Abstract
Background:
Acquired aplastic anemia (AA), the prototypical bone marrow failure syndrome, is inferred to result from immune-mediated destruction of hematopoietic progenitors, as most patients respond to immunosuppressive therapies. Clonal hematopoiesis in AA is evident in the presence of paroxysmal nocturnal hemoglobinuria (PNH) cells in as many as half of patients and by identification of uniparental disomies involving 6p (6pUPD) chromosome in 13% of cases. In addition, "clonal transformation", as defined by the development of myelodysplastic syndromes (MDS) or acute myelogenous leukemia (AML) is a serious long-term complication in 10-15% AA patients.
Methods:
We performed targeted deep sequencing and SNP array-based copy number (CN) analysis of peripheral blood- or granulocyte-derived DNA from 439 patients with AA (280 from US and 159 from Japanese cohorts) for a panel of 103 candidate genes, chosen because they are known to be frequently mutated in myeloid neoplasms. Germline DNA was available for 288 out of 439 patients and was used to confirm the somatic origin of mutations. Whole exome sequencing (WES) was performed in 52 cases. Where serial samples were available, the chronology of detected mutations was also investigated.
Results:
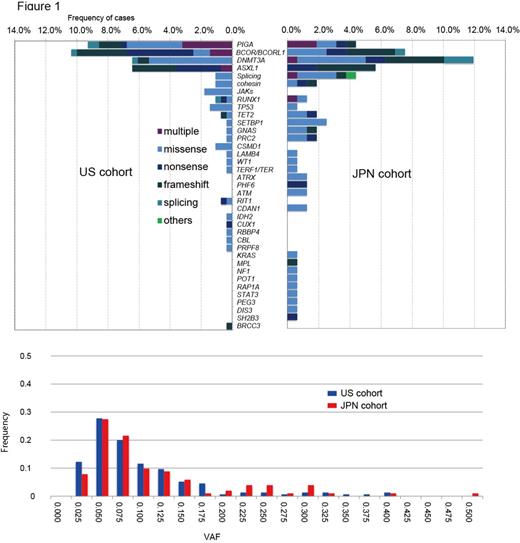
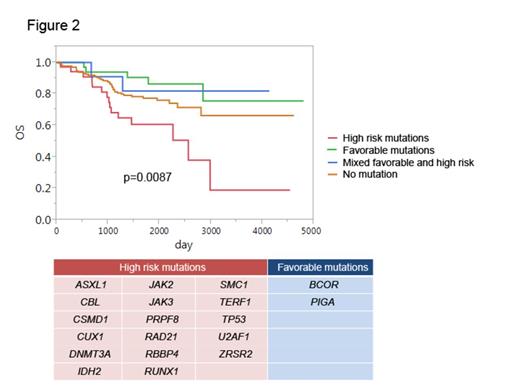
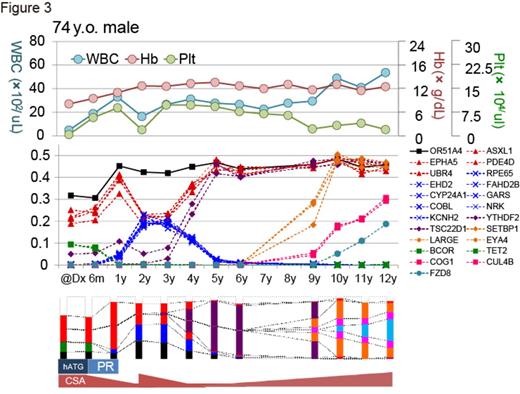
Targeted deep sequencing provided highly concordant results between the US and Japanese cohorts; approximately one third of AA patients had mutations in genes commonly affected in myeloid neoplasms, and about one third of patients in whom mutations were identified had multiple mutations. Multi-lineage involvement of mutations was confirmed in 6 cases using flow-sorted bone marrow samples. However, compared to myeloid neoplasms, mutations in AA were at much lower variant allele frequencies (VAFs) (<10% on average) and most frequently involved 5 genes: PIGA, BCOR/BCORL1, DNMT3A and ASXL1 (Fig.1). Although CN abnormalities were rare, about 13% of AA patients in both cohorts showed 6pUPD. Combined, clonal hematopoiesis was detected in as many as 46.5% and 47.8% of US and Japanese patients, respectively. We focused efforts on the large NIH cohort, due to accessible serial samples and well characterized clinical phenotypes at many time points. For 46 cases for which diagnostic samples were available, mutations were detected from at the time of diagnosis but at very low VAFs. The size of DNMT3A or ASXL1 mutated clones tended to increase over time, regardless of the emergence of chromosomal anomalies or blasts, whereas that of BCOR or PIGA mutated clones was more likely to decrease or remain stable. In both patient cohorts, presence of an acquired mutation was associated with older age, but did not correlate with response to immunosuppressive therapy (IST) or overall survival (OS). Mutations in PIGA and BCOR/BCORL1 were more common in AA than in MDS/AML and when combined, were associated with favorable OS (favorable mutations) (P = 0.044). Conversely, 17 high-risk mutations were extracted to predict poor OS (Fig. 2), which combined with favorable mutations, could be used to stratify AA patients with regard to OS (P = 0.0025). WES allowed capture of more mutations and better characterization of clonal hematopoiesis: more than 60% of AA patients had somatic mutations by combined targeted and whole exome sequencing. In 36 cases, WES was performed for all available serial samples, which enabled comprehensive monitoring of the dynamic chronological behavior of hematopoietic clones for as long as a decade after diagnosis. In many cases, clonal hematopoiesis developed gradually and was unrelated to the severity of cytopenias or to clinical evolution to abnormal cytogenetics, marrow dysplasia, and leukemia. Acquisition of new mutations within founder clones and subsequent selection shaped highly complex clonal structures in some cases (Fig. 3). The emergence of clonal hematopoiesis predated the development of MDS or leukemic transformation, with clones often detectable at time of diagnosis.
Conclusions:
Clonal hematopoiesis in AA was prevalent, associated in about half of cases with mutations in genes recurrently mutated in myeloid neoplasms. The highly biased set of mutated genes associated with clonal hematopoiesis in AA is evidence for Darwinian selection of particular cell clones under in the bone marrow failure environment. Mutations could be used to better predict prognosis of AA patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal