Key Points
NKR-P1B is involved in NK cell tolerance and MHC-I-independent missing-self recognition of Clr-b-deficient target cells.
The NKR-P1B:Clr-b system plays a role in tumor surveillance and immune escape in the Eμ-myc transgenic mouse model of B-cell lymphoma.
Abstract
NKR-P1B is a homodimeric type II transmembrane C-type lectinlike receptor that inhibits natural killer (NK) cell function upon interaction with its cognate C-type lectin-related ligand, Clr-b. The NKR-P1B:Clr-b interaction represents a major histocompatibility complex class I (MHC-I)-independent missing-self recognition system that monitors cellular Clr-b levels. We have generated NKR-P1BB6-deficient (Nkrp1b−/−) mice to study the role of NKR-P1B in NK cell development and function in vivo. NK cell inhibition by Clr-b is abolished in Nkrp1b−/− mice, confirming the inhibitory nature of NKR-P1BB6. Inhibitory receptors also promote NK cell tolerance and responsiveness to stimulation; hence, NK cells expressing NKR-P1BB6 and Ly49C/I display augmented responsiveness to activating signals vs NK cells expressing either or none of the receptors. In addition, Nkrp1b−/− mice are defective in rejecting cells lacking Clr-b, supporting a role for NKR-P1BB6 in MHC-I-independent missing-self recognition of Clr-b in vivo. In contrast, MHC-I-dependent missing-self recognition is preserved in Nkrp1b−/− mice. Interestingly, spontaneous myc-induced B lymphoma cells may selectively use NKR-P1B:Clr-b interactions to escape immune surveillance by wild-type, but not Nkrp1b−/−, NK cells. These data provide direct genetic evidence of a role for NKR-P1B in NK cell tolerance and MHC-I-independent missing-self recognition.
Introduction
The importance of natural killer (NK) cells in host defense against microbial infections and tumors has been highlighted in individuals lacking NK cells or NK cell functions; these individuals suffer from persistent and life-threatening infections of normally benign herpes viruses and tumors.1-3 NK cell function is regulated by integrating activating and inhibitory signals from engaged NK cell receptors.4 The NK cell receptor repertoire in mice includes the Ly49, NKG2D, CD94/NKG2, and NKR-P1 families of receptors, all of which are encoded by genes in the NK gene complex (NKC) on chromosome 6.5,6 The well-characterized Ly49 receptor family is the mouse functional equivalent of the human killer cell immunoglobulin-like receptor (KIR) family, which recognizes class I major histocompatibility complex (MHC-I) molecules.5,6 This NK cell recognition system, termed “missing-self,” involves surveillance of host MHC-I molecules and the response to cells without MHC-I expression.7
NKR-P1 receptors are homodimeric type II transmembrane C-type lectinlike molecules4,6,8 and are conserved across many species.9 This receptor family consists of 5 members in mice (NKR-P1A, NKR-P1B/D, NKR-P1C, NKR-P1F, and NKR-P1G; Nkrp1e is a pseudogene).10-12 NKR-P1A and NKR-P1F are proposed to be activating and are expressed at low levels on all NK cells.13 The activating NK1.1 (NKR-P1C) receptor, a prototypical antigen defining mouse NK cells in the C57BL/6 (B6) mouse strain, is a product of the Nkrp1cB6 gene.14 NKR-P1G has only recently been documented to be inhibitory and primarily involved in mucosal immunity,15 whereas NKR-P1B is a known inhibitory receptor first identified in the Swiss and SJL mouse strains.10,11,16-18 At least 3 different Nkrp1b alleles have been described. The B6 allele has been variably termed Nkrp1d or Nkrp1bB6, and encodes the NKR-P1BB6 receptor, also known as NKR-P1D.11 NKR-P1B is expressed only on a subset of NK cells.13 NKR-P1B+ and NKR-P1B− NK cells differ in their expression of other NK cell receptors in both mice and rats.13,19,20 Ligands for most NKR-P1 receptors have been identified as members of the C-type lectin-related (Clr) family of membrane glycoproteins encoded by the Clec2 genes, which are intermingled among the Nkrp1 (Klrb1) genes within the mouse NKC.12,17,18 The currently known receptor-ligand pairs for these families include NKR-P1B:Clr-b; NKR-P1F:Clr-c,d,g; and NKR-P1G:Clr-d,f,g.17,18,21,22
We have generated an NKR-P1B-deficient B6 mouse strain to study the role of NKR-P1B in NK cell development and function in vivo. We chose to target NKR-P1B for the following reasons: (1) the NKR-P1B:Clr-b system is well characterized in mice, and appropriate reagents, such as specific monoclonal antibodies (mAbs) and a complementary Clr-b gene–deficient mouse strain, are available13,23 ; (2) the NKR-P1B:Clr-b system is analogous to the inhibitory NKR-P1A:LLT1 system in humans, although their expression patterns may vary24,25 ; (3) the existence of 3 significantly different Nkrp1b alleles suggests a possible divergence as a result of pathogen challenge (eg, rat cytomegalovirus encodes a C-type lectinlike protein with homology to rat Clr-11 [Clec2d11] that protects infected cells from NK recognition via the inhibitory rat NKR-P1B receptor)26 ; and (4) in contrast to other, tissue-specific Clr family members, Clr-b, like MHC-I, is broadly expressed on hematopoietic cells, and its expression on transfected cells protects them from NK-mediated lysis.12,17,18,27 In addition, Clr-b expression is often downmodulated on tumor cell lines after virus infection and during genotoxic and cellular stress in vitro.17,26,28 Therefore, NKR-P1B:Clr-b interactions represent an MHC-I-independent missing-self recognition system to monitor cellular levels of Clr-b.17
Materials and methods
Mice
C57BL/6 (B6), β2m-deficient (B2m−/−) (B6.129P2-B2mtm1Unc/J), CMV-cre transgenic (Tg) (B6.C-Tg[CMV-cre]1Cgn/J), and Eµ-myc Tg (B6.Cg-Tg[IghMyc]22Bri/J) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Clr-b-deficient (Ocil−/−) mice were previously described.23 β2m/Clr-b double-deficient (B2m−/−Clrb−/−) mice were produced by breeding Ocil−/− mice with B2m−/− mice. All mice were maintained in the Animal Care and Veterinary Service at the University of Ottawa (Ottawa, Ontario), Sunnybrook Research Institute, or the Donnelly Center for Cellular and Biomolecular Research, University of Toronto (Toronto, Ontario), in accordance with institutional guidelines.
Generation of NKR-P1B-deficient mice
All genetic modifications were performed on the Nkrp1bB6 allele. For clarity and simplicity, this allele will be referred to as Nkrp1b and the receptor as NKR-P1B in the remainder of this article. A targeting vector containing Nkrp1b genomic sequence with a floxed phosphoglycerate kinase (PGK)–neomycin cassette replacing exons 2 to 5 of Nkrp1b was created in a modified pBluescript-SK+ vector by bacterial artificial chromosome recombineering using clone RP23-127M20 in SW106 bacteria with an EcoRV-flanked galactokinase selection cassette, as previously described.29 For a brief description of the strategy, please refer to the supplemental Materials and Methods on the Blood Web site. The pBluescript backbone was removed after AatII and SalI digestion before electroporation into C57BL/6 Bruce-4 embryonic stem (ES) cells, followed by selection in G418. Neomycin-resistant clones were screened by Southern blot analysis, and a targeting efficiency of ∼17% was observed. Chimeric Nkrp1bneo founder mice were produced with Nkrp1b-targeted ES cells by the Clinical Research Institute of Montreal microinjection service (Montreal, Quebec). These mice were bred with B6 females to produce Nkrp1bwt/neo heterozygous mice. Heterozygous mice were interbred to obtain Nkrp1bneo/neo mice. To remove the neomycin cassette, Nkrp1bneo/neo mice were bred with CMV-cre Tg mice on a B6 background (The Jackson Laboratory). The resulting Nkrp1blox/wt mice were interbred to produce Nkrp1blox/lox mice. Mice were genotyped regularly using specific primers (supplemental Materials and Methods). Wild-type (WT) and NKR-P1B-deficient littermates were used in all experiments unless otherwise indicated.
Cells
YAC-1 and CHO cells were purchased from the American Type Culture Collection. CHO cells were stably transfected with pcDNA3-Clr-b expression vector using Lipofectamine (Invitrogen). Lymphokine-activated killer (LAK) cells and bone marrow–derived dendritic cells (BM-DCs) were generated as previously described.30,31
Flow cytometry
For the source of commercially purchased antibodies, please refer to the supplemental Materials and Methods. Anti-Clr-b (4A6) and anti-NKR-P1B (2D9) antibodies have been previously described.13,17,18 Anti-CRACC antibody and anti-NKR-P1B (2D12) hybridoma were kind gifts from Dr André Veillette (Clinical Research Institute of Montreal) and Dr. Koho Iizuka (University of Minnesota, Minneapolis, Minnesota), respectively. Antibody staining for flow cytometry was performed as previously described.32
In vitro NK cell assays
NK cell cytotoxicity was measured using the standard 4-hour 51Cr-release assay as previously described.33 For intracellular interferon (IFN)-γ measurement and CD107a staining, splenocytes from polyinosinic:polycytidylic acid (poly[I:C])-treated mice (150-µg intraperitoneal [IP] injection for 18 hours) or phosphate-buffered saline–treated mice were incubated with YAC-1 cells, plate-bound anti-NKR-P1C (NK1.1), and anti-NKR-P1B (2D12), or with phorbol 12-myristate 13-acetate (PMA; 10 ng/mL) and ionomycin (1 µg/mL) for 5 hours with brefeldin A and monensin (eBioscience). Intracellular IFN-γ staining using the Cytofix/Cytoperm kit (BD Biosciences) and CD107a staining were performed as previously described.34
In vivo splenocyte rejection assay
Splenocytes from WT B6, MHC-I-deficient, Clr-b-deficient, and MHC-I/Clr-b double-deficient mice were labeled with different concentrations of 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (Invitrogen) as previously described.35 A 1:1 mixture of 1 × 107 WT and various gene-deficient splenocytes were injected into the tail vein of recipient mice that were either untreated or treated with 150 µg of IP poly(I:C) for 24 hours or 5 µg of IV CpG-B (ODN 2006, Hycult Biotech) and 30 µg of IV dioleoyltrimethylammoniumpropane (DOTAP) (Invitrogen) for 6 hours prior to splenocyte injection. For NK cell depletion, mice were treated with 200 µg of IP anti-NKR-P1C (NK1.1) antibody 48 hours prior to splenocyte injection. Spleens of recipient mice were harvested 5 to 18 hours later and analyzed as previously described.35
Statistical analysis
Statistical significance was determined by Student t test and log-rank test, where applicable, with a cut-off P value of .05.
Results
Targeted deletion of the Nkrp1b gene
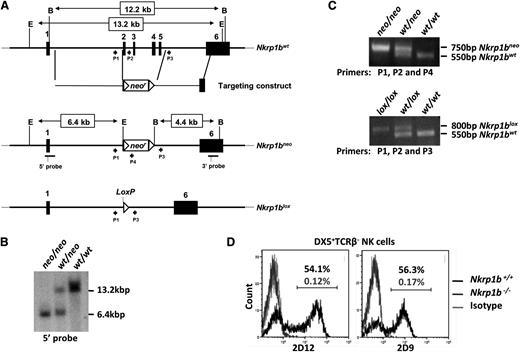
We generated NKR-P1B-deficient mice on a B6 background to study this receptor’s role in NK cell function. A floxed neomycin cassette was inserted into the Nkrp1b gene by homologous recombination, replacing exons 2 to 5 in B6-background ES cells (Figure 1A). Founder mice carrying the Nkrp1bneo allele were bred to WT B6 females. The resulting Nkrp1bwt/neo offspring were genotyped by Southern blot analysis and PCR, and interbred to produce Nkrp1bneo/neo mice (Figure 1B-C). Nkrp1bneo/neo mice were then bred with CMV-cre Tg mice on a B6 background to remove the neomycin cassette through Cre-mediated recombination. The resulting Nkrp1bwt/lox mice were genotyped by PCR and interbred to obtain Nkrp1blox/lox mice (Figure 1C). We confirmed Nkrp1b deletion by flow cytometric analysis using 2 different NKR-P1B-specific mAbs, 2D9 and 2D12.13,18 DX5+TCRβ− NK cells from Nkrp1blox/lox mice lacked any detectable NKR-P1B surface expression (Figure 1D) and are, therefore, referred to as Nkrp1b−/− or NKR-P1B-deficient mice hereafter. NKR-P1B-deficient progeny from heterozygous parents were born with expected Mendelian frequency and without any gross pathological signs. These mice display normal immune development, including normal numbers of T, B, dendritic, NK, and NK T (NKT) cells in central and peripheral immune organs, comparable to B6 WT mice (Table 1).
Generation of NKR-P1B-deficient mice. (A) Nkrp1b deletion strategy. Exons 2 to 5 were replaced with a floxed neomycin (neor) cassette by homologous recombination in ES cells of B6 background. Correctly targeted ES cell clones were selected by Southern blot analysis and used to generate Nkrp1bneo mice. These mice were bred with CMV-cre Tg mice to produce Nkrp1blox mice. Filled boxes denote exons (numbered), and arrowheads represent PCR primers (P) described in the “Materials and methods” section. The location of 5′ and 3′ Southern probes is underlined, and EcoRV (E) and BamHI (B) restriction enzyme sites are shown. (B) Southern blot of EcoRV-digested genomic DNA from mice of the indicated genotypes using the 5′ probe. (C) PCR analysis of tail DNA from Nkrp1bneo and Nkrp1blox mice. (D) Surface expression of NKR-P1B is absent on NK cells from NKR-P1B-deficient mice. Splenocytes from WT and Nkrp1blox/lox mice were stained with mAbs to DX5, TCRβ, and NKR-P1B (2D9 and 2D12). The percentage of NKR-P1B+ NK cells (DX5+TCRβ–) is indicated. PCR, polymerase chain reaction.
Generation of NKR-P1B-deficient mice. (A) Nkrp1b deletion strategy. Exons 2 to 5 were replaced with a floxed neomycin (neor) cassette by homologous recombination in ES cells of B6 background. Correctly targeted ES cell clones were selected by Southern blot analysis and used to generate Nkrp1bneo mice. These mice were bred with CMV-cre Tg mice to produce Nkrp1blox mice. Filled boxes denote exons (numbered), and arrowheads represent PCR primers (P) described in the “Materials and methods” section. The location of 5′ and 3′ Southern probes is underlined, and EcoRV (E) and BamHI (B) restriction enzyme sites are shown. (B) Southern blot of EcoRV-digested genomic DNA from mice of the indicated genotypes using the 5′ probe. (C) PCR analysis of tail DNA from Nkrp1bneo and Nkrp1blox mice. (D) Surface expression of NKR-P1B is absent on NK cells from NKR-P1B-deficient mice. Splenocytes from WT and Nkrp1blox/lox mice were stained with mAbs to DX5, TCRβ, and NKR-P1B (2D9 and 2D12). The percentage of NKR-P1B+ NK cells (DX5+TCRβ–) is indicated. PCR, polymerase chain reaction.
Immune cell populations in lymphoid and nonlymphoid organs from NKR-P1B-deficient mice
| Organ . | Cell populations . | Cell numbers . | |
|---|---|---|---|
| Nkrp1b+/+ . | Nkrp1b−/− . | ||
| Thymus (×106) | Total | 131.6 ± 27.3 | 118.5 ± 32 |
| CD4 SP | 9.9 ± 4.2 | 11.4 ± 3.4 | |
| CD8 SP | 4.63 ± 3.3 | 4.0 ± 1.1 | |
| CD4 CD8 DP | 112.3 ± 22 | 98.9 ± 27.7 | |
| NKT | 0.45 ± 0.22 | 0.5 ± 0.04 | |
| Spleen (×106) | Total | 87.1 ± 10.4 | 83.4 ± 15.4 |
| CD4 | 19.0 ± 2.8 | 18.2 ± 5.3 | |
| CD8 | 12.2 ± 1.3 | 11.3 ± 2.7 | |
| B | 45.1 ± 12.7 | 44.1 ± 14.6 | |
| NK | 3.99 ± 0.8 | 3.68 ± 1.3 | |
| NKT | 1.27 ± 0.24 | 1.29 ± 0.32 | |
| DC | 2.5 ± 0.7 | 2.7 ± 0.7 | |
| pDC | 0.17 ± 0.08 | 0.17 ± 0.06 | |
| Lungs (×105) | Total | 17.0 ± 3.4 | 18.4 ± 2.4 |
| NK | 0.60 ± 0.1 | 0.85 ± 0.3 | |
| NKT | 0.054 ± 0.01 | 0.072 ± 0.01 | |
| Liver (×105) | Total | 26.2 ± 8 | 25.0 ± 6.2 |
| NK | 1.9 ± 0.3 | 2.0 ± 0.4 | |
| NKT | 1.11 ± 0.4 | 1.13 ± 0.39 | |
| Organ . | Cell populations . | Cell numbers . | |
|---|---|---|---|
| Nkrp1b+/+ . | Nkrp1b−/− . | ||
| Thymus (×106) | Total | 131.6 ± 27.3 | 118.5 ± 32 |
| CD4 SP | 9.9 ± 4.2 | 11.4 ± 3.4 | |
| CD8 SP | 4.63 ± 3.3 | 4.0 ± 1.1 | |
| CD4 CD8 DP | 112.3 ± 22 | 98.9 ± 27.7 | |
| NKT | 0.45 ± 0.22 | 0.5 ± 0.04 | |
| Spleen (×106) | Total | 87.1 ± 10.4 | 83.4 ± 15.4 |
| CD4 | 19.0 ± 2.8 | 18.2 ± 5.3 | |
| CD8 | 12.2 ± 1.3 | 11.3 ± 2.7 | |
| B | 45.1 ± 12.7 | 44.1 ± 14.6 | |
| NK | 3.99 ± 0.8 | 3.68 ± 1.3 | |
| NKT | 1.27 ± 0.24 | 1.29 ± 0.32 | |
| DC | 2.5 ± 0.7 | 2.7 ± 0.7 | |
| pDC | 0.17 ± 0.08 | 0.17 ± 0.06 | |
| Lungs (×105) | Total | 17.0 ± 3.4 | 18.4 ± 2.4 |
| NK | 0.60 ± 0.1 | 0.85 ± 0.3 | |
| NKT | 0.054 ± 0.01 | 0.072 ± 0.01 | |
| Liver (×105) | Total | 26.2 ± 8 | 25.0 ± 6.2 |
| NK | 1.9 ± 0.3 | 2.0 ± 0.4 | |
| NKT | 1.11 ± 0.4 | 1.13 ± 0.39 | |
Single-cell suspensions from different organs were stained with antibodies for cell-specific markers and analyzed by flow cytometry. Values are presented as the mean ± SD from 6 to 7 mice of each genotype.
DC, dendritic cells; DP, double positive; SP, single positive; pDC, plasmacytoid dendritic cells.
Developmental and activation marker expression on NKR-P1B-deficient NK cells
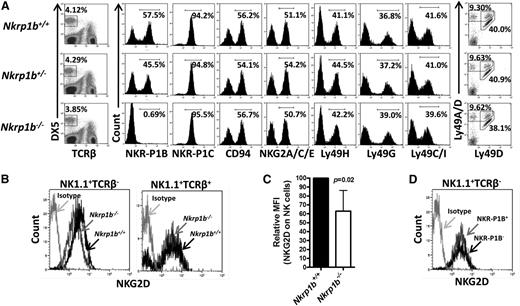
NK cells develop in the bone marrow with sequential acquisition of surface markers at different stages of differentiation.36-38 We determined whether NK cell development and differentiation was affected by the lack of NKR-P1B. We first analyzed the expression of multiple NKC-encoded NK cell receptors in NKR-P1B-deficient mice by flow cytometry and found them expressed at normal levels and percentages on NK cells (Figure 2A). Interestingly, this finding included Ly49 receptors and CD94-NKG2 receptors that are expressed at variable frequencies on NKR-P1B+ and NKR-P1B– NK cell subsets in rodents.13,19,20 One exception was NKG2D, whose expression was moderately downmodulated on NK (NK1.1+TCRβ−), but not NKT (NK1.1+TCRβ+), cells from NKR-P1B-deficient mice (Figure 2B-C). Moreover, NKR-P1B+ and NKR-P1B– NK cells from WT mice had similar surface expression levels of NKG2D (Figure 2D), indicating that NKG2D downregulation was specific to NK cells from NKR-P1B-deficient mice. However, these cells were not impaired in their ability to kill targets expressing NKG2D ligands (data not shown). Interestingly, the percentage of NKR-P1B+ NK cells in heterozygous mice (∼45%) was higher than that deduced from the product rule (∼32%), as was observed for Ly49 expression in heterozygous NKCKD mice.32 This result is consistent with previous studies showing a more codominant vs purely stochastic expression of NKR-P1B in NK cells.13
NKC gene expression in NKR-P1B-deficient mice. (A) Splenic NK cells (DX5+TCRβ–) from WT, NKR-P1B-deficient, and heterozygous littermate mice were analyzed for expression of the indicated NK cell receptors encoded in the NKC. Representative plots from 1 of 6 to 7 mice are shown. The percentage of positively stained NK cells is indicated. (B) Downmodulation of NKG2D on splenic NK cells (NK1.1+TCRβ–) but not NKT cells (NK1.1+TCRβ+) from NKR-P1B-deficient mice. (C) Graphical representation of relative median fluorescence intensity (MFI) of NKG2D on NK cells from WT and NKR-P1B-deficient littermate mice (n = 6 mice). Statistical analysis was performed by Student t test, and the P value is indicated. (D) NKG2D expression on NKR-P1B+ and NKR-P1B− subsets of NK cells from the spleen of a WT B6 mouse.
NKC gene expression in NKR-P1B-deficient mice. (A) Splenic NK cells (DX5+TCRβ–) from WT, NKR-P1B-deficient, and heterozygous littermate mice were analyzed for expression of the indicated NK cell receptors encoded in the NKC. Representative plots from 1 of 6 to 7 mice are shown. The percentage of positively stained NK cells is indicated. (B) Downmodulation of NKG2D on splenic NK cells (NK1.1+TCRβ–) but not NKT cells (NK1.1+TCRβ+) from NKR-P1B-deficient mice. (C) Graphical representation of relative median fluorescence intensity (MFI) of NKG2D on NK cells from WT and NKR-P1B-deficient littermate mice (n = 6 mice). Statistical analysis was performed by Student t test, and the P value is indicated. (D) NKG2D expression on NKR-P1B+ and NKR-P1B− subsets of NK cells from the spleen of a WT B6 mouse.
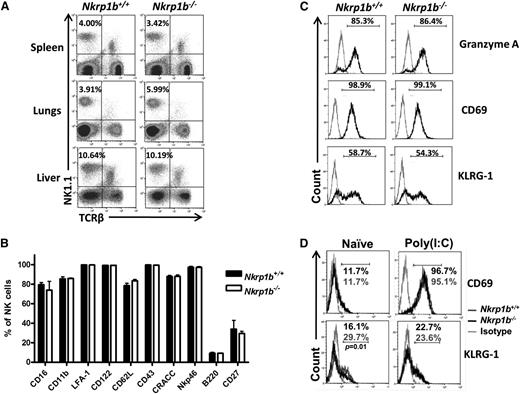
The numbers and frequencies of NK cells in the spleen, lung, and liver were similar in WT and NKR-P1B-deficient mice (Figure 3A; Table 1). Additionally, markers associated with mature NK cells were normally expressed on splenic NK cells (Figure 3B). When cultured with interleukin-2, NKR-P1B-deficient NK cells produced granzyme A and upregulated CD69 and KLRG1 activation markers comparable to WT NK cells (Figure 3C). In addition, IP injection of poly(I:C) resulted in similar CD69 upregulation on NK cells from NKR-P1B-deficient and WT mice (Figure 3D). However, we observed a lower percentage of NK cells expressing KLRG1 in naïve NKR-P1B-deficient mice compared with WT mice (Figure 3D), similar to Ly49-deficient (NKCKD) and MHC-I-deficient mice.32,39,40 Apart from the slight downregulation of NKG2D and KLRG1, development and differentiation of NK cells appears normal in NKR-P1B-deficient mice.
Expression of developmental and activation markers on NK cells from NKR-P1B-deficient mice. (A) Similar proportions of NK cells in the spleen, lungs, and liver of WT and NKR-P1B-deficient littermate mice were observed. A representative plot from each group of mice is shown. The NK cell percentage is indicated. (B) Graphical representation of cell surface marker expression on NK cells from WT and NKR-P1B-deficient littermate mice (n = 6 mice). Splenocytes were stained with mAbs against NK1.1, TCRβ, and various cell surface markers. The mean ± SD of the percentage of positively staining NK cells is depicted. (C) Expression of intracellular granzyme A and activation markers (CD69 and KLRG-1) on NK cells from WT and NKR-P1B-deficient littermates following interleukin-2 treatment in vitro for 3 days. The percentage of positively stained NK cells is indicated. Gray line represents staining with an isotype antibody. (D) Expression of CD69 and KLRG-1 on NK cells from naïve and poly(I:C)-treated WT and NKR-P1B-deficient littermates. The percentage of positively stained NK cells is indicated. Statistical analysis was performed by Student t test, and P values are indicated.SD, standard deviation.
Expression of developmental and activation markers on NK cells from NKR-P1B-deficient mice. (A) Similar proportions of NK cells in the spleen, lungs, and liver of WT and NKR-P1B-deficient littermate mice were observed. A representative plot from each group of mice is shown. The NK cell percentage is indicated. (B) Graphical representation of cell surface marker expression on NK cells from WT and NKR-P1B-deficient littermate mice (n = 6 mice). Splenocytes were stained with mAbs against NK1.1, TCRβ, and various cell surface markers. The mean ± SD of the percentage of positively staining NK cells is depicted. (C) Expression of intracellular granzyme A and activation markers (CD69 and KLRG-1) on NK cells from WT and NKR-P1B-deficient littermates following interleukin-2 treatment in vitro for 3 days. The percentage of positively stained NK cells is indicated. Gray line represents staining with an isotype antibody. (D) Expression of CD69 and KLRG-1 on NK cells from naïve and poly(I:C)-treated WT and NKR-P1B-deficient littermates. The percentage of positively stained NK cells is indicated. Statistical analysis was performed by Student t test, and P values are indicated.SD, standard deviation.
NKR-P1B:Clr-b inhibitory signals are abolished in NKR-P1B-deficient NK cells
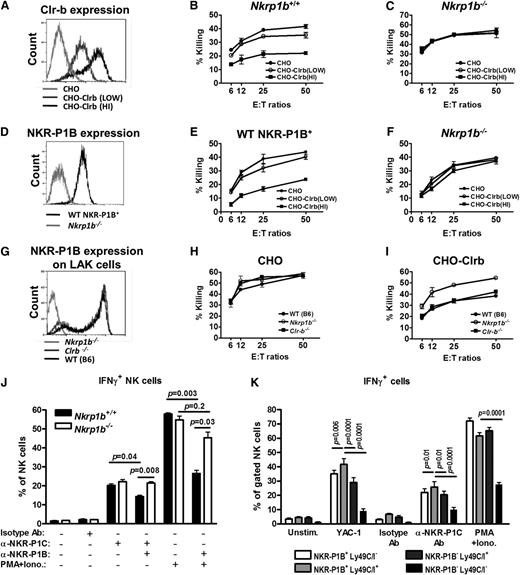
NKR-P1B interacting with Clr-b has been shown to inhibit NK cells in vitro.13,17,18 We performed NK cell cytotoxicity assays against stably transfected CHO target cells expressing high and low levels of Clr-b (Figure 4A). In contrast to the WT LAK cells, which were inhibited by Clr-b-expressing CHO cells (Figure 4B), the NKR-P1B-deficient LAK cells were not inhibited (Figure 4C). Notably, these LAK cells were generated from whole splenocytes, thus the WT LAK cells contained a mixture of NKR-P1B+ (∼60%) and NKR-P1B– (∼40%) cells. For a better comparison, we sorted for WT NKR-P1B+ LAK (NK1.1+TCRβ−) cells on day 3 of culture and further expanded them with interleukin-2 for 6 days. NKR-P1B-deficient LAK (NK1.1+TCRβ−) cells were treated similarly. The sorted WT NKR-P1B+ LAKs maintained NKR-P1B surface expression in culture (Figure 4D) and were inhibited by Clr-b-expressing CHO cells, whereas the NKR-P1B-deficient LAK cells were not inhibited (Figure 4E-F). The sorted WT NKR-P1B– LAK cells often reverted to a mixed population resembling the presort cells (data not shown), suggesting that some of these cells might have expressed NKR-P1B at low levels or induced NKR-P1B during LAK culture. We also performed NK cell cytotoxicity assays using Clr-b-deficient LAK cells, which expressed comparable levels of NKR-P1B (Figure 4G). Similar to WT LAK cells, Clr-b-deficient LAK cells were inhibited by Clr-b-expressing CHO cells (Figure 4H-I), suggesting normal NKR-P1B function despite the absence of its ligand (Clr-b) in these mice.41
NKR-P1B-deficient NK cells are not inhibited by Clr-b on target cells. (A) Flow cytometric analysis of Clr-b expression on Clr-b-transfected CHO cells. Two clones with high (HI) and low expression levels are shown. (B-C) Ability of LAK cells from WT and NKR-P1B-deficient mice to kill CHO target cells was tested by 51Cr-release assay. Data are represented as mean ± SD of percent killing measured in triplicate wells at different effector cell (E) to target cell (T) ratios. (D) NKR-P1B expression on sorted WT NKR-P1B+ and NKR-P1B-deficient LAK cells (NK1.1+TCRβ−), which were further expanded in culture with interleukin-2. (E-F) Ability of sorted WT NKR-P1B+ and NKR-P1B-deficient LAK cells to mediate cytotoxicity toward CHO target cells was tested by 51Cr-release assay. Data are represented as the mean ± SD of percent killing measured in triplicate. (G) NKR-P1B expression on LAK cells from WT, Clr-b-deficient, and NKR-P1B-deficient mice. (H-I) Ability of LAK cells from WT, Clr-b-deficient, and NKR-P1B-deficient mice to kill CHO and Clr-b-expressing CHO (CHO-Clrb) target cells was tested by 51Cr-release assay. Data are represented as mean ± SD of percent killing measured in triplicate wells at different E:T ratios. (J) Splenocytes from NKR-P1B-deficient and WT mice, pretreated with poly(I:C), were incubated with plate-bound isotype control antibody (Isotype Ab), anti-NKR-P1C antibody (NK1.1), or PMA/ionomycin for 5 hours in the presence or absence of plate-bound anti-NKR-P1B antibody (2D12). Intracellular IFN-γ in NK cells was analyzed by flow cytometry, and the mean percentage ± SD of IFN-γ+ NK cells for each stimulation is shown. (K) Splenocytes from WT mice were incubated with YAC-1 cells, plate-bound anti-NKR-P1C antibody (NK1.1), isotype control antibody, or PMA/ionomycin for 5 hours. Intracellular IFN-γ in NK cell subsets, based on the expression of NKR-P1B and Ly49C/I receptors, was analyzed by flow cytometry, and the mean percentage ± SD of IFN-γ+ NK cells for each stimulation is shown. Data in panels A-K are representative data from 1 of multiple independent experiments. Statistical analysis was performed by Student t test, and P values are indicated where applicable. Ab, antibody; PMA+Iono, PMA/ionomycin.
NKR-P1B-deficient NK cells are not inhibited by Clr-b on target cells. (A) Flow cytometric analysis of Clr-b expression on Clr-b-transfected CHO cells. Two clones with high (HI) and low expression levels are shown. (B-C) Ability of LAK cells from WT and NKR-P1B-deficient mice to kill CHO target cells was tested by 51Cr-release assay. Data are represented as mean ± SD of percent killing measured in triplicate wells at different effector cell (E) to target cell (T) ratios. (D) NKR-P1B expression on sorted WT NKR-P1B+ and NKR-P1B-deficient LAK cells (NK1.1+TCRβ−), which were further expanded in culture with interleukin-2. (E-F) Ability of sorted WT NKR-P1B+ and NKR-P1B-deficient LAK cells to mediate cytotoxicity toward CHO target cells was tested by 51Cr-release assay. Data are represented as the mean ± SD of percent killing measured in triplicate. (G) NKR-P1B expression on LAK cells from WT, Clr-b-deficient, and NKR-P1B-deficient mice. (H-I) Ability of LAK cells from WT, Clr-b-deficient, and NKR-P1B-deficient mice to kill CHO and Clr-b-expressing CHO (CHO-Clrb) target cells was tested by 51Cr-release assay. Data are represented as mean ± SD of percent killing measured in triplicate wells at different E:T ratios. (J) Splenocytes from NKR-P1B-deficient and WT mice, pretreated with poly(I:C), were incubated with plate-bound isotype control antibody (Isotype Ab), anti-NKR-P1C antibody (NK1.1), or PMA/ionomycin for 5 hours in the presence or absence of plate-bound anti-NKR-P1B antibody (2D12). Intracellular IFN-γ in NK cells was analyzed by flow cytometry, and the mean percentage ± SD of IFN-γ+ NK cells for each stimulation is shown. (K) Splenocytes from WT mice were incubated with YAC-1 cells, plate-bound anti-NKR-P1C antibody (NK1.1), isotype control antibody, or PMA/ionomycin for 5 hours. Intracellular IFN-γ in NK cell subsets, based on the expression of NKR-P1B and Ly49C/I receptors, was analyzed by flow cytometry, and the mean percentage ± SD of IFN-γ+ NK cells for each stimulation is shown. Data in panels A-K are representative data from 1 of multiple independent experiments. Statistical analysis was performed by Student t test, and P values are indicated where applicable. Ab, antibody; PMA+Iono, PMA/ionomycin.
Intracellular IFN-γ production and degranulation potential of NKR-P1B-deficient NK cells were normal in response to YAC-1 cells, activating NKR-P1C receptor (NK1.1) cross-linking, and PMA/ionomycin treatment, compared to WT NK cells (Figure 4J and supplemental Figure 1). Engagement of NKR-P1B using 2D12 mAb inhibited IFN-γ production by WT, but not NKR-P1B-deficient, NK cells on simultaneous stimulation via NK1.1 cross-linking or PMA/ionomycin (Figure 4J). Additionally, in WT mice, a higher proportion of IFN-γ+ NK cells coexpressed NKR-P1B and Ly49C/I, as compared to those expressing either or none of the inhibitory receptors (Figure 4K). In contrast to previous studies,42 we also observed hyporesponsiveness of NK cells lacking NKR-P1B and Ly49C/I receptors when treated with PMA/ionomycin, perhaps due to our use of a lower concentration of PMA/ionomycin; this difference was abolished on stimulation with higher concentrations of PMA/ionomycin (data not shown). Together, these data reaffirm the inhibitory nature of NKR-P1B and demonstrate that, like the MHC-I-specific Ly49 receptors, NKR-P1B contributes to NK cell functional responsiveness and tolerance.
NKR-P1B-deficient mice exhibit decreased rejection of Clr-b-deficient splenocytes and increased rejection of MHC-I-deficient splenocytes
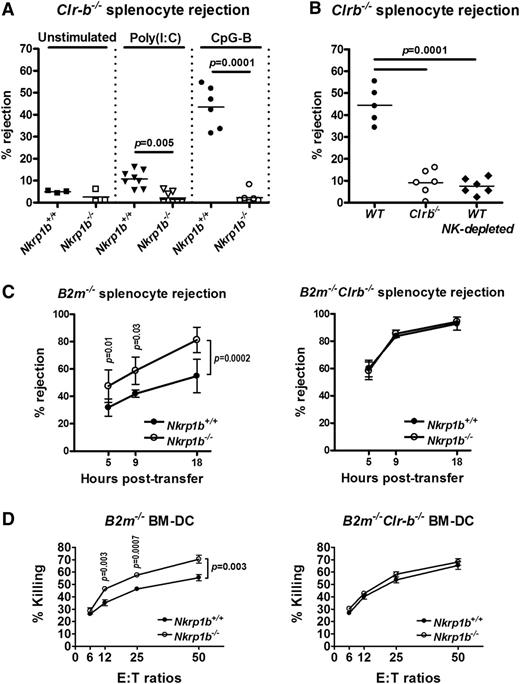
Clr-b is expressed at high levels on mouse hematopoietic cells and frequently downregulated on tumor cell lines cultured in vitro.17 NKR-P1B is thought to be involved in MHC-independent missing-self recognition by monitoring Clr-b expression on host cells. To test this hypothesis in vivo, we performed acute hematopoietic cell rejection assays using Clr-b−/− splenocytes. Splenocytes from Clr-b−/− mice were rejected very weakly within 18 hours by naïve WT and NKR-P1B-deficient mice (Figure 5A), likely a consequence of having normal expression of MHC-I (data not shown). This result is similar to the lower rejection of H-2Kb or H-2Db single-deficient cells compared to that of H-2Kb/Db double-deficient cells.32,35 However, when mice were treated with the Toll-like receptor (TLR) agonists, poly(I:C) or CpG-B ODN, Clr-b−/− splenocytes were efficiently rejected by WT recipient mice, particularly in the CpG-B ODN-treated group, but not by NKR-P1B-deficient mice (Figure 5A). CpG ODN has been shown to directly activate NK cell cytokine responses via activation of TLR9.43 Because Clr-b−/− splenocytes are MHC-I sufficient, their rejection by WT NK cells was due solely to the lack of Clr-b (missing-self), and failure of NKR-P1B-deficient NK cells to reject these cells indicates an inability to detect the presence or absence of Clr-b. Similarly, Clr-b-deficient mice, in which NK cells develop in the absence of the ligand for NKR-P1B, failed to reject Clr-b−/− splenocytes (Figure 5B). Rejection of Clr-b−/− splenocytes by WT mice is mediated by NK cells, because NK cell depletion abrogated this response (Figure 5B).41
NKR-P1B-deficient mice show impaired acute rejection of Clr-b-deficient splenocytes but enhanced rejection of MHC-I-deficient splenocytes. (A) Rejection of 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled splenocytes from Clr-b−/− mice 18 hours after injection. Some mice were treated with poly(I:C) or CpG-B ODN for 24 or 6 hours, respectively, before CFSE-labeled splenocyte injection. Each symbol represents an individual mouse. Small horizontal bars represent mean values. Data are pooled from multiple experiments. (B) Rejection of CFSE-labeled splenocytes from Clr-b−/− mice 18 hours after injection. Some of the WT mice were treated with anti-NKR-P1C antibody (NK1.1) to deplete NK cells (NK-depleted). All mice were treated with CpG-B ODN for 6 hours before CFSE-labeled splenocyte injection. Each symbol represents an individual mouse. Small horizontal bars represent mean values. Data are pooled from 2 independent experiments. (C) Rejection of CFSE-labeled splenocytes from B2m−/− and B2m−/−Clr-b−/− mice at different time points after injection. Mice were not stimulated before splenocyte injection. Mean ± SD of percent rejection at each time point is shown. Data are pooled from multiple experiments. Statistical analysis was performed by Student t test, and P values are indicated. (D) Ability of LAK cells from WT and NKR-P1B-deficient mice to mediate cytotoxicity toward B2m−/− or B2m−/−Clr-b−/− BM-DCs was tested by 51Cr-release assay. Data are represented as the mean ± SD of percent killing measured in triplicate. Statistical analysis was performed by Student t test, and P values are indicated.
NKR-P1B-deficient mice show impaired acute rejection of Clr-b-deficient splenocytes but enhanced rejection of MHC-I-deficient splenocytes. (A) Rejection of 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled splenocytes from Clr-b−/− mice 18 hours after injection. Some mice were treated with poly(I:C) or CpG-B ODN for 24 or 6 hours, respectively, before CFSE-labeled splenocyte injection. Each symbol represents an individual mouse. Small horizontal bars represent mean values. Data are pooled from multiple experiments. (B) Rejection of CFSE-labeled splenocytes from Clr-b−/− mice 18 hours after injection. Some of the WT mice were treated with anti-NKR-P1C antibody (NK1.1) to deplete NK cells (NK-depleted). All mice were treated with CpG-B ODN for 6 hours before CFSE-labeled splenocyte injection. Each symbol represents an individual mouse. Small horizontal bars represent mean values. Data are pooled from 2 independent experiments. (C) Rejection of CFSE-labeled splenocytes from B2m−/− and B2m−/−Clr-b−/− mice at different time points after injection. Mice were not stimulated before splenocyte injection. Mean ± SD of percent rejection at each time point is shown. Data are pooled from multiple experiments. Statistical analysis was performed by Student t test, and P values are indicated. (D) Ability of LAK cells from WT and NKR-P1B-deficient mice to mediate cytotoxicity toward B2m−/− or B2m−/−Clr-b−/− BM-DCs was tested by 51Cr-release assay. Data are represented as the mean ± SD of percent killing measured in triplicate. Statistical analysis was performed by Student t test, and P values are indicated.
We next performed in vivo rejection assays using MHC-I-deficient cells. Strikingly, MHC-I-deficient (B2m−/−) splenocytes were more efficiently rejected by NKR-P1B-deficient mice compared with WT mice (Figure 5C, left). However, WT and NKR-P1B-deficient mice were equally efficient at rejecting MHC-I/Clr-b double-deficient (B2m−/−Clr-b−/−) splenocytes, suggesting that the reduced rejection of MHC-I-deficient splenocytes in WT mice was due to inhibitory NKR-P1B:Clr-b interactions (Figure 5C, right). This finding was also recapitulated in cytotoxicity assays in vitro using B2m−/− and B2m−/−Clr-b−/− BM-DCs as target cells (Figure 5D). Moreover, rejection of B2m−/−Clr-b−/− splenocytes was significantly more efficient than rejection of B2m−/− splenocytes by both WT and NKR-P1B-deficient mice (Figure 5C, left vs right), suggesting that loss of Clr-b synergizes with MHC-I deficiency to activate NK cell killing. Furthermore, this finding may also suggest differential dependence on MHC-I-dependent missing-self recognition by WT and NKR-P1B-deficient NK cells due to altered NK cell education and subset/repertoire formation in the absence of NKR-P1B. Together, these results demonstrate a loss of Clr-b-dependent inhibitory signals and Clr-b-dependent missing-self responses in NKR-P1B-deficient mice.
Involvement of NKR-P1B in tumor surveillance and immune escape
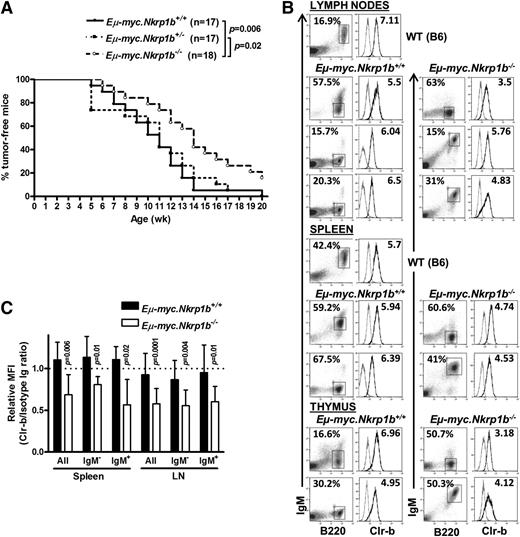
Because NKR-P1B is involved in missing-self recognition of Clr-b, which is often lost or downregulated on tumor cell lines in vitro, we next investigated the role of NKR-P1B in tumor development in vivo using the Eµ-myc Tg mouse model. Eµ-myc Tg mice develop spontaneous B-cell lymphomas due to transgenic expression of the c-myc oncogene in B cells.44 We bred B6-background Eµ-myc Tg mice with NKR-P1B-deficient mice to obtain Eµ-myc+Nkrp1b−/−, Eµ-myc+Nkrp1b+/−, and Eµ-myc+Nkrp1b+/+ littermates. The 3 groups of mice were monitored for 20 weeks for the appearance of palpable tumors (enlarged lymph nodes) and sickness, most often respiratory distress due to an enlarged thymus. The onset of B-cell lymphoma was significantly delayed in NKR-P1B-deficient mice, and these mice stayed tumor-free and healthy longer compared with WT and heterozygous littermates (Figure 6A). Eventually, most Eµ-myc+ NKR-P1B-deficient mice and all of their WT and heterozygous littermates developed tumors by 20 weeks of age. Affected lymphoid organs (thymus, spleen, and lymph nodes) were enlarged and composed mainly of B cells, as reported previously.44,45 Representative examples of immature pre-B cell (B220+IgM–) and mature B cell (B220+IgM+) lymphomas, and Clr-b expression patterns, are shown in Figure 6B. Both the mature B and immature pre-B cell lymphomas from NKR-P1B-deficient mice displayed significantly reduced expression of Clr-b compared to normal mature B cells, whereas lymphomas from WT littermate controls maintained higher Clr-b levels (Figure 6C). Together, these results show that, in NKR-P1B-deficient mice, oncogene-transformed cells may spontaneously lose Clr-b compared to healthy cells (as observed in vitro), yet malignant tumor cells in WT mice may selectively maintain high Clr-b levels to escape immune surveillance by inhibiting NK cells via NKR-P1B.
Delayed onset of B lymphoma in NKR-P1B-deficient mice expressing the Eµ-myc transgene. (A) Kaplan-Meier representation of palpable tumor appearance in Eµ-myc.Nkrp1b+/+, Eµ-myc.Nkrp1b+/−, and Eµ-myc.Nkrp1b−/− mice. Statistical analysis was performed by log-rank test, and the P value is indicated. (B) Flow cytometric analysis of B220, IgM, and Clr-b expression on B lymphoma cells from affected organs (lymph nodes, spleen, and thymus) from Eµ-myc.Nkrp1b+/+ and Eµ-myc.Nkrp1b−/− mice after disease onset and from healthy WT mice. The percentage of gated cells from each organ is indicated. Clr-b expression on gated B lymphoma cells is represented on the histograms by a dark line; isotype control by a gray line. Clr-b median fluorescence intensity (MFI) values are normalized to isotype control IgM MFI (Clr-b/isotype Ig MFI), and are shown on respective plots. (C) Graphical representation of Clr-b expression levels on mature (IgM+), immature (IgM–), and all B lymphoma cells taken together, in the spleen and lymph nodes of Eµ-myc.Nkrp1b+/+ and Eµ-myc.Nkrp1b−/− mice relative to the expression of Clr-b on mature B cells in WT mice. Vertical columns represent Clr-b/isotype Ig MFI ratio, and error bars represent SD. The horizontal dotted line represents the Clr-b/isotype Ig MFI ratio on mature B cells (B220+IgM+) from WT mice (arbitrarily set to 1.0). Statistical analysis was performed by Student t test, and P values are indicated. Ig, immunoglobulin.
Delayed onset of B lymphoma in NKR-P1B-deficient mice expressing the Eµ-myc transgene. (A) Kaplan-Meier representation of palpable tumor appearance in Eµ-myc.Nkrp1b+/+, Eµ-myc.Nkrp1b+/−, and Eµ-myc.Nkrp1b−/− mice. Statistical analysis was performed by log-rank test, and the P value is indicated. (B) Flow cytometric analysis of B220, IgM, and Clr-b expression on B lymphoma cells from affected organs (lymph nodes, spleen, and thymus) from Eµ-myc.Nkrp1b+/+ and Eµ-myc.Nkrp1b−/− mice after disease onset and from healthy WT mice. The percentage of gated cells from each organ is indicated. Clr-b expression on gated B lymphoma cells is represented on the histograms by a dark line; isotype control by a gray line. Clr-b median fluorescence intensity (MFI) values are normalized to isotype control IgM MFI (Clr-b/isotype Ig MFI), and are shown on respective plots. (C) Graphical representation of Clr-b expression levels on mature (IgM+), immature (IgM–), and all B lymphoma cells taken together, in the spleen and lymph nodes of Eµ-myc.Nkrp1b+/+ and Eµ-myc.Nkrp1b−/− mice relative to the expression of Clr-b on mature B cells in WT mice. Vertical columns represent Clr-b/isotype Ig MFI ratio, and error bars represent SD. The horizontal dotted line represents the Clr-b/isotype Ig MFI ratio on mature B cells (B220+IgM+) from WT mice (arbitrarily set to 1.0). Statistical analysis was performed by Student t test, and P values are indicated. Ig, immunoglobulin.
Discussion
NKR-P1B is an inhibitory receptor on ∼60% of NK cells in B6 mice that recognizes the Clr-b ligand.13,17,18 Using a gene-knockout model, we show a loss of Clr-b-mediated inhibition in NKR-P1B-deficient mice. Although the WT NK cells are inhibited by cell surface Clr-b, NKR-P1B-deficient NK cells efficiently kill CHO target cells expressing Clr-b. Additionally, inhibition of IFN-γ production by NKR-P1B receptor cross-linking in WT NK cells is abolished in NKR-P1B-deficient NK cells. Notably, NK-dependent in vivo rejection of MHC-I-deficient cells is elevated in NKR-P1B-deficient mice compared with WT mice, due to the lack of Clr-b-mediated inhibition.
Inhibitory receptors are required for the education (licensing) of developing NK cells to acquire the ability to respond to activation signals. NK cell responsiveness is proportional to the number of self-MHC-specific inhibitory receptors expressed; that is, NK cells that express more inhibitory Ly49 receptors are also more responsive toward stimulation.42,46 Our study demonstrates a higher responsiveness of NK cells expressing the inhibitory receptors NKR-P1B and Ly49C/I, in contrast to those expressing either or none of the receptors. Therefore, like the Ly49 receptors, NKR-P1B seems to contribute to NK cell education and hence higher responsiveness.
It has been shown that lack of MHC-I-dependent NK cell education in full MHC-I-deficient mice results in hyporesponsiveness of the uneducated/unlicensed NK cells toward activating stimuli.47,48 Because our data support a role of the NKR-P1B receptor in NK cell tolerance, one might expect NKR-P1B-deficient NK cells to be hyporesponsive. However, in our study, IFN-γ production and degranulation in response to general activation stimuli appear to be normal in NKR-P1B-deficient NK cells. In contrast, NK cells from Clr-b-deficient mice, like full MHC-I-deficient mice, appear to be hyporesponsive to activating receptor cross-linking and cytokine stimulation,41 despite a complementarity of deficiencies in either the Clr-b ligand or the NKR-P1B receptor. We have encountered a similar situation in Ly49-deficient mice, in which NK cell responses to various general activating stimuli were preserved.32 Similarly, NKR-P1B-deficient mice are as efficient as WT mice regarding in vivo rejection of hematopoietic cells lacking both Clr-b and MHC-I, despite an expected hyporesponsiveness of NKR-P1B-deficient NK cells. This finding may suggest a dominant effect of MHC-I-dependent NK cell education and missing-self responses over those mediated via the single NKR-P1B:Clr-b interaction. Alternatively, a qualitative skewing of NK cell subsets during NK cell education and repertoire formation may result in increased dependence on, and a stronger magnitude of, MHC-I-dependent missing-self responses in NKR-P1B-deficient vs WT mice. In addition, other Clr-b interactions, such as with alternative inhibitory or stimulatory NK receptors recognizing Clr-b alone or in complex with other Clr family members, may impart some education on these cells. Notably, NKR-P1G is another inhibitory member of the NKR-P1 receptor family, and NKR-P1F shares overlapping ligand specificity with NKR-P1G.11,15,21,22,49 A full analysis of Clr-b interactions with other Clr family members remains to be tested.
Educated/licensed NK cells play a role in missing-self recognition, where target cells with reduced expression of inhibitory self-ligands are specifically recognized and eliminated by NK cells.7 Clr-b is broadly expressed on hematopoietic cells and at least some nonhematopoietic tissues, thus overlapping the expression of MHC-I ligands for the inhibitory Ly49 receptors.17,27,50 Clr-b recognition by NKR-P1B provides an MHC-I-independent missing-self recognition mechanism. Although Clr-b-deficient splenocytes are acutely rejected in WT mice treated with TLR agonists, they are not rejected in NKR-P1B-deficient mice in vivo, providing a genetic proof-of-principle for MHC-independent missing-self recognition by NK cells (this study and Chen et al41 ). This complementation resembles the inability of Ly49-deficient mice to reject MHC-I-deficient cells.32 Similarly, Clr-b-deficient mice are unable to reject Clr-b-deficient splenocytes due to self-tolerance mechanisms influencing NKR-P1B:Clr-b-dependent NK cell education.41 Note that MHC-dependent NK cell education and missing-self responses are intact in NKR-P1B-deficient mice, as demonstrated by enhanced rejection of MHC-I-deficient cells. Thus, NKR-P1B:Clr-b-dependent missing-self recognition appears to function independently and in parallel with missing-self recognition of MHC-I ligands by Ly49 receptors.
Clr-b is frequently downregulated on tumor cell lines in vitro.17 However, myc-induced B lymphoma cells in Eµ-myc Tg mice reproducibly maintain high-level expression of Clr-b comparable to normal B cells from WT mice. In contrast, Clr-b was found to be frequently and spontaneously downregulated on B lymphoma cells from NKR-P1B-deficient mice. Taken together, these results suggest that oncogene-transformed cells may face selective pressure from NK cell–mediated immunosurveillance to maintain high Clr-b expression levels, which may in turn represent an effective escape mechanism to inhibit WT (NKR-P1B+), but not NKR-P1B-deficient, NK cells. Thus, NKR-P1B-deficient NK cells, which are resistant to Clr-b inhibition, eliminate tumor cells more efficiently, thereby significantly delaying kinetics and decreasing penetrance of myc-induced spontaneous lymphomas in NKR-P1B-deficient vs WT mice.
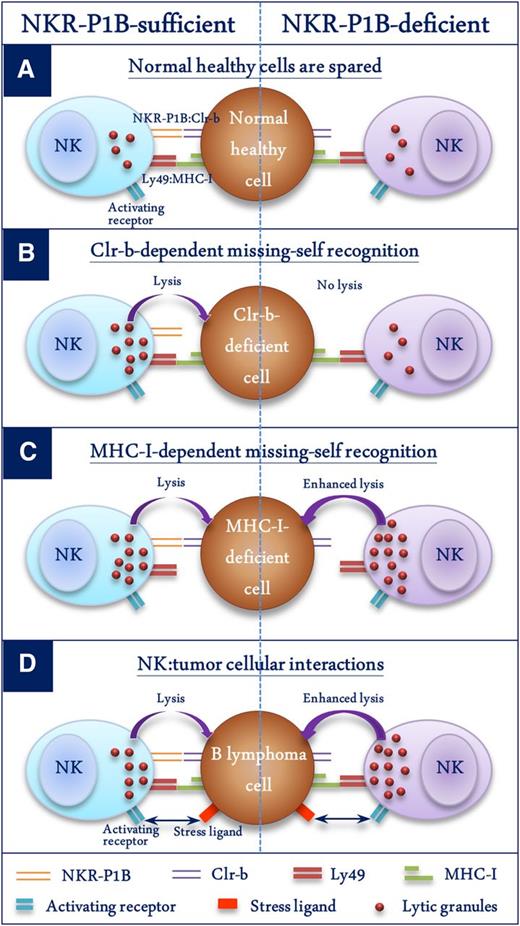
In summary, this study provides in vivo evidence of a negative regulatory role of the NKR-P1B:Clr-b recognition axis in innate immune responses. Figure 7 demonstrates 4 different scenarios and the consequences of NKR-P1B:Clr-b recognition by NK cells: (1) normal healthy cells express both MHC-I and Clr-b and are spared by NK cells (Figure 7A); (2) Clr-b-deficient cells are killed by WT NK cells through an MHC-I-independent missing-self response, and NKR-P1B-deficient NK cells cannot sense the absence of Clr-b, resulting in a loss of Clr-b-dependent missing-self recognition (Figure 7B); (3) MHC-I-deficient cells are killed by WT NK cells through an MHC-I-dependent missing-self response, and NKR-P1B-deficient NK cells are more efficient in killing MHC-I-deficient cells, possibly due to a lack of Clr-b-mediated inhibition and/or a higher dependence on MHC-I-mediated education (Figure 7C); and (4) B lymphoma cells maintain normal surface expression of MHC-I and Clr-b to inhibit NK cells, and NKR-P1B-deficient NK cells are resistant to tumor immune evasion via inhibitory Clr-b ligand (Figure 7D). This latter scenario has potential therapeutic implications. Tumors and infected cells employ various mechanisms to escape immune detection. Expression of ligands specific for inhibitory NK receptors may render such pathological target cells resistant to lysis by NK cells. One could envisage a scenario whereby therapeutic blockade of inhibitory NK receptors would result in augmented NK cell activity. This effect has been demonstrated in the case of T cells by blocking cytotoxic T lymphocyte–associated antigen-4, and in NK cells by blocking inhibitory Ly49C/I, both of which led to augmented antitumor effects.51,52 Our study shows that the absence of NKR-P1B in gene-knockout mice results in a significant advantage over WT mice in controlling myc-induced B-cell lymphoma. This finding lends support to the idea of suppressing inhibitory signals in immune cells to achieve optimal antitumor and antipathogen immunity. NKR-P1B-deficient mice thus represent a useful model for the human NKR-P1A:LLT1 receptor-ligand system to study the role of these receptors on immune cells and their contribution to immunity against tumors and pathogens.
Schematic representation of the role of NKR-P1B:Clr-b recognition in NK cell function. Four different scenarios depicting NK cell function in the presence or absence of NKR-P1B:Clr-b recognition are shown. (A) Normal healthy cells express MHC-I and Clr-b, which are recognized by inhibitory Ly49 and NKR-P1B receptors on NK cells, respectively, resulting in their protection from NK cells. Normal MHC-I levels are sufficient to protect these cells from NKR-P1B-deficient NK cells. (B) WT NK cells kill Clr-b-deficient target cells through an MHC-I-independent missing-self response. NKR-P1B-deficient NK cells cannot sense Clr-b-deficiency on target cells. (C) WT NK cells efficiently kill MHC-I-deficient target cells through MHC-I-dependent missing-self recognition. NKR-P1B-deficient NK cells are more efficient in killing MHC-I-deficient cells, possibly due to lack of Clr-b-mediated inhibition and a higher dependence on MHC-I-mediated education. (D) B lymphoma cells express normal levels of MHC-I and Clr-b. Tumor cells also express ligands for activating NK cell receptors, making them susceptible to NK cells. In the absence of inhibitory NKR-P1B receptor signals, NK cells have improved immunosurveillance capacity against Clr-b-expressing tumor cells, which can escape immune detection by WT NK cells.
Schematic representation of the role of NKR-P1B:Clr-b recognition in NK cell function. Four different scenarios depicting NK cell function in the presence or absence of NKR-P1B:Clr-b recognition are shown. (A) Normal healthy cells express MHC-I and Clr-b, which are recognized by inhibitory Ly49 and NKR-P1B receptors on NK cells, respectively, resulting in their protection from NK cells. Normal MHC-I levels are sufficient to protect these cells from NKR-P1B-deficient NK cells. (B) WT NK cells kill Clr-b-deficient target cells through an MHC-I-independent missing-self response. NKR-P1B-deficient NK cells cannot sense Clr-b-deficiency on target cells. (C) WT NK cells efficiently kill MHC-I-deficient target cells through MHC-I-dependent missing-self recognition. NKR-P1B-deficient NK cells are more efficient in killing MHC-I-deficient cells, possibly due to lack of Clr-b-mediated inhibition and a higher dependence on MHC-I-mediated education. (D) B lymphoma cells express normal levels of MHC-I and Clr-b. Tumor cells also express ligands for activating NK cell receptors, making them susceptible to NK cells. In the absence of inhibitory NKR-P1B receptor signals, NK cells have improved immunosurveillance capacity against Clr-b-expressing tumor cells, which can escape immune detection by WT NK cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by an Operating Grant from the Canadian Institutes of Health Research (CIHR 86630) (A.P.M. and J.R.C.); an Early Researcher Award from the Ontario Ministry of Research and Innovation (J.R.C.); the Victorian Government Operational Infrastructure support program (M.T.G.); and grants from the Biotechnology and Biological Sciences Research Council and Medical Research Council (C.G.B.). A.P.M. holds a Canada Research Chair in Innate Pathogen Resistance; J.R.C. held a Canadian Institutes of Health Research New Investigator Award and holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund, USA.
Authorship
Contribution: M.M.A.R., P.C., J.R.C., and A.P.M. designed the research; M.M.A.R., P.C., A.N.M., A.B.M., M.J.T., and Q.Z. performed the research; C.G.B., V.K., and M.T.G. contributed vital new reagents; M.M.A.R., P.C., A.N.M., A.B.M., and M.J.T. collected the data; M.M.A.R., P.C., A.N.M., J.R.C., and A.P.M. analyzed and interpreted the data; M.M.A.R. and A.N.M. performed the statistical analysis; and M.M.A.R., J.R.C., and A.P.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew P. Makrigiannis, Department of Biochemistry, Microbiology, and Immunology, University of Ottawa, Guindon Hall, Room 4226, 451 Smyth Rd, Ottawa, ON, Canada K1H 8M5; e-mail: amakrigi@uottawa.ca; and James R. Carlyle, Department of Immunology, University of Toronto, Sunnybrook Research Institute, 2075 Bayview Ave, Toronto, ON, Canada M4N 3M5; e-mail: james.carlyle@utoronto.ca.
References
Author notes
J.R.C. and A.P.N. contributed equally to this study.