Abstract
Hematopoietic stem cells (HSCs) are characterized by their ability to execute a wide range of cell fate choices, including self-renewal, quiescence, and differentiation into the many different mature blood lineages. Cell fate decision making in HSCs, as indeed in other cell types, is driven by the interplay of external stimuli and intracellular regulatory programs. Given the pivotal nature of HSC decision making for both normal and aberrant hematopoiesis, substantial research efforts have been invested over the last few decades into deciphering some of the underlying mechanisms. Central to the intracellular decision making processes are transcription factor proteins and their interactions within gene regulatory networks. More than 50 transcription factors have been shown to affect the functionality of HSCs. However, much remains to be learned about the way in which individual factors are connected within wider regulatory networks, and how the topology of HSC regulatory networks might affect HSC function. Nevertheless, important progress has been made in recent years, and new emerging technologies suggest that the pace of progress is likely to accelerate. This review will introduce key concepts, provide an integrated view of selected recent studies, and conclude with an outlook on possible future directions for this field.
Building blocks of transcriptional regulatory networks
The primary components of transcriptional regulatory networks are transcription factor (TF) proteins and the gene regulatory DNA sequences that they bind to.1 By binding to specific DNA sequence motifs within gene regulatory regions, TF proteins are central players for this primary step of decoding gene regulatory instructions. TF proteins typically contain a number of distinct modules, such as DNA binding, transcriptional activation, and protein/protein interaction domains, with the latter 2 being essential for the recruitment of the basal transcriptional machinery and the assembly of higher-order TF complexes. Based on sequence similarity within the DNA binding domain, TF proteins can be categorized into distinct families, such as homeobox, basic helix-loop-helix, or zinc finger TFs. Members of a given TF family often bind to similar DNA sequences, and protein-protein interactions are common both within and between the different TF families.
Individual TFs bind short sequence motifs that are often no longer than 4 to 6 bp. Any given 6-bp sequence will occur on average approximately once every 4000 bp. Consequently, there will be ∼750 000 occurrences just by chance within the 3 000 000 000 bp of the human genome. The number of possible binding sites therefore far exceeds the 20 000 or so human genes, and it has long been assumed that only a small minority of all motif instances play a role in transcriptional regulation. Functional gene regulatory regions should therefore display characteristics that go beyond the mere occurrence of a specific sequence motif. One such characteristic is the presence of clusters of binding sites, typically for up to 5 different TFs. Clustering of binding sites and the prevalence of protein/protein interaction domains within TF proteins facilitate the assembly of higher-order complexes, the formation of which is thought to represent a key step in the decoding of gene regulatory information.
Binding of multiple TFs to individual gene regulatory elements is consistent with the notion of complex networks of interactions, where the locus of each gene participating in the network receives inputs from multiple upstream regulators. Moreover, binding of multiple TFs to a given regulatory element commonly involves synergistic or antagonistic interactions mediated through higher-order complexes. In terms of network structure therefore, inputs from an upstream regulator to its downstream target are rarely simple linear connections, but instead entail synergistic or antagonistic cross-talk between 2 or more upstream regulators. Experimental evidence suggests that some TF complexes exist in solution in the nucleoplasm, and therefore presumably preform (at least partially) before binding to DNA.2 There is also evidence for sequential TF binding, where binding of a so-called “settler” TF requires prior binding of “pioneer” TFs.3 Importantly, a given TF may act as a pioneer TF only on some, but not all, the regulatory elements that it binds to. Rather than being based on generalized assumptions, reconstruction of regulatory network models therefore will require mechanistic knowledge of the transcriptional mechanisms operating at individual gene regulatory elements (Figure 1).
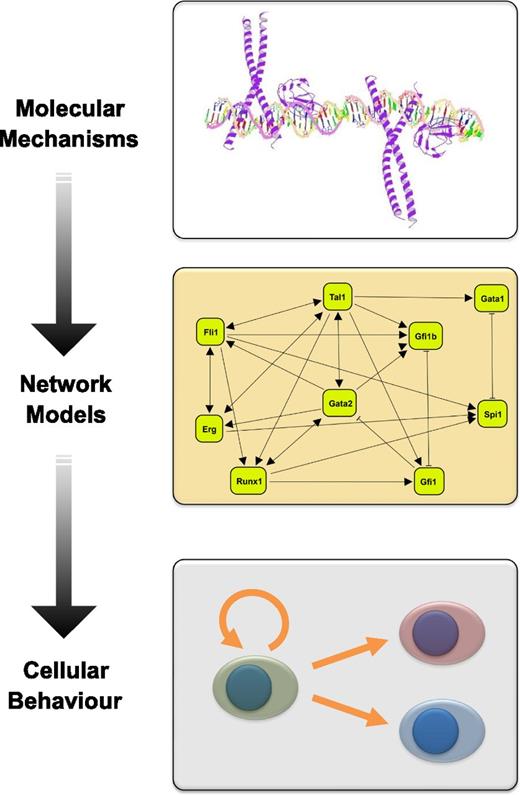
Regulatory network models based on mechanistic insights can advance our understanding of cellular behavior. This diagram illustrates the workflow from experimentally determined molecular mechanisms to regulatory network models that can advance our understanding of cellular decision making processes. Shown are, from top to bottom, a model of the Runx1/CEBP transcription factor complex bound to DNA, a literature curated regulatory network model connecting key HSPC transcription factors, and a diagram illustrating HSC fate decisions including the balance between self-renewal and multilineage differentiation.
Regulatory network models based on mechanistic insights can advance our understanding of cellular behavior. This diagram illustrates the workflow from experimentally determined molecular mechanisms to regulatory network models that can advance our understanding of cellular decision making processes. Shown are, from top to bottom, a model of the Runx1/CEBP transcription factor complex bound to DNA, a literature curated regulatory network model connecting key HSPC transcription factors, and a diagram illustrating HSC fate decisions including the balance between self-renewal and multilineage differentiation.
Cooperative binding of TFs to gene regulatory sequences represents the first major information processing event during the regulation of gene expression.1 This in turn is followed by the recruitment of accessory proteins such as chromatin-modifying enzymes and the multiple components of the RNA polymerase holocomplex.4 Additional points of potential regulation on the route toward the production of functional proteins include transcriptional elongation, RNA processing and nuclear export, translation, and post-translational modifications. In the context of transcriptional regulatory networks, the production of transcription factor proteins is of particular interest, as these will feed back directly into the activity of the network.
Literature-curated hematopoietic transcriptional network models
Given the power of TFs to drive cell fate choice decision making, researchers recognized early on that the construction of models capturing TF interactions might provide a useful tool to improve our understanding of hematopoietic stem and progenitor cell (HSPC) differentiation. However, the generation of detailed functional knowledge on upstream regulator/downstream target gene relationships has traditionally been a laborious task, and therefore no single study would have generated sufficient experimental data to construct comprehensive regulatory network models. In light of these difficulties, several groups recognized the potential power of integrating experimental data from multiple studies to construct hematopoietic regulatory network models, although initial attempts focused on simple 2-factor or 3-factor models.5,6 Although these early efforts might have captured some key properties of core regulatory circuits, they clearly oversimplified the complexities of multifactor regulatory interactions, thus limiting their utility to model regulatory network control of cellular behavior. More recently, a qualitative network model connecting 11 transcription factors controlling the differentiation of common myeloid progenitors (CMPs) was constructed based on a comprehensive survey of the literature.7 The investigators encoded the combinatorial logic governing the interactions between these 11 factors using a Boolean modeling approach, where each factor is represented by a variable that can be either “on” or “off,” and the activating/inhibitory interactions are encoded using combinations of the logical functions “And,” “Or,” and “Not.”
Execution of this model, where simulation is started with an expression state representative of the CMP, not only captured putative intermediate stages of myeloid differentiation but was also able to reach 4 distinct terminal steady states, which corresponded in their expression pattern to megakaryocytes, erythrocytes, granulocytes, and monocytes. Moreover, in silico simulation of gene knockouts recapitulated known lineage depletion results, and in silico overexpression reproduced known TF-mediated reprogramming data. Importantly, the CMP progenitor state itself was unstable in this model and would always differentiate spontaneously into 1 of the 4 terminal myeloid differentiation states. The model therefore did not capture those aspects of blood stem cell regulatory networks that mediate the relative stability of the blood stem/progenitor state. A second literature-curated model based on cross-regulation of 10 hematopoietic stem cell (HSC) TFs, on the other hand, identified HSC-like gene expression as a stable attractor state.8 However, this stable state now required simulation of external triggers to allow exit toward more differentiated states. Of note, this second model focused on cross-regulation between HSC TFs that resulted in numerous positive feedback loops and therefore presumably stabilized the HSC expression state. Importantly, HSCs have the capacity to balance self-renewal and differentiation, which suggests that neither of the 2 modeling approaches outlined above is yet able to capture the complexities inherent in HSC function. In addition to capturing both the self-renewal and differentiation function of HSCs, future modeling efforts should also attempt to connect HSC regulatory network models with literature curated and validated network models for more differentiated cell lineages such as the myeloid and T-lymphoid models generated by the Singh and Rothenberg groups, respectively.9,10
Inferring transcriptional regulatory networks from microarray gene expression profiles
Because the TF proteins that control gene expression are themselves gene products, the levels of TF protein and target gene mRNA abundance will correlate unless post-translational regulation overrides the link between mRNA levels and active protein concentration. Statistical associations between mRNA levels of candidate upstream regulators and downstream target genes can therefore provide clues about possible gene regulatory hierarchies. Following the introduction of microarray technology for genome-wide measurements of mRNA abundance, researchers quickly realized that such datasets could be used to construct more global “gene network” models that encapsulate the regulatory wiring underlying the gene expression identity of a given cell type. Importantly, simple clustering of genes with similar expression across a set of microarray expression profiles cannot distinguish between direct and indirect regulation as the likely cause of coexpression. More sophisticated tools have therefore been developed to reverse engineer network models from expression profiling data, such as graphical modeling,11 integrative methods that use, for example, protein-protein interaction data,12 and statistical/information theoretical methods.13
Particularly promising results for higher eukaryote systems were obtained following the introduction of the Aracne tool,14 which reverse engineers transcriptional networks from microarray data by identifying irreducible statistical dependencies, which cannot be explained by any other statistical dependencies and are therefore much more likely to represent direct regulatory interactions in the network. Aracne has been used to reverse engineer regulatory network models for normal B cells,15 as well as B- and T-cell leukemias.16-18 Of note, Aracne requires hundreds of individual microarray samples to achieve statistical power and therefore has thus far not been used for the construction of HSPC regulatory networks because the required expression datasets do not exist. Moreover, despite their computational accessibility, predictive network models based on correlations between expressed genes are biased toward detecting positive regulatory connections and find it much harder to distinguish between negative regulation and a lack of coincidence for some other reason. However, there is strong evidence from most developmental systems that have been dissected in depth that negative regulatory interactions are key determinants of the decision making processes when multipotent progenitors choose between alternative cell fates. Computational tools designed to specifically uncover inhibitory interactions from gene expression profiling data have been applied to expression datasets from lower eukaryote model systems19 but are not yet widely adopted by the hematopoiesis research community.
An alternative approach to dissect the architecture of gene expression programs is the Module Networks algorithm,20 a probabilistic method that identifies so-called regulatory modules from collections of gene expression profiling data. The identified modules not only report sets of coregulated genes, but also their candidate upstream regulators and the cell types where this regulation occurs. As such, this approach is particularly suited to analyze gene expression profiling data that cover the hematopoietic differentiation hierarchy. Its application by Novershtern et al to expression data for 38 distinct human blood progenitor and mature cell types resulted in the identification of 80 expression modules, several of which were associated with specific expression in HSPCs.21 In parallel, the authors generated network models based on a combination of TF expression and sequence motif content in promoter regions. Of note, the overlap of predicted regulatory interactions generated by these 2 approaches was minimal, because 70% of TFs covered in both the Module Networks and Promoter Motif Analysis showed no significant expression correlation with the module to which they had been assigned by promoter motif scanning.21 One obvious limitation of the promoter-motif based approach is illustrated by previous observations that much of the TF-binding conferring cell type-specific expression occurs at distal sites away from the promoter regions.22 More fundamentally, however, the Novershtern study exemplifies that microarray expression profiling data may yield testable hypotheses about regulatory interactions, but on their own are unlikely to be sufficient for the generation of reliable network models.
Impact of genome-wide TF binding maps for network reconstruction
The apparent problems of reconstructing regulatory networks from expression data alone are possibly most easily explained by the fact that information on actual regulatory elements is missing in this approach. As outlined above, regulatory elements constitute major building blocks of transcriptional regulatory networks, yet traditional approaches to define their location and function have been slow. The recent advent of high-throughput sequencing technology has greatly accelerated the generation of genome-wide TF binding maps, by coupling chromatin immunoprecipitation to high-throughput sequencing (ChIP-Seq).23,24 Due to the rapid decline in sequencing costs, ChIP-Seq technology is now readily available to individual research laboratories, and data integration across laboratories has generated a publicly accessible resource of several hundred TF ChIP-Seq studies across a wide range of hematopoietic cell types.25
However, although ChIP-Seq experiments are increasingly used to complement gene expression profiling,26-34 no major progress has yet been reported toward the generation of validated HSPC regulatory network models. This is at least partly due to the observation that TFs are commonly bound to thousands if not tens of thousands of sites in the genome, often near genes that would not be affected by ablation of the factor in question. This has raised the issue of a need to distinguish functionally significant from nonsignificant binding events.35 However, there are no consensus definitions for these terms, thus leaving the community with no reliable strategy to predict whether or not a given TF binding event is functionally relevant. Robust conclusions from ChIP-Seq studies thus far therefore are largely based on statistically significant associations across a range of binding events, where it is clear that as a group, a specific characteristic is enriched, even if no definitive statement can be made about the nature of any given individual binding event. Of relevance to HSPC regulatory networks, statistical analysis has identified previously unknown combinatorial TF interactions between known HSC regulatory TFs36 and global reorganization of HSPC TF assemblies during the specification of blood progenitors from differentiating embryonic stem cells.37 Moreover, a recent comparative analysis of 10 TFs in HSPCs and mast cells showed that HSPC TFs can actively participate in transcriptional programs of HSPCs, as well as mature hematopoietic lineages, largely by binding to distinct sets of target regions.38
To move beyond generalized statistical associations, ChIP-Seq studies need to be complemented with functional analysis of specific binding events, so that putative regulatory relationships inferred from TF binding are functionally validated. TF knockdown followed by gene expression profiling can identify those TF-bound gene loci where reducing levels of the TF causes either down- or upregulation of the putative target gene. However, this approach is not straightforward because (1) compensatory processes may maintain expression levels of true target genes, (2) expression measurements need to be performed shortly after TF knockdown, because otherwise many expression changes will be due to indirect regulation, and (3) sequence motifs with the highest affinity may retain bound TF the longest in an acute knockdown experiment so that the most important target genes may actually change expression last. High-throughput functional assays of promoter and enhancer regions provide a complementary approach that can assign regulatory function to TF-bound regions from ChIP-Seq studies. Of particular interest are recently reported embryonic stem cell differentiation-based assays,39,40 as they do not rely on immortalized cell line models but instead allow the reporting of regulatory element functionality in a range of primary cell lineages thus approximating transgenic mouse analysis. Although ChIP-Seq technology on its own did not fulfill its original expectation as the silver bullet for comprehensive identification of transcriptional hierarchies, combination with expression profiling and emerging high-throughput enhancer analysis undoubtedly has the potential to make a real impact in the future to accelerate the construction of comprehensive gene regulatory network models.
Reconstructing regulatory hierarchies from single cell analysis
Blood stem cell research has long made use of single cell assays to address fundamental questions such as the self-renewal and differentiation potential of individual cells within heterogeneous cell populations.41 As outlined above, gene expression is controlled by complex gene regulatory networks, and gene expression measurements can be used to infer the potential nature of those underlying networks. However, although gene expression measurements in single mammalian cells were pioneered in the hematopoietic system,42 these early studies relied on Southern blot hybridization of amplified cDNA 3′ ends43 or standard polymerase chain reaction methodology44 and therefore were unable to generate datasets of the scale required for network inference. Recent technological advances in microfluidic technology now readily permit quantification of tens to hundreds of genes in hundreds and even thousands of single cells. In addition to providing insights into population heterogeneity and putative transition stages during blood cell differentiation,45-47 the scale of these datasets provides a substrate for robust statistical analysis of gene expression correlation, as each single cell analyzed represents an independent biological measurement.48 Moignard et al surveyed the expression of 18 transcription factor genes in 690 single blood stem and progenitor cells,47 and went on to exploit pairwise correlation analysis to identify putative regulatory relationships between individual TFs. This analysis highlighted a previously unrecognized triad composed of Gata2, Gfi1, and Gfi1b, which might play an important role in regulating early fate choice decisions of differentiating blood stem cells. Importantly, the authors went on to validate direct regulatory interactions within this triad using a variety of functional assays, thus moving the analysis beyond simple statistical correlation.
A subsequent study profiled >80 TFs in >250 blood stem and progenitor cells49 and again used correlation analysis to predict putative regulatory connections. Integration with previously published ChIP-Seq data for 10 key HSPC TFs36 was used to highlight potentially direct regulatory links, and a central role for the transcription factor Gata2 was further supported by analysis of Gata2+/− HSPCs, which showed altered expression of some Gata2 targets that were consistent with the inferred network model. These early successes in regulatory network inference using single cell expression profiling suggest that this approach may be widely embraced by the scientific community. Future efforts should pay attention to the principles of partial correlation analysis discussed above in the context of the Aracne tool for network reconstruction from microarray expression profiles, because partial correlation analysis measures the degree of association between 2 genes with the potential effects of all other genes removed and therefore focuses analysis on associations that are more likely to be the result of direct regulatory relationships.
High-throughput single cell analysis also offers new opportunities to reconstruct the molecular nature of differentiation trajectories, based on the principle that within a population of differentiating yet unsynchronized cells, each single cell represents a snapshot that corresponds to a particular expression state on a given differentiation pathway. Computer tools have recently been reported that attempt to arrange such single cell measurements into putative differentiation time courses, thus introducing the concept of differentiation pseudotime.50-52 From a network reconstruction perspective, this will offer exciting new opportunities that go beyond simple correlation analysis, because a temporal sequence of expression changes offers potential insights into cause-effect relationships and transcriptional hierarchies. Successful reconstruction of high-quality regulatory network models will require large-scale datasets, probably consisting of thousands of single cells analyzed along a differentiation time course. Further development of algorithms will also be required, for example, to permit accurate reconstruction of pseudotime for branched differentiation trajectories. Moreover, single cell RNA-Seq is rapidly overtaking microfluidics-based quantitative reverse transcriptase-polymerase chain reaction due to falling costs and increased robustness of experimental protocols.53 Significant challenges remain, however, not just in terms of distinguishing experimental noise from true biological heterogeneity,54 but also in the context of network reconstruction where information on thousands of genes is likely to require even larger numbers of single cells to achieve robust inference of regulatory connections.
Outlook
The reconstruction of ever more accurate regulatory network models for HSPCs will require integration of data from all the experimental platforms discussed above (Figure 2). Moreover, availability of additional data types will be essential, particularly with regard to accurate measurements of TF protein abundance and activity. Given the known phenotypic heterogeneity and rarity of HSCs, assay miniaturization to the single cell level will be critical just as it has been for mRNA expression measurements. Mass cytometry,55 as well as recently reported single cell western blot technology,56 offer exciting perspectives, which will be enhanced further once protocols are developed that permit quantification of TF protein and target gene mRNA levels in the same single cell.
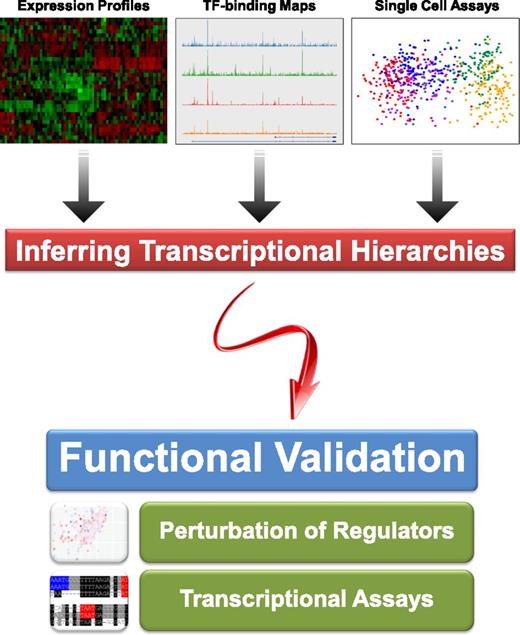
High-throughput technologies can be used to infer transcriptional hierarchies, but reconstruction of regulatory network models requires experimental validation of inferred regulatory connections. Shown at the top are representative examples of microarray expression profiles,73 ChIP-Seq TF binding maps,74 and single cell expression analysis,47 which can all be used to infer regulatory hierarchies. The bottom part of the figure illustrates the importance of follow-up functional experiments such as perturbation and high-throughput transcriptional assays39,40 to validate inferred regulatory connections.
High-throughput technologies can be used to infer transcriptional hierarchies, but reconstruction of regulatory network models requires experimental validation of inferred regulatory connections. Shown at the top are representative examples of microarray expression profiles,73 ChIP-Seq TF binding maps,74 and single cell expression analysis,47 which can all be used to infer regulatory hierarchies. The bottom part of the figure illustrates the importance of follow-up functional experiments such as perturbation and high-throughput transcriptional assays39,40 to validate inferred regulatory connections.
Owing to the central position of gene regulatory elements as building blocks of transcriptional regulatory networks, future progress in network reconstruction is also likely to come from emerging genome-scale “epigenetic” data, because TFs both influence, and are influenced by, the chromatin template at their target regulatory elements. Several recent studies have provided genome-wide DNA methylation maps alongside transcriptome analysis of hematopoietic differentiation and HSC aging,57-61 with the latter study by Goodell and colleagues further interrogating histone modifications, in addition to DNA methylome and transcriptome analysis. Such integrated approaches provide very welcome information on the likely activity and potential dynamic status of individual gene regulatory elements, and as such can be used to examine to what extent emerging network models are consistent with the likely activity of specific gene regulatory elements. However, epigenomic analysis is no substitute for functional dissection of regulatory mechanisms, which remains essential for the generation of validated network models. Of note, recent developments in genome editing (eg, CRISPR/Cas9) provide powerful new means for saturation mutagenesis62 that might become useful for interrogating regulatory elements controlling HSC fate and thereby accelerate our ability to functionally validate regulatory network components.
TF input is particularly important when cells change state from for example a progenitor type to a descendant cell type. However, most of the multigene models and methods discussed in this review identify distinct stable states and not precursor-product relationships or developmental kinetics. Attempts to explain hematopoietic differentiation dynamics are only seen in the small-scale models based on 2 or 3 well-studied genes.5,6 Moreover, all current models are limited by their closed-system scope, and the challenge for whole-genome approaches is to make such explanations fully definitive and inclusive. Large-scale experimental perturbation tests (Figure 2) will be vital, not only to convert the evidence for different stable states into deeper insights into precursor-product relationships and developmental kinetics, but also to make use of new genome-scale datasets, to ultimately generate network models that actually explain the regulation of blood development.
Both experimental approaches and network modeling algorithms will need to take into account the importance of cell-cell interactions. It was shown several years ago that megakaryocyte-derived stimulatory growth factors can promote self-renewal of human HSPCs.63 Using combined genomic and phenotype data, this work has recently been expanded to generate a directional cell-cell communication network between 12 human hematopoietic cell types isolated from umbilical cord blood.64 Microfluidics technology already offers opportunities to monitor single cells responding to an external signal, as well as to quantify signaling molecules secreted by individual HSPCs.65 A blueprint for the integration of inter-cellular communication into regulatory network models has been provided by Eric Davidson’s pioneering work on sea urchin development,1 which is likely to provide an important starting point for the future generation of similarly comprehensive network models for blood cell development.
Genome-scale expression datasets have also been generated for more specialized (but perhaps equally clinically important) fates of HSCs such as mobilization, homing, and the changes that accompany physiological processes such as aging.66-68 More recently, genome-wide epigenomic maps have also been reported comparing young and aged HSCs.61 It will be important to assess the extent to which regulatory network models for the more classical HSC fate choices (eg, lineage choice and self-renewal) can accommodate these additional functions of HSCs. Importantly, emerging genome-scale data coupled with targeted functional experiments will open the door to widen network analysis to encompass the whole spectrum of HSC physiology.
The generation of comprehensive regulatory network models for normal hematopoiesis will have major implications for our understanding of hematological pathologies. Many of the most commonly mutated genes in hematological malignancies encode transcriptional and epigenetic regulators. The functional consequences of such mutations can only be truly appreciated by taking into account the connectivity of a given gene within wider regulatory networks, because this critically determines how a given mutation will impact on the activity of other key regulators and therefore ultimately the behavior of the entire cell. Future therapeutic interventions are likely to increasingly depend on combination therapies for the concurrent targeting of multiple proteins, yet the number of possible permutations increases exponentially with the number of proteins under consideration. Computational modeling of leukemogenic mutations as network perturbations offers exciting opportunities to use computer simulations as a rapid means to identify those combinations of genes and/or regulatory interactions, where therapeutic intervention may have the potential to restore normal network behavior and therefore counteract the detrimental effect of the original mutation.
Finally, much effort in the field is directed at deriving transplantable HSCs from other cells types, via differentiation of induced pluripotent stem cells,69 transdifferentiation of somatic cells such as fibroblasts70 and endothelial cells,71 and also reprogramming/respecification of committed blood cells.72 Ultimately, the success of such efforts is contingent on instating (in the case of directed differentiation or transdifferentiation) or reinstating (in the case of reprogramming from differentiated blood cells) the regulatory networks governing HSC potential onto these other cell types. A more thorough understanding of HSC regulatory networks undoubtedly has the potential to greatly accelerate such research efforts.
Acknowledgments
Research in the author’s laboratory is supported by the Biotechnology and Biological Sciences Research Council, the Medical Research Council (MRC), Leukaemia and Lymphoma Research, Cancer Research UK, the Leukemia and Lymphoma Society, Microsoft Research, and core support grants by the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust–MRC Cambridge Stem Cell Institute.
Authorship
Contribution: B.G. wrote the paper.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Berthold Göttgens, Cambridge Institute for Medical Research, Hills Rd, Cambridge CB2 0XY, United Kingdom; e-mail: bg200@cam.ac.uk.