cells, 2-tailed Student t test). (E) (Upper) Immunoblot of Bcl-6 in Ramos cells transduced with control or miR-217 retrovirus treated for 6 hours with the indicated doses of etoposide. (Lower) Immunoblot of Bcl-6 in miR-217-transduced cells left untreated (−Etoposide) or treated with 10 μM Etposide for 6 hours (+Etoposide) in the presence of increases doses of caffeine (0 mM [−], 2 mM [+], and 5 mM [++]. (F) Analysis of Bcl-6 expression in GC cells by flow cytometry. (Upper) Representative dot plots of Fas, GL7, and Bcl-6 staining of Peyer’s patch B cells. Quantification of Bcl-6 expression levels in B220+Fas+GL7+ GC B cells from Peyer’s patches of littermate controls (white bar) and miR-217KI mice (black bar) (MFI, mean fluorescence intensity; n = 3 mice/genotype, P = .0006, 2-tailed Student t test). (G) Bcl-6 expression in spleens of control and miR-217TG mice analyzed by immunohistochemical staining. A representative picture is shown. The proportion of Bcl-6+ stained in follicle areas was quantified in 205 and 201 spleen follicles of control and 10 miR-217TG mice, respectively. Statistical significance was determined by 2-tailed Student t test (P = .0008).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/2/10.1182_blood-2013-12-543611/4/m_229f5.jpeg?Expires=1769206616&Signature=2Ed19cYsnYvJHvNxRml6WmSUBkjgwfgi7xaSzXExIj1WvNMfXPiQPwRzPAlDb21cg089X-ffsNcDw-yd9HYqUCaM-0JtKXIXJ5dQUspcCUIyEV8NEE71gBYG3HGaeltVMf3NAQMCGLOrA4CqXBuBeePrz51L21lWduXFNMZWEM6CeVM940s6Nl9LSQIrReoOdkBAecj5FyVXKAzhZZKfFSuX6L8UMumkBuL4BdLgv6bn5cTGVRN5lz8xmON0S93bF-SMaiv6wqnEOYGRCXNoSr65dqJ031AlI3X3TzfDSltfedeecNyVXZIVAkTH25ajg2NxGhPE-NHhH-Y-l1C7JA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal