Key Points
JAK3 pseudokinase mutants require JAK1 for their transforming potential.
JAK3 mutants cause T-ALL in a mouse bone marrow transplant model and respond to tofacitinib, a JAK3-selective inhibitor.
Abstract
JAK3 is a tyrosine kinase that associates with the common γ chain of cytokine receptors and is recurrently mutated in T-cell acute lymphoblastic leukemia (T-ALL). We tested the transforming properties of JAK3 pseudokinase and kinase domain mutants using in vitro and in vivo assays. Most, but not all, JAK3 mutants transformed cytokine-dependent Ba/F3 or MOHITO cell lines to cytokine-independent proliferation. JAK3 pseudokinase mutants were dependent on Jak1 kinase activity for cellular transformation, whereas the JAK3 kinase domain mutant could transform cells in a Jak1 kinase–independent manner. Reconstitution of the IL7 receptor signaling complex in 293T cells showed that JAK3 mutants required receptor binding to mediate downstream STAT5 phosphorylation. Mice transplanted with bone marrow progenitor cells expressing JAK3 mutants developed a long-latency transplantable T-ALL–like disease, characterized by an accumulation of immature CD8+ T cells. In vivo treatment of leukemic mice with the JAK3 selective inhibitor tofacitinib reduced the white blood cell count and caused leukemic cell apoptosis. Our data show that JAK3 mutations are drivers of T-ALL and require the cytokine receptor complex for transformation. These results warrant further investigation of JAK1/JAK3 inhibitors for the treatment of T-ALL.
Introduction
The JAK kinases (JAK1, JAK2, JAK3, TYK2) are a family of cytosolic tyrosine kinases that are essential for the signaling of cytokine receptors. All 4 JAK kinases share a common structure with receptor-binding domains at the N-terminus and a regulatory pseudokinase domain and catalytic active kinase domains at the C-terminus. The 2 JAK kinase family members JAK1 and JAK3 are essential components of the heterodimeric interleukin-7 (IL7) receptor, in which the IL7Rα chain is bound by JAK1 and the common γ chain (IL2RG) is bound by JAK3. Activation of the receptor by its ligand results in phosphorylation of both JAK1 and JAK3, with subsequent phosphorylation of STAT5 that translocates to the nucleus to regulate gene expression.1 It is now well established that the correct activation of this IL7 receptor-signaling pathway via JAK1 and JAK3 is critically important for the development of both B cells and T cells. Loss-of-function mutations in IL2RG, IL7R, or JAK3 leads to severe immune deficiency.2 Conversely, activating mutations in JAK1, JAK3, or IL7R have been reported in both B- and T-cell acute lymphoblastic leukemia (ALL) with mutations in any one of these genes found in as much as 25% of T-ALL cases.3-5
Of the JAK family kinase mutations identified in hematologic malignancies, JAK1 mutations are rare in childhood ALL but are more frequent in adult ALL cases.6,7 Alternatively, JAK3 mutations have been identified in acute megakaryoblastic leukemia, T-cell prolymphocytic leukemia, and more recently in juvenile myelomonocytic leukemia and natural killer T-cell lymphoma (NK/T-lymphoma).8-13 The transformative potential of these JAK3 mutations has been confirmed in cell-based assays in vitro, and the JAK3 A572V mutation identified in acute megakaryoblastic leukemia was also shown to confer features of megakaryoblastic leukemia and transform murine lymphoid cells in vivo.10,14 However, systematic studies using both in vitro and in vivo models of newly identified JAK3 mutations are lacking, as is knowledge on their potential mechanism of transformation.
In this work, 12 JAK3 pseudokinase and kinase domain mutations identified in T-ALL from massive parallel sequencing projects were studied using in vitro and in vivo experiments to assess their transformation potential. We report that the majority of these JAK3 mutants transform cells to cytokine-independent growth in vitro, and that JAK1 kinase activity is required downstream of most JAK3 mutants. Moreover, we show that expression of JAK3 mutants in hematopoietic stem/progenitor cells leads to the development of a T-ALL–like disease in a mouse bone marrow transplant model. We finally demonstrate that the use of tofacitinib, a JAK3-selective inhibitor, can reduce the leukemic burden within our in vivo bone marrow transplant model.
Materials and methods
Expression plasmids
JAK3 wild-type and mutant sequences were synthesized by GenScript. Kinase-deficient JAK1 was provided by Dr Haan.15 The IL2RG expression plasmid was provided by Dr Barata. JAK1 and IL7R constructs were described previously.16 All constructs were cloned into the murine stem cell virus–green fluorescent protein (GFP) or murine stem cell virus–puro vectors.
Cell culture, virus production, and retroviral transduction
Two-hundred ninety-three T cells were cultured in Dulbecco’s modified Eagle medium with 10% fetal calf serum (FCS). Virus production and retroviral transduction were performed as previously described.17 Ba/F3 and MOHITO18 cells were cultured in RPMI 1640 with 10% to 20% FCS and IL3 (10 ng/μL; Peprotech), IL-2 (25 ng/μL; Peprotech), and IL7 (50 ng/μL; Peprotech), respectively. For electroporation, 1.5 × 106 Ba/F3 cells were resuspended in 400 μL of serum-free medium containing 200 nmol siRNA duplexes and then transferred to 4-mm cuvettes (Biorad). The electroporated cells were transferred to 12-well plates containing 2 mL of prewarmed RPMI supplemented with 10% FCS. Growth reduction was analyzed using the guava easyCyte Flow Cytometer (Millipore) 1 hour and 48 hours after electroporation. Human T-ALL cells were cultured as described.19 Ethical approval and informed consent was obtained for the use of human T-ALL cells.
Western blotting
Cells were lysed in cold lysis buffer containing 5 mM NA3VO4 and protease inhibitors (Complete, Roche). The proteins were separated on NuPAGE NOVEX Bis-Tris 4% to 12% gels (Invitrogen). Western blot analysis was performed using antibodies against JAK1, anti–phospho-tyrosine(4G10) (Millipore); AKT, phospho-AKT, phospho-ERK, JAK3, phospho-JAK3, STAT1, phospho-STAT1, STAT3, phospho-STAT3 phospho-STAT5, SRC, phospho-SRC (Cell Signaling); ERK, phospho-JAK1, STAT5 (Santa Cruz Biotechnology); and β-actin (Sigma-Aldrich); secondary antibodies were conjugated with horseradish peroxidase (GE Healthcare). Anti–phospho-JAK1 antibody was used to detect phosphorylated JAK1 and JAK3. Bands were visualized using a cooled charge-coupled device camera (ImageQuant LAS-4000; GE Healthcare).
Murine bone marrow transplantation
BALB/c mice were purchased from Charles River Laboratories. Male BALB/c mice were sacrificed and bone marrow cells were harvested from the femur and tibia. Lineage-negative cells were enriched (STEMCELL Technologies) and cultured in RPMI with 20% FCS, IL3 (10 ng/mL), IL6 (10 ng/mL), SCF (50 ng/mL) (all from Peprotech), and penicillin-streptomycin. 1 × 106 cells were transduced by spinoculation (90 minutes at 2500 rpm; 8 μg/mL polybrene). Cells were washed in phosphate-buffered saline and injected (1 × 106 cells/0.3 mL) into the lateral tail vein of sublethally-irradiated (5 Gy) female BALB/c mice. Mice were housed in individually ventilated cages and monitored daily. We obtained approval from the local ethics committee.
Inhibitor experiments
Ba/F3 or MOHITO cells were seeded in 96-well plates (1 × 105 cells/mL) and treated with tofacitinib, ruxolitinib, or vehicle (dimethylsulfoxide [DMSO]). A quantitative evaluation of proliferation was done after 24 hours using ATPlite (PerkinElmer) and measured on the VICTOR X4 Reader (PerkinElmer). For in vivo experiments, BALB/c mice were treated through oral gavage with tofacitinib (20-40 mg/kg per day) or vehicle.
Flow cytometry analyses
Single-cell suspensions were prepared from peripheral blood, bone marrow, spleen, thymus, and lymph nodes. Cells were analyzed on a FACSCanto flow cytometer (BD Biosciences). Data were analyzed with FlowJo software (Tree Star).
Results
JAK3 mutants identified in T-ALL transform Ba/F3 and MOHITO cells to cytokine-independent growth
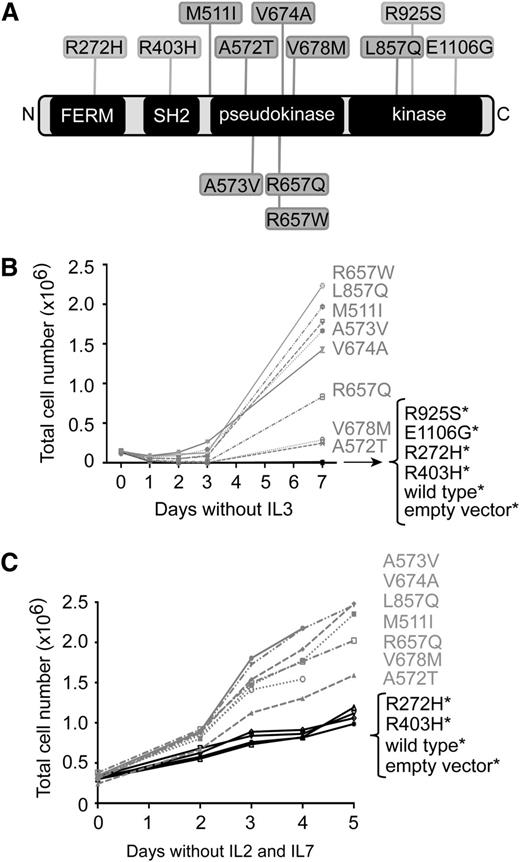
We recently reported the results from exome, transcriptome, and targeted resequencing of 89 patients with T-ALL.4,20,21 In this cohort of patients, we identified 12 mutations affecting the JAK3 protein in 10 of 89 diagnostic samples. To determine whether JAK3 mutations identified in T-ALL are oncogenic driver mutations, we selected 12 mutants to study in vitro using the cytokine-dependent Ba/F3 and MOHITO cell lines. These mutations included FERM/SH2 domain mutations (R272H, R403H), pseudokinase domain mutations (M511I, A572T, A573V, R657W, R657Q, V674A, V678M), and kinase domain mutations (L857Q, R925S, E1106G) (Figure 1A and supplemental Table 1, available on the Blood Web site).
JAK3 mutants transform Ba/F3 and MOHITO cells to cytokine-independent growth. (A) Schematic representation of JAK3 protein and its main domains; the four-point-one, Ezrin, Radixin, Moesin (FERM) domain; the Src homology-2 (SH2) domain; and the pseudokinase and kinase domains. Mutations studied in this work are shown. (B-C) The proliferation curve of Ba/F3 (B) or MOHITO (C) cells expressing various JAK3 mutants, JAK3 wild-type, or empty vector, in the absence of cytokines. Mutations that did not stimulate proliferation more than wild-type JAK3 were considered nontransforming mutants and are indicated with asterisk (*).
JAK3 mutants transform Ba/F3 and MOHITO cells to cytokine-independent growth. (A) Schematic representation of JAK3 protein and its main domains; the four-point-one, Ezrin, Radixin, Moesin (FERM) domain; the Src homology-2 (SH2) domain; and the pseudokinase and kinase domains. Mutations studied in this work are shown. (B-C) The proliferation curve of Ba/F3 (B) or MOHITO (C) cells expressing various JAK3 mutants, JAK3 wild-type, or empty vector, in the absence of cytokines. Mutations that did not stimulate proliferation more than wild-type JAK3 were considered nontransforming mutants and are indicated with asterisk (*).
The Ba/F3 and MOHITO cells were transduced by retroviral vectors encoding either JAK3 wild-type or JAK3 mutants that co-express GFP. In all cases, transduction efficiency exceeded 80% as measured by GFP expression. Of the JAK3 mutations that were screened, 8 of 12 were able to transform the Ba/F3 cells to IL-3–independent growth (Figure 1B). Of these 8 JAK3 mutants, 2 mutants (V678M and A572T) were weaker in their ability to transform Ba/F3 cells to IL-3–independent growth. The remaining 4 JAK3 mutants, including 2 kinase domain mutations (R925S and E1106G), and 2 FERM/SH2 domain mutants (R272H, R403H) did not transform Ba/F3 cells. Similarly, JAK3 wild-type and the empty vector–transduced cells were not able to induce autonomous cell growth. Similar results were observed in the MOHITO cell line, because the same JAK3 mutants were able to stimulate the proliferation and survival of the cells in the absence of IL-2 and IL-7 cytokines (Figure 1C). Transformation was further demonstrated by the fact that the transformed MOHITO cells out-competed the nontransduced cells, because only GFP+ cells were able to grow in the absence of cytokines (data not shown).
JAK3 mutants activate JAK1, STAT5, and ERK
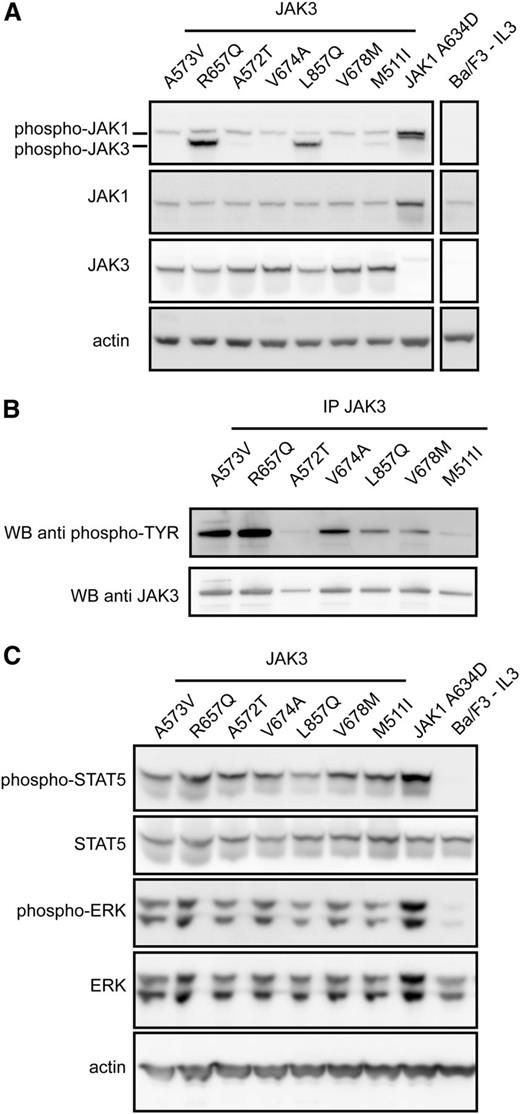
We next sought to identify the downstream signaling pathways activated by JAK3 mutants in the transformed Ba/F3 cell lines. We first analyzed the levels of JAK3 phosphorylation by western blot using a phospho-JAK antibody which recognizes both pJAK1 and pJAK3. This showed that JAK3 phosphorylation was variable between the different JAK3 mutants, with some mutants (ie, R657Q, L857Q) showing a clear phosphorylation of JAK3 and others showing only weak JAK3 phosphorylation (Figure 2A). To determine whether other tyrosine residues were potentially phosphorylated, we carried out JAK3 immunoprecipitation followed by immunoblot detection with a general antiphosphotyrosine antibody. This confirmed that all JAK3 mutants had phosphorylated tyrosine residues present, with strong tyrosine phosphorylation evident for A573V and R657Q; moderate for V674A, L857Q, and V678M; and weak for A572T and M511I mutants (Figure 2B).
JAK3 mutants signal through JAK1, STAT5, and ERK in a cytokine-independent manner. (A) Western blot analysis of whole-cell lysates of Ba/F3 cells transformed by JAK3 mutants. Phosphorylation of JAK1 was detected for all JAK3 mutants. JAK3 phosphorylation was clearly detected for some, but not all, JAK3 mutant proteins, most likely because of the specificity of the used antibody. JAK3 protein expression was detected with a human-specific antibody, not recognizing the endogenous JAK3 expression. (B) JAK3 phosphorylation could be detected for all JAK3 mutants after immunoprecipitation and detection with a phosphotyrosine antibody (4G10). (C) All transforming JAK3 mutants were able to phosphorylate downstream signaling components STAT5 and ERK.
JAK3 mutants signal through JAK1, STAT5, and ERK in a cytokine-independent manner. (A) Western blot analysis of whole-cell lysates of Ba/F3 cells transformed by JAK3 mutants. Phosphorylation of JAK1 was detected for all JAK3 mutants. JAK3 phosphorylation was clearly detected for some, but not all, JAK3 mutant proteins, most likely because of the specificity of the used antibody. JAK3 protein expression was detected with a human-specific antibody, not recognizing the endogenous JAK3 expression. (B) JAK3 phosphorylation could be detected for all JAK3 mutants after immunoprecipitation and detection with a phosphotyrosine antibody (4G10). (C) All transforming JAK3 mutants were able to phosphorylate downstream signaling components STAT5 and ERK.
We also observed that JAK1 phosphorylation was present in Ba/F3 cells expressing the different JAK3 mutants. The level of JAK1 phosphorylation was lower in the JAK3 mutant–transformed cells compared with cells transformed by the constitutively active JAK1 A634D mutant (Figure 2A). In addition, phosphorylation of STAT5 and ERK was detected in all Ba/F3 cells expressing JAK3 mutants (Figure 2C). Notably, we did not observe phosphorylation of STAT1, STAT3, or AKT in any of the transformed Ba/F3 cells expressing JAK3 mutants (supplemental Figure 1). These results were also confirmed in the MOHITO cell line (data not shown). Taken together, these results show that cytokine-independent growth of Ba/F3 or MOHITO expressing JAK3 mutants was concomitant with increased phosphorylation of JAK1, STAT5, and ERK.
JAK1 is essential for the transforming properties of JAK3 pseudokinase domain mutants
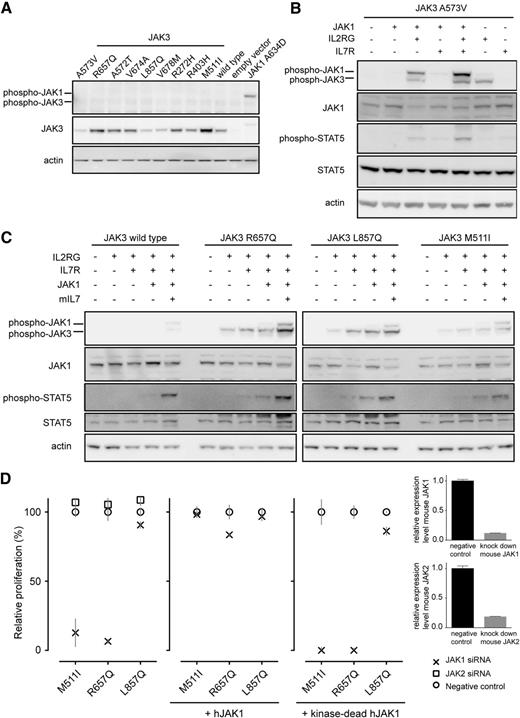
We then investigated whether the transforming capacity of JAK3 mutants was dependent on the presence of the cytokine receptor complex (eg, IL7R complex). To test this, we used 293T cells that lack endogenous cytokine receptor expression. Cells transfected with JAK3 mutants alone did not result in JAK3 phosphorylation (Figure 3A). Co-expression of JAK3 mutants with IL2RG was sufficient to obtain JAK3 phosphorylation (Figure 3B-C), whereas activation of STAT5 required the coexpression of JAK3 mutants with IL2RG and JAK1 (Figure 3B) or IL2RG and IL7R (Figure 3C). The reconstitution of the entire receptor complex yielded the strongest activation of both JAK3 and STAT5. These data indicate that JAK3 mutants cannot function in the absence of additional receptor components. For wild-type JAK3, co-expression of the complete receptor complex (IL7R, IL2RG, JAK1, JAK3) with IL7 ligand stimulation was required to obtain STAT5 phosphorylation (Figure 3C). JAK3 mutants unable to transform Ba/F3 cells did not cause JAK1 or JAK3 phosphorylation in 293T cells and were predicted as “neutral” variants using PROVEAN22 analysis (supplemental Figure 2), supporting evidence that the R925S and E1106G are nontransforming passenger mutations.
JAK1 kinase activity is essential for the transforming properties of JAK3 mutants. (A) Western blot detection of JAK3 mutants expressed in 293T cells. (B-C) Western blot analysis of whole-cell lysates after reconstitution of the IL7-receptor signaling complex in 293T cells. The 293T cells were transiently transfected with the constructs as indicated. (D) The graph shows relative proliferation of Ba/F3 cells expressing JAK3 M511I, R657Q, or L857Q 48 hours after knockdown of endogenous Jak1 or Jak2 compared with scrambled siRNA. Rescue of the proliferation of cells with Jak1 knockdown was achieved by expression of human JAK1, but not by expression of kinase dead JAK1. Relative proliferation is shown 48 hours after knockdown of endogenous Jak1. Knockdown efficiency was determined by quantitative polymerase chain reaction for all cell lines; results are shown for one cell line but are representative of all cell lines.
JAK1 kinase activity is essential for the transforming properties of JAK3 mutants. (A) Western blot detection of JAK3 mutants expressed in 293T cells. (B-C) Western blot analysis of whole-cell lysates after reconstitution of the IL7-receptor signaling complex in 293T cells. The 293T cells were transiently transfected with the constructs as indicated. (D) The graph shows relative proliferation of Ba/F3 cells expressing JAK3 M511I, R657Q, or L857Q 48 hours after knockdown of endogenous Jak1 or Jak2 compared with scrambled siRNA. Rescue of the proliferation of cells with Jak1 knockdown was achieved by expression of human JAK1, but not by expression of kinase dead JAK1. Relative proliferation is shown 48 hours after knockdown of endogenous Jak1. Knockdown efficiency was determined by quantitative polymerase chain reaction for all cell lines; results are shown for one cell line but are representative of all cell lines.
To confirm the essential role of JAK1 for the signaling of the JAK3 mutants, we downregulated the expression of endogenous murine Jak1 or Jak2 in the Ba/F3 cells. Knockdown of Jak expression was confirmed by quantitative polymerase chain reaction and consistently showed knockdown efficiency of >90% (Figure 3D). For the pseudokinase domain mutants, siRNA-mediated knockdown of Jak1 expression resulted in a >90% decrease in cell proliferation. This was not the case for cells transformed by the kinase domain JAK3 L857Q mutant, where loss of JAK1 did not significantly alter cell proliferation. For both pseudokinase and kinase domain mutants, loss of endogenous Jak2 expression did not affect cell proliferation (Figure 3D).
To determine whether the observed dependence on Jak1 was caused by the complete loss of Jak1 protein or only Jak1 kinase activity, we performed rescue experiments with either wild-type human JAK1 or a kinase-dead JAK1 mutant. Expression of the siRNA-resistant human JAK1, but not the kinase dead JAK1, could rescue the block in proliferation upon Jak1 knockdown (Figure 3D). Taken together, these results indicate that JAK1 kinase activity is essential for the transforming capacity of JAK3 pseudokinase domain mutants, but not for the kinase domain mutant L857Q.
JAK3 mutant–transformed cells are sensitive to JAK kinase inhibitors
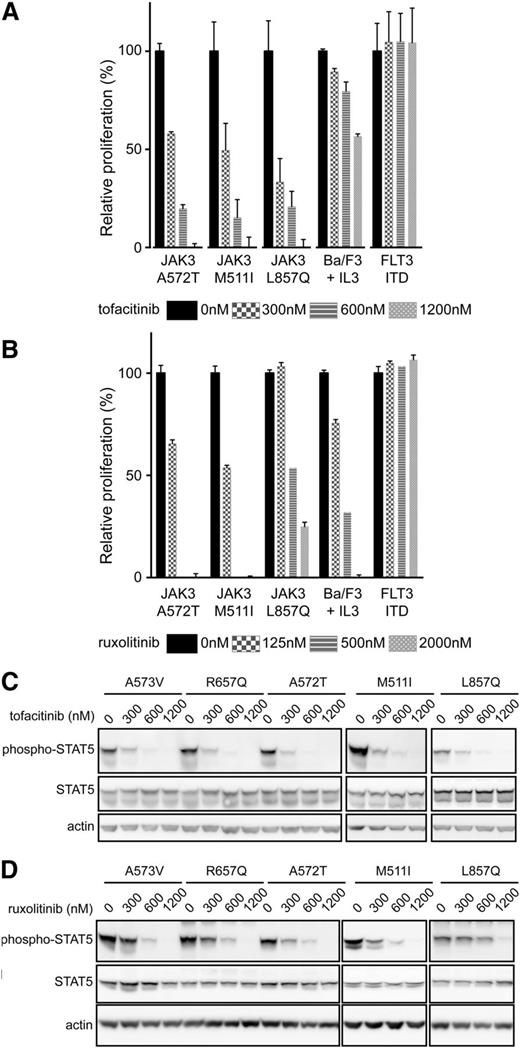
To determine the sensitivity of the different JAK3 mutants to JAK kinase inhibitors, and to further confirm the requirement of JAK1, we treated the transformed Ba/F3 and MOHITO cells with tofacitinib, a JAK3-selective inhibitor,23 or ruxolitinib, a JAK2/JAK1-selective inhibitor.24 Cells were first treated with increasing concentrations of the JAK3-selective inhibitor tofacitinib or vehicle (DMSO), with a quantitative evaluation of proliferation performed at 24 hours posttreatment. All Ba/F3 cells transformed by JAK3 mutants were sensitive to tofacitinib treatment, with IC50 values ranging between 246 nM and 408 nM (Figure 4A). Wild-type Ba/F3 cells, which are dependent on IL3/JAK2 signaling, had a significantly higher IC50 of 1400 nM, with these high concentrations speculated to also have an inhibitory effect on JAK2 kinase activity. The proliferation of cells transformed by FLT3-ITD kinase, which are independent of JAK activity, was not affected by tofacitinib treatment. Similar results were obtained for transformed MOHITO cells (supplemental Figures 3 and 4).
Cells dependent on JAK3 mutants are sensitive to JAK3 and JAK1 inhibition. (A-B) Relative proliferation of Ba/F3 cells transformed by JAK3 mutants or FLT3 ITD or wild-type Ba/F3 cells stimulated with IL3 after treatment with tofacitinib or ruxolitinib, respectively. Proliferation was compared with proliferation of cells after vehicle (DMSO) treatment. Full dose-response curves for all JAK3 mutants are shown in the supplemental material (supplemental Figures 3-6). (C-D) Western blot analysis of Ba/F3 cells expressing JAK3 mutants after 90 minutes of treatment with tofacitinib or ruxolitinib.
Cells dependent on JAK3 mutants are sensitive to JAK3 and JAK1 inhibition. (A-B) Relative proliferation of Ba/F3 cells transformed by JAK3 mutants or FLT3 ITD or wild-type Ba/F3 cells stimulated with IL3 after treatment with tofacitinib or ruxolitinib, respectively. Proliferation was compared with proliferation of cells after vehicle (DMSO) treatment. Full dose-response curves for all JAK3 mutants are shown in the supplemental material (supplemental Figures 3-6). (C-D) Western blot analysis of Ba/F3 cells expressing JAK3 mutants after 90 minutes of treatment with tofacitinib or ruxolitinib.
Confirmation that JAK1 is required for the transforming capacities of JAK3 mutant proteins was also evident after cells were treated with the JAK1-selective inhibitor ruxolitinib. Ba/F3 cells dependent on the expression of JAK3 pseudokinase domain mutants were sensitive for low doses of ruxolitnib treatment, with IC50 values ranging from 120 nM to 398 nM. This is in agreement with our earlier results, where all pseudokinase domain mutants require JAK1 kinase activity for their transforming capacities. The cells expressing the JAK3 L857Q kinase domain mutant were less sensitive, with an IC50 value of 855 nM, confirming that JAK3 L857Q is less dependent on JAK1 kinase activity. The inhibitory effects observed at higher ruxolitinib concentration in the L857Q cells is speculated to be a result of inhibition of the JAK3 kinase by ruxolitinib.24 Similar results were obtained for transformed MOHITO cells (supplemental Figures 5 and 6). Finally, we also treated the Ba/F3 cells with low doses of tofacitinib, ruxolitinib, or a combination of both drugs. This resulted in the synergistic inhibition of cells dependent on JAK1 cells but not in cells independent of JAK1 activity (eg, L857Q mutant) (supplemental Figure 7), confirming again that pseudokinase domain mutants are dependent on JAK1 activity.
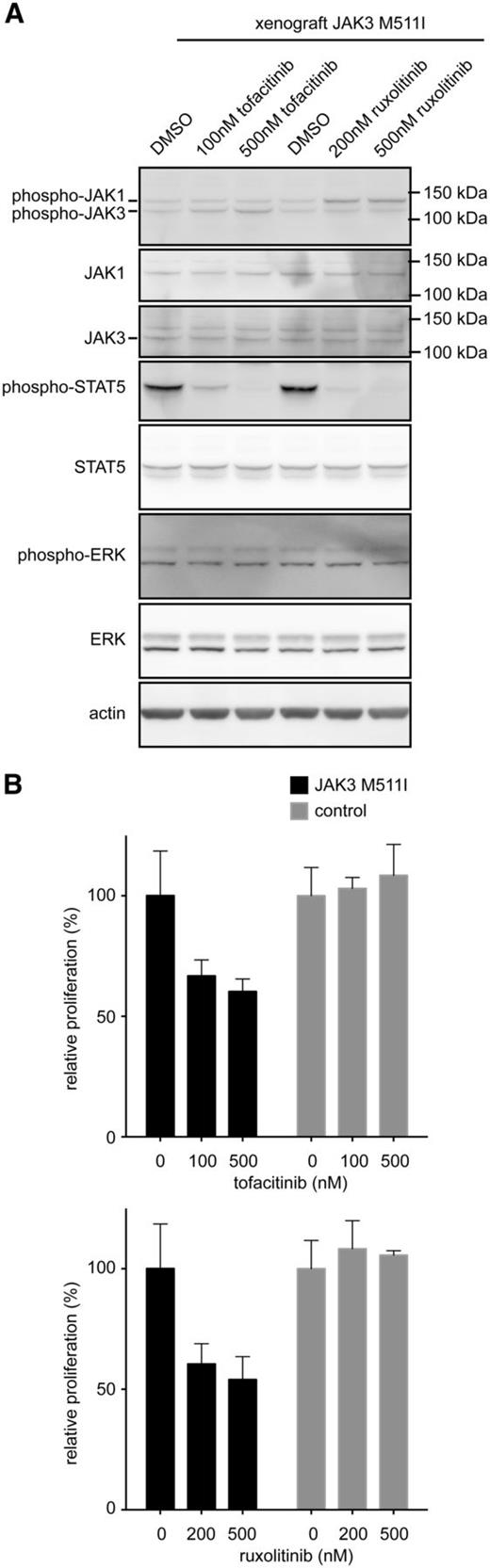
To determine whether treatment of JAK3 mutant–transformed Ba/F3 cells also decreases downstream signaling, cells were treated with an increasing concentration of tofacitinib (300, 600, 1200 nM), ruxolitinib (125, 500, 2000 nM), or vehicle (DMSO) for 90 minutes before cell lysis. In all cases, STAT5 phosphorylation significantly decreased with increasing tofacitinib concentration (Figure 4C). Once again, changes in STAT5 phosphorylation for the kinase domain mutant L857Q were less marked after ruxolitinib treatment, confirming that this mutant can signal to STAT5 independent of Jak1 (Figure 4D). We used here STAT5 phosphorylation as a read-out, because effects on JAK1/JAK3 phosphorylation by JAK kinase inhibitors are generally modest, as also previously described.25 In primary human T-ALL cells with the JAK3 M511I mutation, we observed inhibition of STAT5 phosphorylation upon treatment with tofacitinib or ruxolitinib, but no effect was seen on phosphorylation of ERK (Figure 5A). This may be caused by the presence of other mutations in the T-ALL sample, which leads to the activation of the ERK pathway in a JAK-independent manner. These data are also in line with our observation that the proliferation of the JAK3 M511I mutant T-ALL cells was inhibited by tofacitinib or ruxolitinib, whereas the proliferation of human T-ALL cells that did not express mutant JAK3 were not affected by the presence of JAK inhibitors (Figure 5B).
Ex vivo treatment of primary human T-ALL cells with JAK3 M511I mutation. (A) Western blot analysis of xenograft-derived cells that express JAK3 M511I were cultured ex vivo and treated for 90 minutes with tofacitinib, ruxolitinib, or vehicle (DMSO). (B) Graph shows relative proliferation after 48 hours of treatment with JAK inhibitors of human T-ALL xenograft–derived cells.
Ex vivo treatment of primary human T-ALL cells with JAK3 M511I mutation. (A) Western blot analysis of xenograft-derived cells that express JAK3 M511I were cultured ex vivo and treated for 90 minutes with tofacitinib, ruxolitinib, or vehicle (DMSO). (B) Graph shows relative proliferation after 48 hours of treatment with JAK inhibitors of human T-ALL xenograft–derived cells.
JAK3 mutants cause T-ALL in a bone marrow transplant mouse model
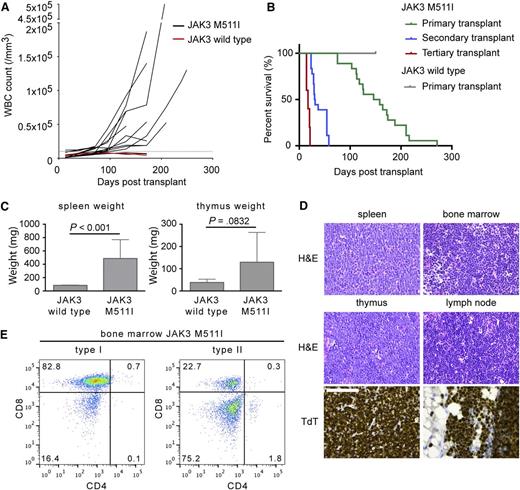
Having established that the JAK3 mutants could transform cells in vitro, we used a mouse bone marrow transplant model to determine whether they were also transforming in vivo. Our initial assessment focused on the JAK3 M511I pseudokinase domain mutation because it was the most common JAK3 mutation identified in T-ALL. In this model, hematopoietic progenitor cells transduced with retroviral constructs expressing JAK3 M511I and GFP were injected in the tail vein of irradiated female recipient BALB/c animals. Animals initially developed a lymphoproliferative disease over the first 12 weeks characterized by moderate increased white blood cell (WBC) counts (range, 10 000-20 000/μL) (Figure 6A). After this initial lymphoproliferative stage, all animals expressing JAK3 M511I progressed to an acute phase. The acute phase was fatal and was characterized by a rapid rise in WBC counts (range, 50 000-300 000/μL) between 14 and 28 weeks posttransplant. Mice transplanted with wild-type JAK3-expressing cells did not develop disease (Figure 6A-B).
Expression of JAK3 M511I in the bone marrow cells of Balb/c mice cause a T-lymphoproliferative disease that progresses to T-ALL. (A) Evolution of the WBC count in JAK3 M511I animals. WBC count was measured every 2 weeks. The upper limit of normal WBC count (10 000/mm3) is indicated by a dashed line. (B) Kaplan-Meier survival curve of BALB/c mice receiving bone marrow transplantation of lineage-negative cells expressing JAK3 M511I. Mice transplanted with cells expressing wild-type JAK3 had neither hematologic abnormalities nor signs of disease. (C) The spleen and thymus weights of mice transplanted with cells expressing JAK3 M511I compared with mice transplanted with JAK3 wild-type cells. Significance was determined by Student t test. (D) Hematoxylin and eosin (H&E) staining of the spleen, bone marrow, thymus, and lymph node. Terminal deoxynucleotidyl transferase (TdT) staining of the thymus and lymph node. The scale bar represents 100 μm. (E) Analysis of the bone marrow cells of diseased animals by flow cytometry with anti-CD4 and anti-CD8 antibodies. Pathology images were obtained using a Leica DM2500 microscope and a Leica DFC290HD camera and analyzed using the Leica Application suite LAS v4.1 software. Images were processed with Adobe Photoshop CS5.
Expression of JAK3 M511I in the bone marrow cells of Balb/c mice cause a T-lymphoproliferative disease that progresses to T-ALL. (A) Evolution of the WBC count in JAK3 M511I animals. WBC count was measured every 2 weeks. The upper limit of normal WBC count (10 000/mm3) is indicated by a dashed line. (B) Kaplan-Meier survival curve of BALB/c mice receiving bone marrow transplantation of lineage-negative cells expressing JAK3 M511I. Mice transplanted with cells expressing wild-type JAK3 had neither hematologic abnormalities nor signs of disease. (C) The spleen and thymus weights of mice transplanted with cells expressing JAK3 M511I compared with mice transplanted with JAK3 wild-type cells. Significance was determined by Student t test. (D) Hematoxylin and eosin (H&E) staining of the spleen, bone marrow, thymus, and lymph node. Terminal deoxynucleotidyl transferase (TdT) staining of the thymus and lymph node. The scale bar represents 100 μm. (E) Analysis of the bone marrow cells of diseased animals by flow cytometry with anti-CD4 and anti-CD8 antibodies. Pathology images were obtained using a Leica DM2500 microscope and a Leica DFC290HD camera and analyzed using the Leica Application suite LAS v4.1 software. Images were processed with Adobe Photoshop CS5.
All mice showed accumulation of CD8 single-positive cells in the peripheral blood, with the majority of mice succumbing to the disease between 100 and 200 days posttransplant (Figure 6B). At end-stage disease, there was a significant increase in spleen and thymus weight (Figure 6C). Spleen, thymus, lymph nodes, and bone marrow were infiltrated by TdT+ immature T cells (Figure 6D). GFP+ blast cells in all organs also had variable expression of TCRβ and CD3. Specific phenotypic analysis of the bone marrow showed that 5 of 16 mice had >50% blasts that were CD8 single-positive, whereas in 11 of 16 mice, the bone marrow contained a CD4/CD8 double-negative population (Figure 6E), corresponding to an accumulation of more immature T cells. Mouse leukemia cells showed phosphorylation of JAK1/JAK3, STAT5, and ERK (supplemental Figure 8).
Notably, the leukemic cells were also transplantable and caused rapid development of T-ALL and earlier onset of disease in the secondary and tertiary transplanted animals (Figure 6B). In 4 of 7 JAK3 M511I secondary transplant experiments, recipient mice developed an acute leukemia within 100 days posttransplant. All secondary transplanted mice had an accumulation of TCRβ/CD3– cells, indicating that the acute phase seen in primary transplant experiments was associated with loss of differentiation and accumulation of more immature T cells (supplemental Figures 9-12). Leukemic cells with high TCRβ expression failed to induce leukemia in recipient mice, indicating that more differentiated cells are not able to induce leukemia in a secondary transplant. Analysis of TCRβ rearrangement showed oligoclonal origin of the primary leukemias, whereas TCRβ was not rearranged in secondary leukemias, in agreement with the immunophenotyping (supplemental Figures 11-13). Three heterozygous mutations (Notch1 L1668P, Notch1 indel(R2361), Pten G165E) were identified in primary leukemias (supplemental Figure 13). Together, these data support a clonal nature of the mouse leukemias.
In addition to JAK3 M511I, other JAK3 mutations (A573V, L857Q, V674A, and R657Q) were tested in the BALB/c bone marrow transplant model. Mice transplanted with cells expressing JAK3 A573V or V674A showed a gradual increase in the WBC count and developed a similar T-ALL–like disease as observed for the JAK3 M511I mutant. In contrast to JAK3 A573V and V674A, mice transplanted with cells expressing JAK3 L857Q or R657Q showed no increase in WBC count compared with the control mice but did show severe splenomegaly and lymphadenopathy. Expression of JAK3 L857Q caused severe thymus hyperplasia, whereas the JAK3 R657Q mutant caused B-cell leukemia. Detailed results are presented in supplemental Figure 14.
JAK3 M511I mutant cells respond to tofacitinib treatment in vivo
Our in vitro data showed that tofacitinib, a selective JAK3 inhibitor, potently inhibited all tested JAK3 mutants in cell-based assays. Tofacitinib does not show potent activity against JAK2 and was recently approved by the Food and Drug Administration for the treatment of rheumatoid arthritis. We therefore selected tofacitinib to determine the in vivo response of JAK3-driven leukemia to JAK inhibitor treatment. We used the primary JAK3 M511I transplant model and the secondary JAK3 M511I transplant model and treated those animals by oral gavage.
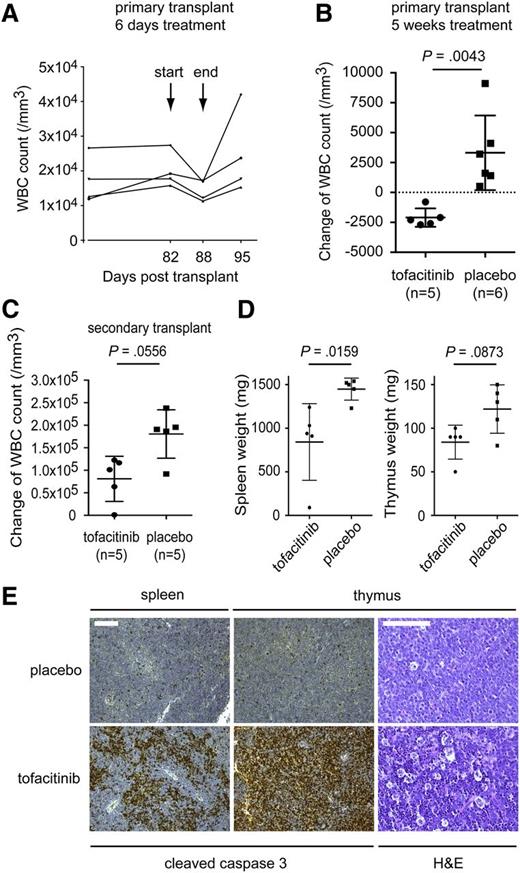
In a first experiment, primary transplanted animals were treated with tofacitinib (30 mg/kg per day) for 6 consecutive days. Treatment started 82 days after transplantation, at the time the mice had increased WBC counts. All mice showed a decrease in WBC count during treatment and an increase in WBC count after treatment was stopped, demonstrating a clear effect of tofacitinib on the proliferation of the leukemia cells in vivo (Figure 7A). In a second experiment, primary transplanted mice were treated with tofacitinib (40 mg/kg per day, n = 5) or placebo (n = 6) for 5 weeks. Treatment started 48 weeks after bone marrow transplant, at which time WBC values had already increased. Mice receiving tofacitinib treatment showed a decrease in WBC count at the end of the experiment, whereas WBC counts of placebo-treated mice were all increased (Figure 7B).
JAK3 M511I cells are sensitive to the JAK3 inhibitor tofacitinib in vivo. (A) Four BALB/c mice that received a primary transplant of cells expressing JAK3 M511I were treated for 6 days with tofacitinib, 2 times per day with a total concentration of 30 mg/kg per day. WBC count was determined before, during, and after treatment. The graph shows WBC count over time, indicating days post-transplant. (B) Eleven BALB/C mice received a primary transplant of cells expressing JAK3 M511I. Mice were randomly divided into 2 groups. One group was treated with tofacitinib for 5 weeks, twice daily, with a total concentration of 40 mg/kg per day. The other group received vehicle (DMSO) treatment. WBC count was determined at the start and end of treatment. The graph shows differences in WBC counts over the 5 weeks of treatment. (C) Ten BALB/c mice received a secondary transplant of cells expressing JAK3 M511I. Afterward, mice were randomly divided into 2 groups. Both groups were treated for 14 days, one group with tofacitinib and the second group with vehicle (DMSO). Treatment was performed 2 times per day with a total concentration of 20 mg/kg per day. WBC count was determined at the start and the end of the experiment. The graph shows the difference in WBC count over the 14 days of treatment. (D) Graphs show the spleen, thymus, and lymph node weights of secondary transplanted mice with JAK3 M511I after 18 days of treatment with tofacitinib or vehicle. Significance was determined by Student t test. (E) Cleaved caspase-3 staining of the spleen and thymus samples retrieved from the experiment shown in (C). H&E staining of the thymus after treatment with placebo or tofacitinib. The scale bars represent 100 μm. Pathology images were obtained using a Leica DM2500 microscope and a Leica DFC290HD camera and analyzed using the Leica Application suite LAS v4.1 software. Images were processed with Adobe Photoshop CS5.
JAK3 M511I cells are sensitive to the JAK3 inhibitor tofacitinib in vivo. (A) Four BALB/c mice that received a primary transplant of cells expressing JAK3 M511I were treated for 6 days with tofacitinib, 2 times per day with a total concentration of 30 mg/kg per day. WBC count was determined before, during, and after treatment. The graph shows WBC count over time, indicating days post-transplant. (B) Eleven BALB/C mice received a primary transplant of cells expressing JAK3 M511I. Mice were randomly divided into 2 groups. One group was treated with tofacitinib for 5 weeks, twice daily, with a total concentration of 40 mg/kg per day. The other group received vehicle (DMSO) treatment. WBC count was determined at the start and end of treatment. The graph shows differences in WBC counts over the 5 weeks of treatment. (C) Ten BALB/c mice received a secondary transplant of cells expressing JAK3 M511I. Afterward, mice were randomly divided into 2 groups. Both groups were treated for 14 days, one group with tofacitinib and the second group with vehicle (DMSO). Treatment was performed 2 times per day with a total concentration of 20 mg/kg per day. WBC count was determined at the start and the end of the experiment. The graph shows the difference in WBC count over the 14 days of treatment. (D) Graphs show the spleen, thymus, and lymph node weights of secondary transplanted mice with JAK3 M511I after 18 days of treatment with tofacitinib or vehicle. Significance was determined by Student t test. (E) Cleaved caspase-3 staining of the spleen and thymus samples retrieved from the experiment shown in (C). H&E staining of the thymus after treatment with placebo or tofacitinib. The scale bars represent 100 μm. Pathology images were obtained using a Leica DM2500 microscope and a Leica DFC290HD camera and analyzed using the Leica Application suite LAS v4.1 software. Images were processed with Adobe Photoshop CS5.
In a third experiment, secondary transplanted mice were treated for 14 days with tofacitinib (20 mg/kg per day, n = 5) or placebo (n = 5). The WBC count of placebo-treated mice increased significantly during the 14 days of treatment, whereas the overall increase in WBC count for tofacitinib-treated mice was lower compared with the placebo-treated mice (Figure 7C), indicating that tofacitinib has an inhibitory effect on the leukemia cells. In addition, mice treated with tofacitinib showed reduced spleen and thymus weight compared with mice treated with placebo (Figure 7D). Histopathologic analysis of spleen and thymus documented clear induction of apoptosis in the tofacitinib-treated animals, as observed by anticleaved caspase-3 staining and the typical “starry sky” pattern in hematoxylin and eosin (H&E)–stained tissue sections caused by the increased number of macrophages clearing the apoptotic cells (Figure 7E).
Discussion
Protein tyrosine kinases are attractive targets for therapy in oncology, because these proteins are often mutated and activated in cancer, and the tumor cells become addicted to these activated signaling pathways for their proliferation and survival. In addition, in leukemia, tyrosine kinases such as ABL1, FLT3, PDGFR, and JAK2 have been extensively studied as possible targets for therapy, with the successful application of kinase inhibitors for the treatment of CML as one of the most important breakthroughs in this area.26
The JAK3 tyrosine kinase was recently found to be recurrently mutated in juvenile myelomonocytic leukemia, NK cell lymphoma, and T-ALL, with mutations mainly clustering in the pseudokinase and kinase domains. Earlier studies had also reported rare mutations of JAK3 in AML. Together, these studies identify JAK3 as a possible new target for therapy in various hematologic malignancies. To further study the role of JAK3 as a possible therapeutic target, we performed a detailed analysis of the in vitro and in vivo transforming properties of a selection of 12 JAK3 mutations identified in T-ALL.
Using in vitro and in vivo models, we have shown that most JAK3 mutants identified in T-ALL patient samples were capable of conferring cytokine-independent growth to cell lines in vitro and to cause leukemia in vivo. Few of the mutants were lacking transforming potential in these experiments, illustrating that results from sequencing always need to be confirmed by functional assays to distinguish driver mutations from passenger mutations.
Based on the Ba/F3 and MOHITO data, we can also distinguish strong JAK3 mutants from weaker transforming mutants such as the A572T and V678M mutants. It is of interest to note that these 2 weaker alleles were identified together with a strong JAK3 mutation M511I or L857Q, respectively (supplemental Table 1). The fact that some T-ALL cases harbor 2 transforming JAK3 mutations suggests that T-ALL leukemia cells that are dependent on a JAK3 mutation can obtain an additional proliferation advantage by mutating the second JAK3 allele, similar to what has been observed for the JAK2 V617F mutation in myeloproliferative neoplasms.27
Several lines of evidence suggest that JAK1 is an essential kinase required downstream of JAK3 mutants. Indeed, a close relationship exists between JAK1 and JAK3 at the normal IL7 receptor (and other type I cytokine receptors), where both kinases phosphorylate and activate each other upon stimulation of the receptor. We observed that in the Ba/F3 cells transformed by JAK3 mutants, JAK1 was always phosphorylated, and that transformation by JAK3 mutants was lost upon knock-down of Jak1. Remarkably, the dependency on JAK1 was observed for all pseudokinase domain JAK3 mutants, but not for the JAK3 kinase domain mutant (L857Q). These observations are also in line with the data obtained with JAK kinase inhibitors, which show that the JAK3 pseudokinase mutants are more sensitive to ruxolitinib, a kinase inhibitor targeting JAK2 and JAK1, than the JAK3 kinase domain mutant (L857Q).24 In contrast, all JAK3 mutants showed equal sensitivity to tofacitinib, a JAK3-selective inhibitor.23 These findings indicate that different JAK3 mutations may have different signaling properties, which may affect their sensitivity to JAK kinase inhibitors.
Several JAK kinase inhibitors are currently under development for the treatment of myeloproliferative neoplasms with JAK2 mutation or for the treatment of autoimmune diseases. Our data show that both JAK1-selective and JAK3-selective inhibitors can have inhibitory effects on JAK3 mutation–positive leukemias, but that different JAK3 mutants show different signaling properties that affect their sensitivity to the various JAK inhibitors. Further preclinical and clinical studies are warranted and may bring new treatment opportunities for hematologic malignancies with JAK3 mutation.11,13,20,28,29
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Emilie Bittoun for assistance with histopathology and Drs Barata and Haan for providing constructs.
This study was supported by grants from the Belgian government (cancer plan), the FWO-Vlaanderen and the Foundation Against Cancer; a European Research Council-starting grant; the Interuniversity Attraction Poles granted by the Federal Office for Scientific, Technical and Cultural Affairs, Brussels, Belgium; and the Agency for Innovation by Science and Technology in Flanders, Belgium (S.D.).
Authorship
Contribution: S. Degryse, L.C., C.E.d.B., O.G., N.M., K.J., E.G., and V.G. performed experiments and analyzed data; T.T. supervised histopathology and analyzed data; S. Demeyer, G.H., M.F., and S.A. contributed to sequence analysis; J.P.M. contributed valuable reagents; S.D., C.E.d.B., and J.C. wrote the manuscript; and J.C. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Cools, Herestraat 49, Box 602, Leuven, 3000 Belgium; e-mail: jan.cools@cme.vib-kuleuven.be.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal