Key Points
IRF4 regulates MYC expression in ALCL.
ALCL survival depends on IRF4/MYC signaling.
Abstract
Anaplastic large cell lymphoma (ALCL) is a distinct entity of T-cell lymphoma that can be divided into 2 subtypes based on the presence of translocations involving the ALK gene (ALK+ and ALK− ALCL). The interferon regulatory factor 4 (IRF4) is known to be highly expressed in both ALK+ and ALK− ALCLs. However, the role of IRF4 in the pathogenesis of these lymphomas remains unclear. Here we show that ALCLs of both subtypes are addicted to IRF4 signaling, as knockdown of IRF4 by RNA interference was toxic to ALCL cell lines in vitro and in ALCL xenograft mouse models in vivo. Gene expression profiling after IRF4 knockdown demonstrated a significant downregulation of a variety of known MYC target genes. Furthermore, our analyses revealed that MYC is a primary target of IRF4, identifying a novel regulatory mechanism of MYC expression and its target gene network in ALCL. MYC, itself, is essential for ALCL survival, as both knockdown of MYC and pharmacologic inhibition of MYC signaling were toxic to ALCL cell lines. Collectively, our results demonstrate that ALCLs are dependent on IRF4 and MYC signaling and that MYC may represent a promising target for future therapies.
Introduction
ALCL is a distinct subtype of T-cell lymphoma and accounts for 2% to 3% of all malignant lymphoma cases.1,2 Within the current World Health Organization classification of lymphoid malignancies, 2 different subtypes of ALCL are distinguished based on the presence or absence of translocations involving the anaplastic lymphoma kinase (ALK) gene.3 The vast majority of ALK+ ALCLs are characterized by the t(2;5) (p23;q35) translocation that leads to the fusion of ALK to the nucleophosmin (NPM) gene resulting in the expression of the NPM-ALK chimeric transcript.4 ALK fusions are associated with activation of the catalytic domain of ALK, leading to constitutive activity of various downstream signaling pathways, such as JAK/STAT or PI3K/AKT.5-7 Despite these advances in the understanding of the molecular pathogenesis of ALK+ cases, ALCL lymphomagenesis is still not fully understood. In roughly 40% of systemic ALCLs, ALK translocations are not detectable, suggesting that not yet identified molecular aberrations may be responsible for ALCL development.8
Interferon regulatory factor 4 (IRF4) is a member of the IRF family of transcription factors that consists of 9 members.9 IRF4 is expressed in most cell types of the immune system and is induced in T-cells by T-cell receptor stimulation.10 IRF4 binds only weakly to DNA and thus interacts in T-cells with other transcription factors such as JUN and basic leucine zipper transcription factor (ATF-like) to either activate or repress gene expression.10-12 Different studies reported high IRF4 protein expression measured by immunohistochemistry in the vast majority of primary ALCL cases.13-15 However, its role in the molecular pathogenesis of these lymphomas remains unknown. To this end, we functionally investigated the role of IRF4 in the biology of ALCL. We detected that ALCLs are addicted to IRF4 by upregulating the oncogenic transcription factor MYC. Knockdown of both IRF4 and MYC induced toxicity in ALCL models, suggesting that the IRF4-MYC circuit may represent a promising molecular target for future therapies.
Material and methods
Cell culture, retroviral constructs, and transduction
The human ALK+ ALCL cell lines, K299, JB6, and SU-DHL-1, the ALK− ALCL line FE-PD, the Sézary syndrome (SS)-derived cell line HuT 78, the T-acute lymphoblastic leukemia (T-ALL) cell lines Jurkat and KE-37, and the multiple myeloma (MM) cell line H929 were cultured in RPMI 1640 with 10% fetal calf serum. DEL (ALK+ ALCL) and Mac-2A (ALK− ALCL) cells, as well as the MM cell line U266, were cultured in Iscove modified Dulbecco medium supplemented with 20% human plasma. All cell lines were maintained at 37°C with 5% CO2.
For efficient retroviral transductions, cell lines were engineered to express the murine ecotropic receptor as previously described.16,17 Additionally, these cell lines were engineered to express the bacterial tetracycline repressor allowing doxycycline-inducible small hairpin RNA (shRNA) or complementary DNA (cDNA) expression.16,17 The shRNA-mediated RNA interference and toxicity assays were performed as described.16,17 In brief, to assess toxicity of the shRNA, retroviruses that coexpress green fluorescent protein (GFP) were used. Flow cytometry was performed 2 days after shRNA transduction to determine the initial GFP-positive proportion of live cells for each shRNA. Subsequently, cells were cultured with doxycycline to induce shRNA expression and sampled over time. The GFP-positive proportion at each time was normalized to that of the negative control shRNA and further normalized to the day 2 fraction. The targeting sequence of IRF4 shRNAs #1 and #2 were CCGCCATTCCTCTATTCAAGA and GTGCCATTTCTCAGGGAAGTA as described.18 As a negative control, shRNA, we used a previously described nontoxic shRNA directed against MSMO1.16 Each shRNA experiment was completely reproduced at least 2 times for each cell line. For the shRNA rescue experiment, an IRF4 cDNA (NM_002460.3) and an MYC cDNA (NM_002467.2) were created and the experiment was performed as described.16 The rescue experiments were reproduced at least 2 times.
Patient samples and immunohistochemistry
Conventional full tissue slides of 82 ALCL samples, 38 ALK+ and 44 ALK−, were analyzed within the present study. MYC and IRF4 staining was performed as previously described.19,20 A cutoff level of ≥30% positive tumor cells was applied to define positivity of a sample as previously described for IRF4 and MYC.21,22
Clinical data were available for 14 of 82 patients analyzed. For these patients, treatment consisted of anthracycline-based polychemotherapy with or without radiotherapy.
In vivo xenograft mouse studies
For in vivo testing of the K299 and JB6 xenograft mouse models, 6- to 8-weekold female NOD.Cg-Prkdc severe combined immunodeficiency (SCID) Il2rgtm1Wjl/SzJ (NSG, Jackson Laboratory) mice were used. To induce either IRF4 shRNA #1 or the nontoxic shRNA directed against MSMO1, mice received drinking water supplemented with 1 mg/mL doxycycline (Genaxxon) and 5% sucrose at day 5 of palpable tumors of approximately 30 mm3. Tumor size was measured 3 times weekly in 2 dimensions using caliper and tumor volume was calculated according to the following formula: 1/2 × (length × width2 ). All animal experiments were conducted in accordance with the United Kingdom Co-ordinating Committee on Cancer Research guidelines for the welfare of animals and the German animal protection law. Detailed protocols are available in the supplemental Material and Methods on the Blood web site.
Gene expression profiling
Gene expression profiling was performed as previously described.23 Detailed protocols are available in the supplemental Material and Methods.
Quantitative PCR
Quantitative polymerase chain reaction (PCR) was performed as previously described using predesigned assays (Applied Biosystems).16 IRF4/MYC messenger RNA (mRNA) expression was normalized to the expression of GAPDH.
Western blotting
Protocols are available in the supplemental Material and Methods on the Blood Web site.
Chromatin immunoprecipitation
Detailed protocols are available in the supplemental Material and Methods. Previously described primer pairs were used for chromatin immunoprecipitation (ChIP) real-time PCR to investigate IRF4 binding in the MYC promoter and in a control locus.18,24,25 The applied primer pairs are summarized in supplemental Table 1.
In vitro viability assay and inhibitor studies
Cell viability of ALCL cell lines and U266 cells after JQ1 (Tocris Bioscience) and 10058-F4 (Sigma-Aldrich) inhibitor treatment was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) after 3 and 4 days of incubation, respectively, as described.16 Each experiment was reproduced 3 times for each cell line. Inhibitor studies using the ALK inhibitor crizotinib were performed methodically as described.16
Results
IRF4 is highly expressed in ALCL
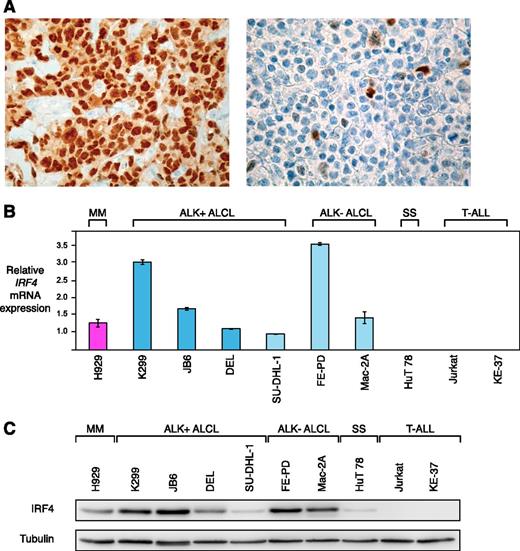
Previous work suggested that IRF4 is expressed in the majority of primary ALCL cases.13-15 To confirm these results, we assessed IRF4 protein expression in 82 primary ALCL patient samples by immunohistochemistry. Of these, 38 samples were ALK+, whereas 44 were ALK−. Overall, 75 of 82 (91%) cases stained positive for IRF4 (Figure 1A). Forty-three of 44 (98%) ALK− ALCLs expressed IRF4, whereas 32 of 38 (84%) ALK+ ALCLs had detectable IRF4 expression levels (P = .045; IRF4 expression in ALK− vs ALK+ ALCLs; 2-tailed Fisher’s exact test).
IRF4 is highly expressed in ALCL. (A) Immunohistochemical IRF4 staining of an IRF4-positive ALCL case (left) with internal negative controls (tumor-infiltrating macrophages and endothelia), and an IRF4-negative ALCL case (right) (original magnification ×320). (B) IRF4 mRNA is highly expressed in ALCL compared with the SS cell line HuT 78 and T-ALL cell lines as measured by quantitative PCR. IRF4 mRNA levels were normalized to expression of GAPDH. Error bars indicate standard deviation. (C) Western blot analysis of IRF4 expression in MM, ALCL, SS, and T-ALL cell lines. Five of 6 ALCL cell lines have high IRF4 protein expression.
IRF4 is highly expressed in ALCL. (A) Immunohistochemical IRF4 staining of an IRF4-positive ALCL case (left) with internal negative controls (tumor-infiltrating macrophages and endothelia), and an IRF4-negative ALCL case (right) (original magnification ×320). (B) IRF4 mRNA is highly expressed in ALCL compared with the SS cell line HuT 78 and T-ALL cell lines as measured by quantitative PCR. IRF4 mRNA levels were normalized to expression of GAPDH. Error bars indicate standard deviation. (C) Western blot analysis of IRF4 expression in MM, ALCL, SS, and T-ALL cell lines. Five of 6 ALCL cell lines have high IRF4 protein expression.
IRF4-positive and IRF4-negative cases could not be distinguished morphologically or pathologically. For a total of 14 patients, we were able to obtain clinical follow-up data. The median follow-up for these 14 patients was 49 months (range 6-288 months). In this cohort, 7 events occurred during follow-up (3 patients relapsed and 4 patients died [2 due to treatment-resistant progressive disease and 2 due to secondary malignancies]). Expression of none of the investigated immunohistochemical markers (IRF4 [P = .78] and MYC [P = .37] see below) were associated with differences in survival, albeit given the small sample size, one has to be very cautious in drawing definitive conclusions from these analyses.
Next, we analyzed whether established ALCL cell lines represent adequate models for functional analyses of IRF4. To this end, we determined IRF4 mRNA expression levels by quantitative PCR in 6 ALCL cell lines (including the 4 ALK+ cell lines K299, JB6, DEL, SU-DHL-1, and the 2 ALK− cell lines, FE-PD and Mac-2A). The MM cell line H929 served as a positive control, whereas the SS-derived cell line HuT 78 and 2 T-ALL cell lines (Jurkat and KE-37) were used as negative controls. All ALCL cell lines expressed higher IRF4 mRNA levels compared with the negative control lines and similar or higher levels compared with the levels detected in H929 cells (Figure 1B). These data were confirmed when IRF4 protein expression was investigated by western blotting. Five of 6 ALCL cell lines had high IRF4 expression levels (K299, JB6, DEL, FE-PD, and Mac-2A). Only SU-DHL-1 cells were characterized by low IRF4 expression (Figure 1C). In contrast, HuT 78 cells had low IRF4 expression levels, whereas Jurkat and KE-37 did not have any detectable IRF4 levels. Collectively, these data suggest that ALCL cell lines represent adequate models for functional analyses of IRF4.
Downregulation of IRF4 is toxic to ALCL in vitro and in vivo
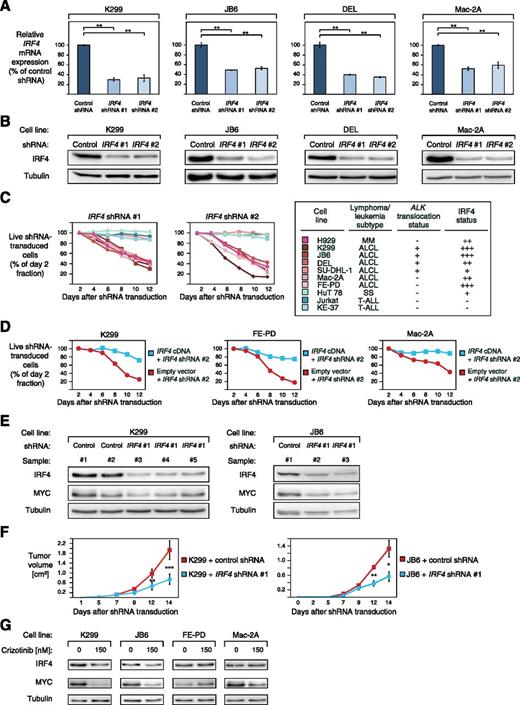
To elucidate the functional significance of IRF4 in ALCL, we knocked down its expression using 2 previously described specific shRNAs.18 Both IRF4 shRNAs significantly decreased IRF4 expression on mRNA level after 48 hours of shRNA induction and on protein level after 96 hours (Figure 2A-B). Transduction of these shRNAs induced toxicity in the positive MM control cell line H929, as described,18 and in ALCL cell lines expressing high levels of IRF4 (K299, JB6, DEL, FE-PD, and Mac-2A) (Figure 2C), whereas viability of SU-DHL-1 cells that are characterized by low IRF4 expression (Figure 1C) was not affected, despite significant IRF4 knockdown (Figure 2C; supplemental Figure 1A). Likewise, IRF4 downregulation did not affect survival of SS (HuT 78) or T-ALL cell lines (Jurkat and KE-37) that were used as negative controls (Figure 2C). A previously described, nontoxic shRNA directed against MSMO116 that we used as a negative control, did not induce any toxicity in our panel of cell lines (supplemental Figure 1B).
ALCLs are addicted to IRF4. (A) IRF4 shRNA #1 and #2 downregulate IRF4 mRNA in K299, JB6, DEL, and Mac-2A cells 48 hours after shRNA induction measured by quantitative PCR. IRF4 mRNA levels were normalized to expression of GAPDH. Error bars indicate standard deviation. (B) IRF4 shRNA #1 and #2 downregulate IRF4 protein in K299, JB6, DEL, and Mac-2A cells 96 hours after shRNA induction measured by western blotting. (C) IRF4 knockdown by two independent shRNAs is toxic to IRF4-positive ALCL cell lines. In contrast, the IRF4 low expressing ALCL cell line SU-DHL-1, the SS cell line HuT 78, and IRF4-negative T-ALL cell lines are not affected by IRF4 downregulation. Representative results from at least 2 independent replicates are shown. Baseline expression of IRF4 in the investigated cell lines is indicated based on western blotting (Figure 1C) and the ALK translocation status is indicated. (D) Exogenous expression of IRF4 cDNA rescues K299, FE-PD, and Mac-2A cells from IRF4 shRNA-induced toxicity. Representative results from 3 independent replicates are shown. (E) IRF4 and MYC knockdown is detectable by western blotting in mouse xenograft (K299 and JB6) tumor biopsies from cells transduced with IRF4 shRNA #1 compared with control shRNA transduced cells (shRNA directed against MSMO1). (F) Tumor growth curve of K299 and JB6 xenograft mouse models that inducibly express IRF4 shRNA #1 (blue) or a control shRNA directed against MSMO1 (red). IRF4 knockdown significantly reduced in vivo tumor growth (P = .0007 for K299, IRF4 shRNA vs control shRNA; P = .03 for JB6, IRF4 shRNA vs control shRNA; paired Student t test). Error bars indicate standard error of the mean. (G) Treatment of ALK+ (K299 and JB6) and ALK− (FE-PD and Mac-2A) ALCL cell lines with 150 nM crizotinib for 24 hours leads to IRF4 and MYC downregulation as measured by western blotting. *P < .05, **P < .01, ***P < .001.
ALCLs are addicted to IRF4. (A) IRF4 shRNA #1 and #2 downregulate IRF4 mRNA in K299, JB6, DEL, and Mac-2A cells 48 hours after shRNA induction measured by quantitative PCR. IRF4 mRNA levels were normalized to expression of GAPDH. Error bars indicate standard deviation. (B) IRF4 shRNA #1 and #2 downregulate IRF4 protein in K299, JB6, DEL, and Mac-2A cells 96 hours after shRNA induction measured by western blotting. (C) IRF4 knockdown by two independent shRNAs is toxic to IRF4-positive ALCL cell lines. In contrast, the IRF4 low expressing ALCL cell line SU-DHL-1, the SS cell line HuT 78, and IRF4-negative T-ALL cell lines are not affected by IRF4 downregulation. Representative results from at least 2 independent replicates are shown. Baseline expression of IRF4 in the investigated cell lines is indicated based on western blotting (Figure 1C) and the ALK translocation status is indicated. (D) Exogenous expression of IRF4 cDNA rescues K299, FE-PD, and Mac-2A cells from IRF4 shRNA-induced toxicity. Representative results from 3 independent replicates are shown. (E) IRF4 and MYC knockdown is detectable by western blotting in mouse xenograft (K299 and JB6) tumor biopsies from cells transduced with IRF4 shRNA #1 compared with control shRNA transduced cells (shRNA directed against MSMO1). (F) Tumor growth curve of K299 and JB6 xenograft mouse models that inducibly express IRF4 shRNA #1 (blue) or a control shRNA directed against MSMO1 (red). IRF4 knockdown significantly reduced in vivo tumor growth (P = .0007 for K299, IRF4 shRNA vs control shRNA; P = .03 for JB6, IRF4 shRNA vs control shRNA; paired Student t test). Error bars indicate standard error of the mean. (G) Treatment of ALK+ (K299 and JB6) and ALK− (FE-PD and Mac-2A) ALCL cell lines with 150 nM crizotinib for 24 hours leads to IRF4 and MYC downregulation as measured by western blotting. *P < .05, **P < .01, ***P < .001.
To demonstrate that IRF4 shRNA-mediated toxicity was specifically caused by IRF4 knockdown, we performed a rescue experiment by transducing K299, FE-PD, and Mac-2A cells with a vector that carries the IRF4 coding region, which is not targeted by IRF4 shRNA #2. Indeed, exogenous IRF4 expression rescued all ALCL cells from shRNA-mediated toxicity, indicating the specificity of our approach (Figure 2D).
We next determined if IRF4 dependency translates into an in vivo setting. To this end, we created ALCL xenograft mouse models using K299 and JB6 cells that were transduced with vectors that encode either IRF4 shRNA #1 or a control shRNA. Correspondingly, shRNA-mediated IRF4 knockdown was detectable in tumor samples from IRF4 shRNA-transduced cells compared with control shRNA transduced tumor samples (Figure 2E). In both JB6 and K299 models, IRF4 knockdown significantly impaired lymphoma growth over 14 days compared with ALCL cells transduced with the previously described nontoxic, control shRNA directed against MSMO116 (P = .0007 for K299, IRF4 shRNA vs control shRNA; P = .03 for JB6, IRF4 shRNA vs control shRNA; paired Student's t test) (Figure 2F), indicating that IRF4 promotes ALCL lymphoma growth. Collectively, our results indicate that ALCLs are dependent on the function of the transcription factor IRF4. Furthermore, this addiction is detectable in both ALK+ and ALK− ALCLs, indicating that the same pathways can be used by these 2 lymphoma subtypes.
NPM-ALK signaling upregulates IRF4 in ALCL
Previous work suggested that NPM-ALK signaling promotes IRF4 expression.26 To investigate if NPM-ALK signaling contributes to upregulated IRF4 expression in our panel of ALCL cell lines, we used the specific ALK inhibitor crizotinib. Treatment of ALK+ (K299 and JB6) and ALK− (FE-PD and Mac-2A) cells with 150 nM crizotinib for 24 hours significantly downregulated IRF4 expression in the 2 ALK+ cell lines, whereas the IRF4 levels in FE-PD and Mac2A cells were unaffected (Figure 2G). These results suggest that NPM-ALK signaling contributes to IRF4 upregulation in ALK+ ALCL.
IRF4 promotes MYC expression in ALCL
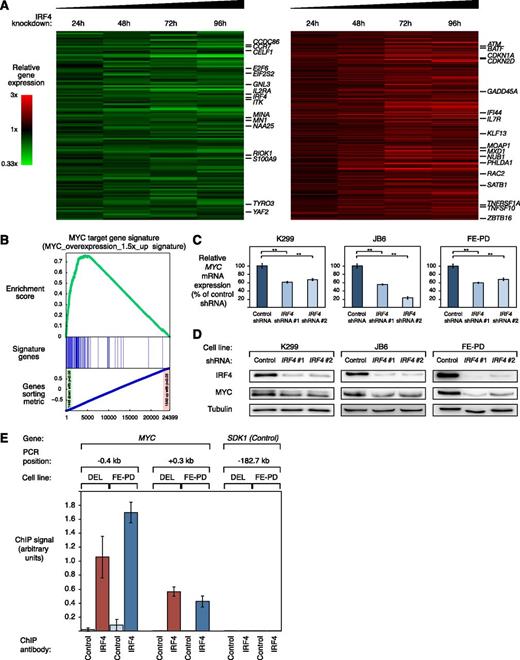
To investigate which biologic processes are regulated by IRF4 in ALCL, we profiled gene expression changes after 24, 48, 72, and 96 hours after IRF4 knockdown in the 2 ALK+ cell lines K299, and DEL and in the ALK− cell line FE-PD. To identify IRF4 regulated genes in both ALCL subtypes, we created a common gene expression signature consisting of genes that were consistently downregulated or upregulated across all time points in all 3 cell lines (Figure 3A; supplemental Table 2). Thus, we identified 115 genes that were significantly downregulated and 211 genes that were significantly upregulated (P < .0025) after IRF4 silencing (Figure 3A; supplemental Table 2).
IRF4 downregulation suppresses the gene expression network of MYC in ALCL. (A) Gene expression profiling after IRF4 knockdown in K299, DEL, and FE-PD cells. Changes of gene expression were profiled at the indicated time points after induction of IRF4 shRNA #1. Gene expression changes are depicted according to the color scale shown. Genes that are involved in critical biological processes are highlighted. (B) Gene set enrichment analysis of a previously described MYC gene expression signature. The MYC signature is significantly enriched with genes that are downregulated after IRF4 knockdown. (C) IRF4 shRNA #1 and #2 downregulate MYC mRNA in K299, JB6, and FE-PD cells 48 hours after shRNA induction as measured by quantitative PCR. MYC mRNA levels were normalized to expression of GAPDH. Error bars indicate the standard deviation. (D) IRF4 shRNA #1 and #2 downregulate MYC protein in K299, JB6, and FE-PD cells 48 hours after shRNA induction as measured by western blotting. (E) Chromatin immunoprecipitation quantitative PCR analysis of IRF4 binding in the MYC promoter and at a control locus in DEL (ALK+) and FE-PD (ALK−) cells. The control locus is a region on chromosome 7, used as a negative control for IRF4 binding, due to the lack of observable IRF4 binding in previous studies.24,25 Error bars depict standard error of the mean. **P < .01.
IRF4 downregulation suppresses the gene expression network of MYC in ALCL. (A) Gene expression profiling after IRF4 knockdown in K299, DEL, and FE-PD cells. Changes of gene expression were profiled at the indicated time points after induction of IRF4 shRNA #1. Gene expression changes are depicted according to the color scale shown. Genes that are involved in critical biological processes are highlighted. (B) Gene set enrichment analysis of a previously described MYC gene expression signature. The MYC signature is significantly enriched with genes that are downregulated after IRF4 knockdown. (C) IRF4 shRNA #1 and #2 downregulate MYC mRNA in K299, JB6, and FE-PD cells 48 hours after shRNA induction as measured by quantitative PCR. MYC mRNA levels were normalized to expression of GAPDH. Error bars indicate the standard deviation. (D) IRF4 shRNA #1 and #2 downregulate MYC protein in K299, JB6, and FE-PD cells 48 hours after shRNA induction as measured by western blotting. (E) Chromatin immunoprecipitation quantitative PCR analysis of IRF4 binding in the MYC promoter and at a control locus in DEL (ALK+) and FE-PD (ALK−) cells. The control locus is a region on chromosome 7, used as a negative control for IRF4 binding, due to the lack of observable IRF4 binding in previous studies.24,25 Error bars depict standard error of the mean. **P < .01.
IRF4 knockdown in ALK+ and ALK− ALCLs affected expression of various genes known to be involved in critical cellular processes such as cell cycle control (eg, CDKN1A, CDKN2D, E2F6), cell proliferation (eg, BAG2, CELF1, TYRO3), or DNA repair (eg, ATM, LIG3) (Figure 3A). In addition, we identified several known MYC target genes (eg, GNL3, MINA) that were deregulated after IRF4 knockdown. Moreover, MYC mRNA itself was significantly repressed by IRF4 knockdown (P = .01; paired t test), suggesting that IRF4 silencing leads to suppression of MYC and its target gene network. To test this hypothesis and obtain a better understanding of the gene expression changes in an unbiased manner, we performed a gene set enrichment analysis using previously described gene expression signatures.27,28 This analysis revealed that the most significantly downregulated gene signature was a previously described MYC target gene set (“Myc_overexpression_1.5×_up”; P < .001; false discovery rate [FDR] = 0.001) (Figure 3B; supplemental Figure 2A and supplemental Table 3). In addition, 2 other independent MYC target gene sets were significantly enriched with downregulated genes, indicating that MYC activity is inhibited by IRF4 downregulation (“Myc_RNAi_OCILy3”; P < .001; FDR = 0.001 and “Myc_ChIP_PET_Expr_Up”; P < .001; FDR = 0.001) (supplemental Table 3). To further confirm these results, we obtained several MYC target gene signatures from the Molecular Signatures Database.29 Indeed, all 5 MYC signatures were enriched with downregulated genes (supplemental Figure 2B; supplemental Table 4). Thus, our data indicate that IRF4 regulates MYC and its target gene network in both ALK+ and ALK− ALCL.
Besides MYC target gene sets, several signatures involved in critical cellular processes such as cell proliferation, HIF1A signaling, and Notch signaling were significantly downregulated after IRF4 silencing, indicating a potentially important role of IRF4 in regulating these processes and pathways (supplemental Table 3).
To confirm the gene expression data that MYC expression is controlled by IRF4 in ALCL cells, we determined MYC mRNA and protein expression levels after IRF4 shRNA-mediated knockdown. Real-time PCR confirmed that MYC mRNA is significantly downregulated in ALK+ (K299 and JB6) and ALK− (FE-PD) ALCL cell lines 48 hours after IRF4 knockdown (Figure 3C). Next, we investigated if IRF4 silencing represses MYC protein levels. Indeed, western blotting corroborated that IRF4 knockdown results in MYC protein downregulation in K299, JB6, and FE-PD cells 48 hours after shRNA induction (Figure 3D). These data were confirmed in vivo in our xenograft models, as MYC protein knockdown was detectable in tumors derived from cells transduced with IRF4 shRNA #1 compared with cells transduced with a control shRNA (Figure 2E). At last, we determined MYC protein levels 24 hours after crizotinib treatment using western blot analysis. Crizotinib treatment significantly downregulated both IRF4 and MYC protein expression, supporting our data of an IRF4-MYC circuit (Figure 2G).
To investigate whether MYC is a primary or secondary IRF4 target, we performed conventional IRF4 ChIP of the MYC promoter in DEL (ALK+) and FE-PD (ALK−) cells. In both cell lines, we detected IRF4 binding in the MYC promoter using previously described primer pairs,24 involving the regions −0.4 to +0.3 kb with respect to the MYC transcriptional start site (Figure 3E). Thus, our data indicate that MYC is a primary IRF4 target in ALK+ and ALK− ALCL cells and that IRF4, therefore, directly controls the MYC-driven gene expression network in these lymphomas.
MYC is critical for ALCL survival and may represent a novel target for ALCL therapy
As our analyses identified MYC as one of the predominant IRF4 target genes, we investigated its role in mediating ALCL cell survival. First, we determined MYC expression in our panel of ALCL cell lines and in our cohort of primary ALCL samples. All ALCL cell lines expressed MYC levels as determined by western blotting (Figure 4A), whereas 69 of 82 (84%) primary ALCL samples were MYC-positive measured by immunohistochemistry (Figure 4B). MYC expression was irrespective of the ALK status. Thirty-eight of 44 (86%) primary ALK− ALCLs expressed MYC, whereas 31 of 38 (82%) ALK+ ALCLs had detectable MYC expression (P = .76; MYC expression in ALK+ vs ALK− ALCLs; 2-tailed Fisher’s exact test). In 67 of 82 (82%) samples, both IRF4 and MYC were coexpressed and thus IRF4-positive samples were more frequently MYC positive compared with IRF4 negative cases (P = 8.3 × 10−4; Fisher’s exact test).
ALCLs depend in their survival on MYC signaling. (A) Western blot analysis of MYC expression in 6 ALCL cell lines and in the MM cell line U266. All ALCL cell lines have detectable MYC expression compared with the negative control U266. (B) Immunohistochemical MYC staining of an MYC-positive ALCL case (left) and an MYC-negative ALCL case (right) (original magnification ×320). (C) MYC shRNA #1 and #2 significantly downregulate MYC protein 96 hours after induction measured by western blotting in K299, DEL, FE-PD, and Mac-2A cells. (D) shRNA-mediated MYC knockdown is toxic to ALCL cell lines. Baseline expression of MYC in the investigated cell lines based on western blotting (seen in panel A) and the ALK translocation status are indicated. Representative results are shown. (E) Expression of an MYC cDNA rescues DEL, JB6, and FE-PD cells, transduced with MYC shRNA #2 (targeting the 3′UTR [untranslated region] of MYC) from toxicity. Representative results from 2 independent replicates are shown. (F) Expression of an MYC cDNA rescues DEL cells, transduced with IRF4 shRNA #1 and #2 from toxicity. In contrast, JB6 and FE-PD cells are not rescued from shRNA-induced toxicity. Representative results from at least 2 independent replicates are shown. (G) ALCL cell lines are sensitive to MYC inhibition using the BET family inhibitor JQ1 after 3 days of incubation. Viability of the negative control cell line U266 is not affected by JQ1. Baseline expression of MYC in the investigated cell lines based on western blotting (seen in panel A) is indicated. Representative results of 3 replicates are shown.
ALCLs depend in their survival on MYC signaling. (A) Western blot analysis of MYC expression in 6 ALCL cell lines and in the MM cell line U266. All ALCL cell lines have detectable MYC expression compared with the negative control U266. (B) Immunohistochemical MYC staining of an MYC-positive ALCL case (left) and an MYC-negative ALCL case (right) (original magnification ×320). (C) MYC shRNA #1 and #2 significantly downregulate MYC protein 96 hours after induction measured by western blotting in K299, DEL, FE-PD, and Mac-2A cells. (D) shRNA-mediated MYC knockdown is toxic to ALCL cell lines. Baseline expression of MYC in the investigated cell lines based on western blotting (seen in panel A) and the ALK translocation status are indicated. Representative results are shown. (E) Expression of an MYC cDNA rescues DEL, JB6, and FE-PD cells, transduced with MYC shRNA #2 (targeting the 3′UTR [untranslated region] of MYC) from toxicity. Representative results from 2 independent replicates are shown. (F) Expression of an MYC cDNA rescues DEL cells, transduced with IRF4 shRNA #1 and #2 from toxicity. In contrast, JB6 and FE-PD cells are not rescued from shRNA-induced toxicity. Representative results from at least 2 independent replicates are shown. (G) ALCL cell lines are sensitive to MYC inhibition using the BET family inhibitor JQ1 after 3 days of incubation. Viability of the negative control cell line U266 is not affected by JQ1. Baseline expression of MYC in the investigated cell lines based on western blotting (seen in panel A) is indicated. Representative results of 3 replicates are shown.
To functionally analyze the role of MYC for ALCL survival, we knocked down its expression using 2 different specific shRNAs that mediate significant MYC downregulation 96 hours after shRNA induction (Figure 4C). MYC silencing induced cytotoxicity in all ALCL cell lines, confirming that MYC plays a pivotal role for ALCL survival (Figure 4D). As expected, expression of an MYC cDNA rescued ALCL cells from MYC knockdown-induced toxicity, confirming the specificity of our shRNA approach (Figure 4E).
To evaluate the degree to which MYC downregulation contributes to IRF4 knockdown-induced toxicity, we performed a rescue experiment in which we introduced an MYC cDNA or an empty vector in IRF4 #1 and #2 shRNA-transduced DEL, JB6, and FE-PD cells. We detected a complete MYC-induced rescue in DEL cells indicating that MYC knockdown significantly contributes to the lethal effect of IRF4 downregulation in these cells. In contrast, in JB6 and FE-PD cells, no MYC cDNA-induced rescue was detectable (Figure 4F), indicating that deregulation of additional signaling pathways contributes to the IRF4 knockdown-induced toxicity in these cells.
Finally, we tested whether the addiction to MYC signaling can be exploited therapeutically. To this end, we treated ALCL cell lines with the BET family inhibitor JQ1 that downregulates MYC expression, and we determined cell viability after 3 days of incubation.30,31 The MM cell line U266 that does not express MYC (Figure 4A), but MYCL,32 was used as a negative control. JQ1 significantly reduced cell viability of all ALCL cell lines irrespective of their ALK translocation status, whereas viability of U266 was not affected (Figure 4G). These data were supported when we treated our ALCL cell line panel and U266 with the small molecule inhibitor 10058-F4 that inhibits MYC-MAX heterodimerization.33 Viability of most ALCL cell lines was strongly affected by MYC-MAX inhibition compared with the control cell line (supplemental Figure 3). Collectively, our data indicate an important role of MYC in maintaining ALCL survival that can be blocked pharmacologically, suggesting that MYC inhibition may represent a novel therapeutic approach for ALCL patients.
Discussion
Within the present study, we have identified an unexpected role of IRF4 in the biology of ALK+ and ALK− ALCLs. Our data demonstrate that both subtypes of ALCL are addicted to the IRF4 gene expression network, as IRF4 knockdown induced toxicity in ALCL models both in vitro and in vivo. The molecular mechanisms leading to IRF4 upregulation in ALCL remain not to be completely understood. Translocations involving IRF4 have been described in peripheral T-cell lymphoma such as cutaneous ALCL, but are very rare in systemic ALCL.15 The NPM-ALK chimeric protein and IL-2 signaling have been shown to upregulate IRF4 protein by activating the transcription factors STAT3 and STAT5 in T-cell lymphoma cells.26 The role of NPM-ALK signaling in mediating IRF4 expression was confirmed in our study, as treatment with the ALK inhibitor crizotinib downregulated IRF4 expression in ALK+ ALCL cell lines. However, as both ALK+ and ALK− ALCLs express IRF4, potentially other molecular mechanisms promote IRF4 expression in these lymphomas. In the activated B-cell–like (ABC) subtype of diffuse large B-cell lymphoma (DLBCL) it was recently shown that IRF4 is a target gene of the oncogenic nuclear factor-κB (NF-κB) signaling pathway.34,35 In ALCL, previous studies showed that the NPM-ALK fusion protein suppresses NF-κB activation in ALK+ ALCL.36 However, it is possible that in ALK− ALCLs, NF-κB signaling might be involved in IRF4 regulation, and this should be addressed in future work.
Our gene expression analyses revealed that IRF4 knockdown affects a number of critical cellular processes such as proliferation, apoptosis, and DNA damage, and potentially oncogenic signaling pathways such as Notch indicating a central role of IRF4 in the molecular pathogenesis of ALCL. The predominant primary IRF4 target that emerged from our analyses is the oncogenic transcription factor MYC. MYC is aberrantly expressed in a variety of different forms of cancer and plays an important role in the biology of various lymphoma subtypes such as Burkitt lymphoma or DLBCL.37 MYC regulates roughly 15% of all genes in the human genome and has known activating and repressing functions on its target genes. It is involved in the regulation of cell cycle control, cell growth, protein synthesis, angiogenesis, and apoptosis.37 Previous work by Raetz et al38 suggested that MYC is a downstream target of ALK and thus it is predominantly expressed in ALK+ ALCLs. In this study, all ALK− ALCLs did not express MYC protein as measured by immunohistochemistry.38 However, our results indicate that IRF4 upregulates MYC in both ALK+ and ALK− ALCLs. Accordingly, we could not confirm a difference in MYC expression in ALK+ and ALK− ALCLs, as MYC was detectable in cell lines and in primary patient samples of both subtypes. The discrepant immunohistochemistry results may be caused by the use of different antibodies and differences in the staining techniques.38
Previous work identified MYC as a direct transcriptional target of IRF4 in MM cells.18 In contrast, IRF4, which is also involved in the biology of ABC DLBCL, does not bind to the MYC promoter in these lymphomas.25 Given that IRF4 upregulates MYC expression in ALCL as indicated by our results, it seems conceivable that IRF4 regulates specific target genes and gene expression networks in a context-dependent manner. One possible explanation is that IRF4, which only binds weakly to DNA by itself, interacts with different DNA binding partners in these different hematologic malignancies. However, additional analyses in the future are required to decipher these specific IRF4 functions.
IRF4 and MYC expression measured by immunohistochemistry were significantly associated with each other and both proteins were coexpressed in 82% of the samples. However, in some cases, IRF4 and MYC expression did not correlate, potentially indicating that additional molecular mechanisms regulate MYC expression in ALCL. Translocations affecting MYC are infrequent in these lymphomas and seem to be associated with an aggressive clinical course.39-41 In contrast, a recent study using array comparative genomic hybridization on primary ALCL samples reported recurrent MYC gains.42 In addition, other MYC upregulating mechanisms may be used by ALCL.
Despite the evident role of IRF4 in mediating MYC expression in ALCL, we detected a MYC-induced rescue of IRF4-induced toxicity in only 1 of 3 ALCL cell lines. Potentially, these data can be explained by different mechanisms. It is possible that the survival of the 2 cell lines that were not rescued by MYC depend on additional signaling pathways that are deregulated by IRF4 knockdown such as HIF1A or Notch. Alternatively, we cannot completely rule out technical reasons such as inadequate MYC expression in these cells.
Finally, we evaluated if addiction to MYC signaling can be exploited therapeutically in ALCL. Despite improvements in therapy, there is still a significant fraction of patients diagnosed with ALCL who succumb to their disease.43 Two different MYC inhibitors induced cytotoxicity in all ALK+ and ALK− ALCL cell line models. These data suggest that MYC inhibition may offer a promising target and a novel therapeutic strategy to overcome therapy resistance in patients diagnosed with ALCL, irrespective of their ALK status.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by research grants from the Deutsche Krebshilfe and the Swiss National Science Foundation (Sinergia grant) (G.L.); the Deutsche Forschungsgemeinschaft, DFG EXC 1003 Cells in Motion - Cluster of Excellence, Münster, Germany, and a Doctoral Scholarship from the Philipps-University Marburg (M.G.).
Authorship
Contribution: A.W., U.S., I.A., M.H., A.S., C.H., H.M., and B.W.-W. performed experiments; A.W. and G.L. designed research; A.W. and G.L. wrote the manuscript; M.G. performed bioinformatic and biophysical analyses; T.E., U.S., I.A., M.H., and A.S. analyzed experiments; and O.M., I.F., B.D., S.D., S.M., M.J., N.C.T.E., A.R., G.O., P.L., A.T., and G.L. analyzed data.
Conflict-of -interest disclosure: The authors declare no competing financial interest.
Correspondence: Georg Lenz, Medical Department A, University Hospital Münster, Translational Oncology, Albert-Schweitzer Campus 1, 48149 Münster, Germany; e-mail: georg.lenz@ukmuenster.de.

![Figure 4. ALCLs depend in their survival on MYC signaling. (A) Western blot analysis of MYC expression in 6 ALCL cell lines and in the MM cell line U266. All ALCL cell lines have detectable MYC expression compared with the negative control U266. (B) Immunohistochemical MYC staining of an MYC-positive ALCL case (left) and an MYC-negative ALCL case (right) (original magnification ×320). (C) MYC shRNA #1 and #2 significantly downregulate MYC protein 96 hours after induction measured by western blotting in K299, DEL, FE-PD, and Mac-2A cells. (D) shRNA-mediated MYC knockdown is toxic to ALCL cell lines. Baseline expression of MYC in the investigated cell lines based on western blotting (seen in panel A) and the ALK translocation status are indicated. Representative results are shown. (E) Expression of an MYC cDNA rescues DEL, JB6, and FE-PD cells, transduced with MYC shRNA #2 (targeting the 3′UTR [untranslated region] of MYC) from toxicity. Representative results from 2 independent replicates are shown. (F) Expression of an MYC cDNA rescues DEL cells, transduced with IRF4 shRNA #1 and #2 from toxicity. In contrast, JB6 and FE-PD cells are not rescued from shRNA-induced toxicity. Representative results from at least 2 independent replicates are shown. (G) ALCL cell lines are sensitive to MYC inhibition using the BET family inhibitor JQ1 after 3 days of incubation. Viability of the negative control cell line U266 is not affected by JQ1. Baseline expression of MYC in the investigated cell lines based on western blotting (seen in panel A) is indicated. Representative results of 3 replicates are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/1/10.1182_blood-2014-08-594507/4/m_124f4.jpeg?Expires=1765899626&Signature=TwWIms~b7wyuUfbXhGzlIgXCl4-JfDMj2om9GcTGAB7N91-VKmtfrU922DCEjX0aJgrJYd2hygvxKQ6JscSmR--rDJ13C3-5ON3de0w3t7tr4l0G-fZ2EzKJKG9dLv5xQEzLxSlMqFiBl~ywxbg-hSzu5o6vxVTgF~NBzVOiOXND2UYOmifLXkRxcCocZM81BVSbzhzeKyXFxn0xT6F7jJOCQ~YHATbaB1z1dBq3LX9UaXXOsy~nt-ikJUtSHukaPPpB-LCRM~hrDMC0jO2QCjyfD3Nt-zjWd3A47dQMvoPg0eR3poO8XcrCMWDOKieIE8WjLdcO4oHxfCaU8WqJ9Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)