Key Points
APTs as miRNA targets provide a novel molecular mechanism for how primary CLL cells escape from CD95-mediated apoptosis.
Palmitoylation as a novel posttranslational modification in CLL might also impact on survival signaling, proliferation, and migration.
Abstract
Resistance toward CD95-mediated apoptosis is a hallmark of many different malignancies, as it is known from primary chronic lymphocytic leukemia (CLL) cells. Previously, we could show that miR-138 and -424 are downregulated in CLL cells. Here, we identified 2 new target genes, namely acyl protein thioesterase (APT) 1 and 2, which are under control of both miRs and thereby significantly overexpressed in CLL cells. APTs are the only enzymes known to promote depalmitoylation. Indeed, membrane proteins are significantly less palmitoylated in CLL cells compared with normal B cells. We identified APTs to directly interact with CD95 to promote depalmitoylation, thus impairing apoptosis mediated through CD95. Specific inhibition of APTs by siRNAs, treatment with miRs-138/-424, and pharmacologic approaches restore CD95-mediated apoptosis in CLL cells and other cancer cells, pointing to an important regulatory role of APTs in CD95 apoptosis. The identification of the depalmitoylation reaction of CD95 by APTs as a microRNA (miRNA) target provides a novel molecular mechanism for how malignant cells escape from CD95-mediated apoptosis. Here, we introduce palmitoylation as a novel posttranslational modification in CLL, which might impact on localization, mobility, and function of molecules, survival signaling, and migration.
Introduction
Chronic lymphocytic leukemia (CLL) cells are characterized by a distinct resistance toward CD95 (Fas/APO-1).1-4 CD95, as prototypical member of the tumor necrosis factor superfamily, is able to transmit a death signal to the cell upon binding of the CD95 ligand (CD95L).5 CD95 is involved in fundamental processes like the maturation or homeostasis of lymphocytes, as well as the elimination of virus-infected or malignant cells. Malignant cells might develop resistance mechanisms to escape from CD95-mediated apoptosis, either by silencing its expression,6 by mutation,7 or by overexpression of antiapoptotic proteins.2 Prior studies emphasized that proper CD95-mediated apoptotic signaling might depend on posttranslational modifications of CD95.8-11 This is of particular interest because the nature of molecular events downstream of the death-inducing signaling complex is well investigated, but the knowledge of initial events, especially at the plasma membrane, remains poorly defined. It was shown that palmitoylation, a common posttranslational modification, is crucial for localization of CD95 to specialized membrane microdomains and for formation of stable receptor aggregates.8,9
Palmitoylation is the only reversible lipid modification, indicating that it is involved in dynamic processes.12-14 Overall, proteome analyses identified >1000 proteins to be palmitoylated. These proteins are involved in central biological processes, like differentiation, activation, immune responses, proliferation, and migration.13,14 Cycles of depalmitoylation and repalmitoylation govern the transient membrane association of peripheral membrane proteins and thereby regulate their steady-state localization and function. Palmitoylation is catalyzed by protein acyltransferases, which are represented by the 23 members of the DHHC protein family,15 and reversed by acyl protein thioesterases (APTs) 1 and 2. APT1 and APT2 currently are the only thioesterases known to depalmitoylate in vivo.16-18 Although we could recently show that APTs interact with each other, there is no evidence of a direct interaction between APTs and their different substrates so far.19
Regulation of protein expression is controlled by microRNAs (miRNAs),20 small noncoding RNAs that negatively regulate expression of genes, either by degradation of messenger RNA (mRNA) or by translational silencing.21 We previously reported that miRNAs are deregulated in primary CLL cells, thus causing differential expression of oncogenes.22,23 It remains to be clarified whether CD95 resistance in CLL is a result of deregulated gene expression which finally impacts on the palmitoylation status and signaling of CD95. Indeed, our data support the prior finding of CD95 palmitoylation8,9 in primary CLL cells. Above all, we approached how and by which enzymes palmitoylation of CD95 is altered. Second, we investigated the potential regulation of CD95-modifying enzymes by miRNAs. These novel findings provide an opportunity to specifically interfere with the depalmitoylation process of CD95 and thereby increase sensitivity toward CD95-mediated apoptosis in CLL.
Materials and methods
Patient samples
After informed consent according to the Helsinki protocol was given, blood was obtained from CLL patients. CLL cells were purified as described before.24-26 This study was approved by the Ethics Committee of the University of Cologne.
Detection of palmitoylated proteins
Novel nonradioactive biochemical approaches (detection of palmitoylated proteins via click chemistry and acyl-biotin exchange assays) were applied to analyze palmitoylation of membrane proteins and CD95 according to the protocols by Martin et al,13 Martin and Cravatt,27 and Wan et al.28 Detailed protocols can be found in the supplemental Methods (see supplemental Data available on the Blood Web site) under “Analysis of palmitoylation by click chemistry” and “Acyl-biotin exchange assay”.
miRNA, RNA extraction, and cDNA synthesis
RNA and miRNA were extracted using TriFast (PEQLAB). Therefore, cells were lysed with TriFast. Chloroform was added for extraction, followed by centrifugation. RNA was precipitated with isopropanol and then washed with 70% ethanol; afterward, it was resuspended in RNAse-free water and incubated at 55°C for 10 minutes. Synthesis of complementary DNA (cDNA) was performed with the Transcriptor First Strand cDNA Synthesis kit (Roche Applied Sciences) following the manufacturer’s instructions.
Transfections
For APT1 and APT2 knockdown, 2 × 107 CLL cells were nucleofected with 80 pM specific ON-TARGETplus SMARTpool siRNA (Dharmacon). Nontargeting ON-TARGETplus siCONTROL siRNA (Dharmacon) was used as a negative control. The nucleofection was performed using program U13 at the Amaxa Nucleofector (Lonza). HeLa cells (4 × 103 to 5 × 105 ) were transfected with 100 pM specific ON-TARGETplus SMARTpool siRNA (Dharmacon) or nontargeting ON-TARGETplus siCONTROL siRNA (Dharmacon) for RNA interference (RNAi) experiments, whereas they were transfected with 30 ng to 2.5 µg of plasmid DNA for other experiments (eg, luciferase assays, fluorescence recovery after photobleaching [FRAP]) using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions.
Quantitative reverse transcription PCR
The following specific primers were used for polymerase chain reaction (PCR) with the LightCycler TaqMan Master kit (Roche Applied Sciences): APT1 no. 52, forward CAGAATTATTTTGGGAGGGTTTT; APT1 no. 52, reverse CAGTTTCTGCTGTGTGGTAAGG; APT2 no. 9, forward CAAGTACATCTGTCCCCATGC; APT2 no. 9, reverse GACTCAGCCCCATCAGGTC. β-actin was applied as housekeeping gene standard. For miRNA expression analysis, TaqMan MicroRNA assays (Applied Biosystems) for miR-138 and miR-424 were used. SNORD48 served as a control. All experiments were performed in replicates and crossing points were determined by second derivative maximum method. Relative quantification analysis was performed using standard curves.
Luciferase assays
The luciferase assays were performed in HeLa cells as described previously.22 Cells were cotransfected with wild-type or mutated 3′-untranslated region (3′-UTR) clones and specific miRIDIAN miRNAs (Dharmacon) as described in “Transfections.” MiRIDIAN miRNA hsa-miR-107 and hsa-miR-34 (Dharmacon) were used as negative controls. The experiment was conducted using the Dual-Glo Luciferase Assay System (Promega), according to the manufacturer’s instructions.
FLIM-FRET
For fluorescence lifetime imaging microscopy–Förster resonance energy transfer (FLIM-FRET) experiments, HeLa cells were transfected with CD95-enhanced yellow fluorescent protein (eYFP), CD95-eYFP (C199S) (CD95-eYFP mut), APT1-mCherry, and/or APT2-mCherry as described in “Transfections.” Cells were imaged on a TCS SP5 (Leica) equipped with Picoquant FLIM modules (LSM Upgrade kit/SMD Module; Picoquant) and a PL APO 63×/1.4 objective (Leica) at 37°C. Cells expressing eYFP-tagged proteins were excited at 514 nm with a pulsed supercontinuum laser (80 MHz). Photons were detected using avalanche photodiodes and timed using a time-correlated single-photon counting module (PicoHarp 300; Picoquant). The average photon arrival times for each pixel were computed in presence and absence of mCherry fusion proteins as FRET acceptors. Simultaneously, intensity images of the donor and acceptor channels were acquired on photomultiplier tubes with LAS-AF (Leica). FLIM datasets were analyzed as previously described.19,29 Details about additional controls and analysis can be found in the supplemental Methods under “FLIM-FRET details.”
FRAP
To investigate the dynamics of CD95 on the plasma membrane, HeLa cells were transfected with CD95-eYFP or CD95-eYFP mut as described in “Transfections.” Four hours after transfection, 10 µM ZVAD was added to all samples. After 24 hours, cells were analyzed by the UltraView Vox (Perkin Elmer) equipped with an EMCCD camera and an Apo TIRF 60×/1.49 objective (Nikon) at 37°C. Samples were bleached with an argon laser (514 nm, YFP, 100% at maximum speed) in a manually defined region of interest (ROI). Images were acquired with the program Volocity (Perkin Elmer). Data acquisition rate was 1 frame every 30 seconds and 3 prebleach images were collected at maximum speed before the bleach. Data analysis was performed by using the easyFRAP software.30
XTT
Cell viability of HeLa cells was evaluated using XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) assays. HeLa cells were seeded and transfected as defined in “Transfections” and/or treated as described. XTT assays were then performed using the Cell Proliferation kit II (Roche Applied Sciences) according to the manufacturer’s instructions.
Annexin V-FITC/7-AAD staining
Apoptosis rates of CLL cells were determined by flow cytometry using a FACSCanto (BD). Therefore, cells were harvested and then washed once with Cell Wash (BD). In the next step, they were stained with Annexin V–fluorescein isothiocyanate (FITC) (Immuno Tools) and 7-aminoactinomycin D (7-AAD) (eBioscience) in Annexin V–binding buffer (BD), subsequently followed by further analysis.
Results
miRNAs miR-138 and miR-424 are deregulated and target APT1 and APT2 in CLL
As we and others have shown, miRNAs play a central regulatory role in the pathogenesis of CLL.22,23,31-34 Based on our previous studies where we identified differential miRNA and mRNA profiles in primary CLL cells, we now aimed to connect both datasets.22,23,35 We found miR-138 and -424 to be significantly downregulated in CLL cells. However, both miRNAs were not functionally described in CLL thus far. At the same time, we could show that most genes involved in the lipid metabolism were upregulated in CLL.35 Therefore, we performed in silico analyses comparing conserved binding sites for both miRNAs (http://www.targetscan.org) and the 3′-UTRs of our upregulated genes, which are involved in lipid metabolism.22,23,35 For miR-138, we found a conserved binding site within the 3′-UTR of the depalmitoylating enzyme APT1. Interestingly, the analysis also revealed an as-yet-unknown conserved binding site for miR-424 within the 3′-UTR of APT2, a protein we recently identified as a second depalmitoylating enzyme.36 According to our bead array data, we first investigated whether both miRNAs (miR-138 and miR-424) are downregulated in CLL cells compared with B cells from healthy donors by applying quantitative reverse transcription PCR (Figure 1A). In detail, miR-138 was 5-fold overexpressed in normal B cells vs primary CLL cells (P = .0027), whereas miR-424 was even 25-fold overexpressed in normal B cells compared with CLL cells (P = .0025), where it was hardly detectable.
APT1 and APT2 are regulated by miR-138 and miR-424. Quantitative PCR was used to compare expression of miRNAs potentially regulating APT1 and APT2 in CLL cells (n = 19 [miR-138] or n = 7 [miR-424]) and healthy B cells (n = 9 [miR-138] or n = 5 [miR-424]). miR-138 and miR-424 are significantly downregulated in primary CLL cells (A). Luciferase assays were performed to analyze the binding of both miRNAs to the respective 3′-UTR of APT1 or APT2. It could be shown that both miRNAs are in principle able to regulate APT1 (n = 4) or APT2 expression (n = 3) (B). By western blotting it could be demonstrated that miR-138 regulates APT1 (n = 3) and miR-424 regulates APT2 protein expression (n = 3) (C). MiR-138 and miR-424 regulate APT1- and APT2-mRNA levels by reducing mRNA levels (D). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SEM, standard error of the mean.
APT1 and APT2 are regulated by miR-138 and miR-424. Quantitative PCR was used to compare expression of miRNAs potentially regulating APT1 and APT2 in CLL cells (n = 19 [miR-138] or n = 7 [miR-424]) and healthy B cells (n = 9 [miR-138] or n = 5 [miR-424]). miR-138 and miR-424 are significantly downregulated in primary CLL cells (A). Luciferase assays were performed to analyze the binding of both miRNAs to the respective 3′-UTR of APT1 or APT2. It could be shown that both miRNAs are in principle able to regulate APT1 (n = 4) or APT2 expression (n = 3) (B). By western blotting it could be demonstrated that miR-138 regulates APT1 (n = 3) and miR-424 regulates APT2 protein expression (n = 3) (C). MiR-138 and miR-424 regulate APT1- and APT2-mRNA levels by reducing mRNA levels (D). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SEM, standard error of the mean.
Next, we aimed to provide proof of whether both miRs directly regulate APT1 and APT2. By using luciferase reporter assays with subsequent mutagenesis of predicted binding sites, we could show that miR-138 bound specifically to APT1–3′-UTR (Figure 1B left panel), whereas miR-424 interacted specifically with the APT2–3′-UTR (Figure 1B right panel).
Based on these findings, we performed western blots after transfection of each miRNA to determine their impact on protein expression of APT1/2. Transient transfection of miR-138 reduced protein expression of APT1 to ∼60%, whereas miR-424 halved APT2 expression in comparison with scrambled miR controls (Figure 1C). Simultaneously, we analyzed mRNA levels of APT1/2 to investigate whether mRNAs are degraded by miRNAs. Indeed, we found that APTs were not only downregulated by miRNAs on protein but also on the mRNA level (Figure 1D). These results clearly indicate that miR-138 and miR-424 regulate expression of APT1 and APT2.
APT1 and APT2 are overexpressed in primary CLL cells
Taking into consideration that APTs were upregulated in gene array data of CLL cells, we next aimed to quantify and compare the expression patterns of both APTs on mRNA and the protein level between CLL cells and normal B cells by quantitative reverse transcription PCR and immunoblotting. We could show that APT1 and APT2 were significantly overexpressed on mRNA and the protein level in primary CLL cells. Meanwhile, APT2 was hardly detectable in normal B cells on the protein level; a high APT2 expression could be detected in CLL cells (Figure 2B). Moreover, APT1 and APT2 were equally expressed with respect to the immunoglobulin variable heavy (IgVH) mutational status (supplemental Figure 3).
APT1 and APT2 are overexpressed in primary CLL cells ex vivo. APT1 and APT2 mRNA expression was analyzed in healthy B cells (n = 4) and CLL cells (n = 7 [APT1] or n = 8 [APT2]) by quantitative PCR (A). Western blotting showed that APT1 and APT2 are overexpressed not only on mRNA, but also on protein level in CLL cells (n = 7 [APT1] or n = 9 [APT2]) compared with healthy B cells (n = 4 [APT1] or n = 5 [APT2]) (B-C). Global palmitoylation was analyzed by click chemistry. Comparison of CLL cells (n = 10), healthy PBMCs (n = 6), and healthy B cells (n = 6) revealed that the global palmitoylation level is significantly lower in CLL cells than in the healthy controls (D). Palmitoylation of proteins can be enhanced by the use of the depalmitoylation inhibitor PB (n = 5) (E). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.
APT1 and APT2 are overexpressed in primary CLL cells ex vivo. APT1 and APT2 mRNA expression was analyzed in healthy B cells (n = 4) and CLL cells (n = 7 [APT1] or n = 8 [APT2]) by quantitative PCR (A). Western blotting showed that APT1 and APT2 are overexpressed not only on mRNA, but also on protein level in CLL cells (n = 7 [APT1] or n = 9 [APT2]) compared with healthy B cells (n = 4 [APT1] or n = 5 [APT2]) (B-C). Global palmitoylation was analyzed by click chemistry. Comparison of CLL cells (n = 10), healthy PBMCs (n = 6), and healthy B cells (n = 6) revealed that the global palmitoylation level is significantly lower in CLL cells than in the healthy controls (D). Palmitoylation of proteins can be enhanced by the use of the depalmitoylation inhibitor PB (n = 5) (E). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.
Taking these findings into consideration, we hypothesized that overexpression of depalmitoylating enzymes should result in lower global protein palmitoylation. Therefore, we investigated whether global palmitoylation of membrane proteins is altered in primary CLL cells in comparison with B cells and peripheral blood mononuclear cells (PBMCs) of healthy volunteers by applying click chemistry (supplemental Figure 1A).13,37 Our results uncover that CLL cells have significantly less palmitoylation compared with normal B cells (P = .0176) and normal PBMCs (P = .0207), which show the most highly palmitoylated membrane proteome (Figure 2D, supplemental Figure 1B).
To determine the impact of depalmitoylation for lower levels of membrane protein palmitoylation in primary CLL cells, we applied Palmostatin B (PB) to inhibit APT1 and APT2.36 We designed PB as a specific small-molecule inhibitor for APT1 and APT2.36 Indeed, inhibition of depalmitoylation by PB significantly increased the global palmitoylation of membrane proteins in primary CLL cells (n = 5) in comparison with dimethylsulfoxide (DMSO)–treated controls (P = .015) (Figure 2E, supplemental Figure 1C). These findings reveal that differences in the protein-palmitoylome are most likely due to overexpressed APTs in primary CLL cells.
APT1 and APT2 interact with CD95
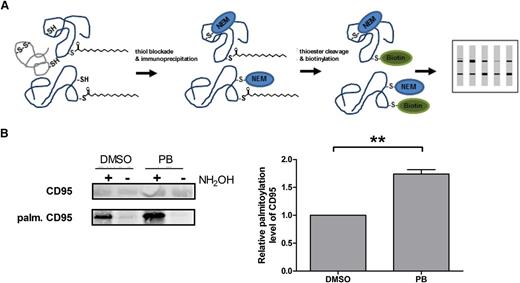
Coming from these general findings, we focused on CD95. CLL cells are resistant toward CD95-mediated apoptosis, independent of CD95 expression patterns on their surface or mutations of the CD95 gene, but mostly due to unknown downstream mechanisms.3,38-44 Interestingly, CD95 was shown to be palmitoylated at cysteine 199 in humans. This palmitoylation site is important for apoptosis induction via the CD95:CD95L interaction. However, the regulatory mechanism is unknown.8,9 Therefore, we first investigated the CD95 palmitoylation status in primary CLL cells with the help of acyl-biotin exchange (ABE) chemistry. Indeed, we could detect palmitoylated CD95. Additionally, inhibition of APT1/APT2 by PB resulted in significantly increased palmitoylation of CD95 (P = .0053) (Figure 3).28
CD95 is depalmitoylated by APTs. Principle of ABE assays according to Wan et al28 (A). Palmitoylation of CD95 was analyzed by ABE assays. Therefore, CD95 of primary CLL cells was immunoprecipitated. Inhibition of depalmitoylation by PB significantly increased palmitoylation levels of CD95 (n = 3) (B).
CD95 is depalmitoylated by APTs. Principle of ABE assays according to Wan et al28 (A). Palmitoylation of CD95 was analyzed by ABE assays. Therefore, CD95 of primary CLL cells was immunoprecipitated. Inhibition of depalmitoylation by PB significantly increased palmitoylation levels of CD95 (n = 3) (B).
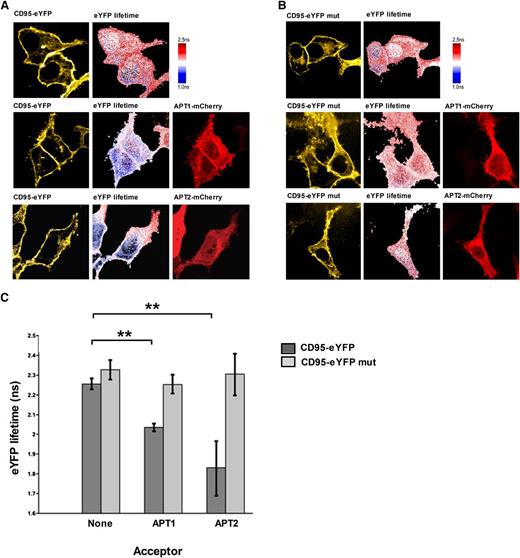
Given that inhibition of depalmitoylating enzymes APT1/APT2 results in significantly increased palmitoylation of CD95, we hypothesized that CD95 and APT1/2 might interact with each other. Due to the highly dynamic nature of palmitoylation, we expected interactions between APTs and their substrates to be transient and hence performed FLIM-FRET assays. Because APTs are partially cytosolic proteins and therefore might interact with CD95 either at the membrane or in the cytosol, we performed experiments in HeLa cells, which are much larger than small lymphocytes and easier to transfect. Analyses of the FLIM-FRET data identified a direct interaction of both APT1 and APT2 with CD95, but not with the CD95 (C199S) palmitoylation mutant (Figure 4). These results reveal an interaction taking place directly at the palmitoylation site of CD95 with most significance not only at the plasma membrane, but also in the cytoplasm. So far, this is the first report proving a direct interaction between depalmitoylating enzymes and their substrate.
CD95 and APTs interact at the plasma membrane. FLIM-FRET experiments of CD95 with APT1 and APT2 demonstrate that CD95 directly interacts with both proteins (A). The interaction is dependent on the palmitoylation of cysteine 199 (B). Lifetime of the donor (CD95-eYFP) at the plasma membrane and in the cytoplasm is significantly reduced after incubation with the acceptor (APT1- or APT2-mCherry) (C). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.
CD95 and APTs interact at the plasma membrane. FLIM-FRET experiments of CD95 with APT1 and APT2 demonstrate that CD95 directly interacts with both proteins (A). The interaction is dependent on the palmitoylation of cysteine 199 (B). Lifetime of the donor (CD95-eYFP) at the plasma membrane and in the cytoplasm is significantly reduced after incubation with the acceptor (APT1- or APT2-mCherry) (C). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.
Palmitoylation controls mobility of CD95 within membranes
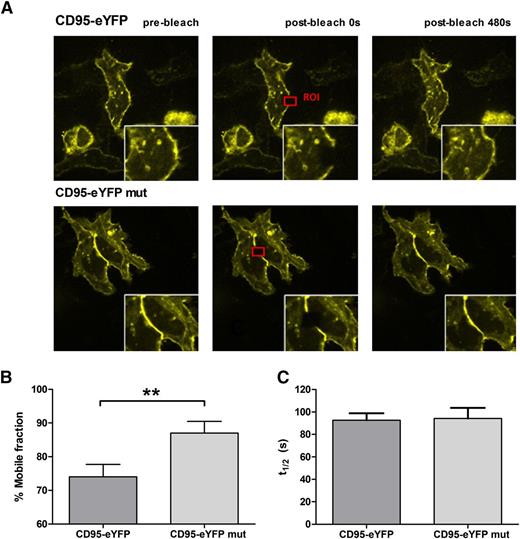
Receptor function depends on the distribution within the membrane and on the interaction with other proteins that are either membrane-localized or present in the cytoplasm or the extracellular space. Because CD95 predominantly interacts with APTs at the plasma membrane, we asked how palmitoylation impacts on the distribution of CD95 in the membrane. Therefore, we determined the mobility of CD95 in correlation to the status of its palmitoylation site by applying FRAP. Figure 5A shows representative prebleach and postbleach images with a highlighted ROI. Interestingly, our results show that palmitoylation reduces the mobile fraction of CD95 molecules significantly in comparison with the palmitoylation-deficient CD95 mutant (P = .0072) (Figure 5B), whereas time to recovery was not affected (Figure 5C).
Mobility of CD95 within the plasma membrane is regulated by palmitoylation. FRAP assays were performed to investigate the impact of palmitoylation on the receptor’s mobility on the plasma membrane. A typical experiment is shown for CD95-eYFP and CD95-eYFP mut (A). Comparison of wild-type (palmitoylated) CD95 with mutated (C199S, unpalmitoylated) CD95 showed that the mobile fraction is significantly higher in cells expressing the unpalmitoylated receptor (n = 15), while t1/2 is unaffected (B-C). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.
Mobility of CD95 within the plasma membrane is regulated by palmitoylation. FRAP assays were performed to investigate the impact of palmitoylation on the receptor’s mobility on the plasma membrane. A typical experiment is shown for CD95-eYFP and CD95-eYFP mut (A). Comparison of wild-type (palmitoylated) CD95 with mutated (C199S, unpalmitoylated) CD95 showed that the mobile fraction is significantly higher in cells expressing the unpalmitoylated receptor (n = 15), while t1/2 is unaffected (B-C). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.
APT1 and APT2 control CD95-mediated cell death in different malignant entities
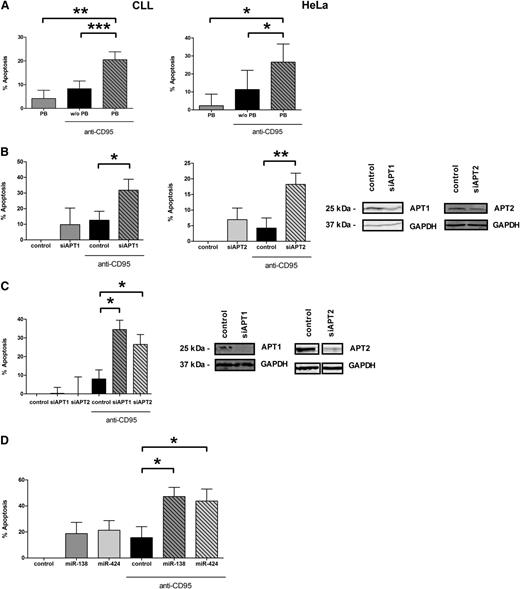
In this study, we identified that palmitoylation of CD95 is regulated by APT1 and APT2. As a consequence, we asked whether apoptosis via CD95 is also controlled by APTs. We applied 3 different settings to determine whether APT1 or APT2 regulate apoptosis via CD95: (1) pharmacologic inhibition of APTs via PB, (2) specific knockdown by siRNAs against APT1/2, and (3) specific knockdown by overexpression of corresponding miRNAs. To elucidate whether control of CD95-mediated apoptosis by APTs is limited to CLL cells only or is detectable in other cancer cells as well, we also performed those experiments in HeLa cells. CLL cells were cocultivated on CD40L-expressing feeder cells to increase CD95 expression on their cell surface, whereas HeLa cells already express high amounts of CD95. Here, we could show that treatment with PB does not change expression of CD95 on the surface of CLL and HeLa cells (supplemental Figure 2). Single treatment with anti-CD95 or PB did not result in significant apoptosis (Figure 6A). Importantly, inhibition of APTs via PB concomitant with anti-CD95 treatment resulted in significant CD95-mediated cell death independent of the IgVH mutational status, increasing from 5.4% to 20% in CLL cells (P = .0004) and from 11.3% to 26.6% in HeLa cells (P = .0473), compared with solely anti-CD95–treated cells (Figure 6A). To elucidate whether this is a specific effect mediated by APTs and to investigate whether both APTs are involved, we used specific siRNAs against APT1 and APT2. Indeed, knockdown of both APTs in addition to treatment with anti-CD95 significantly sensitized CLL cells and HeLa cells toward CD95-mediated apoptosis. Apoptosis in CLL cells was increased from 12.6% to 31.4% after APT1 knockdown (P = .0354) and from 4.2% to 18.4% after APT2 knockdown (P = .0043). In HeLa cells, apoptosis was increased from 8% to 34.5% (APT1 knockdown, P = .0181) or 26.5% (APT2 knockdown, P = .0267) (Figure 6B-C). To investigate the specificity of the proposed effect between APTs and CD95, primary CLL cells were treated with BH3 mimetics (ABT-737 and ABT-199) or fludarabine and PB. In contrast to CD95 and APT inhibition, we could not find any synergism between PB and the aforementioned drugs in primary CLL cells (supplemental Figure 4). Moreover, we extended our observations to HeLa cells, by targeting APTs via PB or siRNAs and additional treatment with ABT-737, ABT-199, and fludarabine. Again, we could not find additional or synergistic apoptosis (data not shown). These data support the relevance of APTs for CD95-mediated apoptosis.
APT1 and APT2 control CD95-mediated apoptosis. PB treatment significantly enhanced apoptosis mediated through anti-CD95 (CH11) in CLL cells after 24 hours (n = 14). Apoptosis of CLL cells was determined by Annexin V/7-AAD staining and subsequent flow cytometry. Apoptosis was also significantly increased after treatment of HeLa cells with PB and CD95 after 24 hours (n = 3). Survival of HeLa cells was determined by XTT assays (A). Specific siRNA knockdown of APT1 (n = 3) or APT2 (n = 5) for 48 hours was able to mimic the PB effect in CLL cells (B) and in HeLa cells (n = 3) (C). Knockdown efficiency was determined by western blotting (B-C). In addition, overexpression of miRNAs regulating APT1 and APT2 for 48 hours also led to increased apoptosis in HeLa cells (n = 4) (D). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.
APT1 and APT2 control CD95-mediated apoptosis. PB treatment significantly enhanced apoptosis mediated through anti-CD95 (CH11) in CLL cells after 24 hours (n = 14). Apoptosis of CLL cells was determined by Annexin V/7-AAD staining and subsequent flow cytometry. Apoptosis was also significantly increased after treatment of HeLa cells with PB and CD95 after 24 hours (n = 3). Survival of HeLa cells was determined by XTT assays (A). Specific siRNA knockdown of APT1 (n = 3) or APT2 (n = 5) for 48 hours was able to mimic the PB effect in CLL cells (B) and in HeLa cells (n = 3) (C). Knockdown efficiency was determined by western blotting (B-C). In addition, overexpression of miRNAs regulating APT1 and APT2 for 48 hours also led to increased apoptosis in HeLa cells (n = 4) (D). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.
Because we could elucidate that miRNAs control expression of APTs, we finally asked whether overexpression of miR-138 and miR-424 also influences CD95-mediated apoptosis. Indeed, both miRNAs sensitized malignant cells toward CD95-mediated apoptosis (Figure 6D), but not toward BH3 mimetic-mediated or fludarabine-mediated apoptosis (data not shown). Compared with treatment with anti-CD95 only (15.6%), overexpression of miR-138 increased apoptosis to 47.2% (P = .0142), whereas overexpression of miR-424 led to 43.7% apoptosis (P = .01885) (Figure 6D).
Discussion
Posttranslational modifications are essential for regulation of intracellular signaling. With this study, we introduce palmitoylation as a novel and relevant modification regulating CD95 signaling in CLL cells. We identified membrane proteins in CLL cells to be mainly depalmitoylated, while palmitoylation levels are high in healthy PBMCs and healthy B cells. In fact, we found that both depalmitoylating enzymes, APT1 and APT2, are overexpressed in primary CLL cells compared with the healthy counterpart. This is especially the case for APT2, which could hardly be detected in normal B cells. This finding is of special interest because so far nothing is known about the expression patterns and the distribution of APT1 and APT2 in normal tissues or malignant disease. These results allow us to hypothesize that APTs might act as oncoproteins in CLL cells. In light of global palmitoylome studies, which show that palmitoylated proteins are involved in central cellular processes, this emphasizes the importance of palmitoylation and might put it on par with modifications like phosphorylation.13,28,45,46
Indeed, we uncovered that APT1 and APT2 control CD95-mediated apoptotic signaling by a direct interaction with CD95. To date, only a few substrates of APTs are known. However, a direct interaction has not been identified thus far.17,18,47 This is the first study that could experimentally prove a direct interaction between a depalmitoylating enzyme and its substrate. Interestingly, our findings show that the interaction between APTs and CD95 takes place at the palmitoylated cysteine. In general, there is not much known about the mechanism of substrate recognition by APT1 and APT2. One study concerning Ras shows that the protein is only depalmitoylated after isomerization of a proline next to the region with the palmitoylated cysteine by FK506-binding protein 12 (FKPB12).48 Not only is CD95 a predominantly apoptotic receptor, but it is also known to activate nonapoptotic pathways.49,50 There are indications that expression of the receptor on the cell surface is important for proper apoptotic signaling in CLL.51 Here, we show that high expression on the cell surface is not sufficient for sensitizing resistant cells to death receptor-induced apoptosis. This emphasizes the importance of APT1/2 for CD95 signaling. We provide functional data that inhibition of APTs leads to sensitization of primary CLL cells and HeLa cells to CD95-mediated apoptosis, but not toward apoptosis which is mediated by fludarabine or BH3 mimetics. Thereby, we revealed that APTs directly control death receptor-mediated apoptosis independent of the mutational status (IgVH). Occurring differences in apoptosis rates between CLL cells and HeLa cells might be due to heterogeneity of CLL patients, the amount of CD95 on the surface, or knockdown efficacy of APTs.
Based on our results, we asked how APTs are regulated and showed that differential expression of both APTs is controlled by miRNAs. In line with recent findings in neurons, we could show that APT1 is regulated by miR-138 in malignant cells.52 Both observations (ours and those of Siegel et al52 ) underscore that miR-138 is a true regulator of APT1. However, we additionally identified miR-424 as a potent regulator of APT2. Both miRNAs have been found, by others and us, to be associated with tumor-suppressing functions.22,53,54 Indeed, after re-expression of both miRNAs we were able to restore CD95-mediated apoptosis by downregulating APTs. Additionally, no other protein that is associated with death receptor signaling is known to be regulated by 1 of the identified miRNAs.55,56 Nevertheless, miRNAs might mediate functional changes through coregulating function-related genes.57 Therefore, we cannot exclude that, to some extent, apoptosis is induced by targeting non-death receptor pathways. For instance, in lung cancer, it was shown that miR-138 inhibits proliferation by regulating 3-phosphoinositide–dependent protein kinase-1 (PDK1), a kinase critical for protein kinase B (PKB) signaling.54 Downregulation of miR-424 results in Chk1-mediated progression of cervical cancer.53
In summary, this study identifies a new sequence of events leading to resistance toward CD95-mediated apoptosis in CLL, beginning first with downregulation of specific miRNAs (miR-138/miR-424), which second leads to overexpression of APTs and results in nonapoptotic CD95 signaling.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Julia Claasen (University Hospital Cologne, Cologne, Germany) for excellent technical support.
This work was supported by German Cancer Aid (DKH109159: Lipid metabolism and signaling as new targets in CLL [C.P.P. and C.-M.W.]) and supported in part by grants from the Deutsche Forschungsgemeinschaft (DFG: Excellence Cluster 229: Cellular Stress Responses in Aging-Associated Diseases/CECAD, Bonn, Germany [M.H., C.-M.W., and L.P.F.] and KFO-286 [M.H. and C.P.P.]). C.-M.W. is supported by the CLL Global Research Foundation (Houston, TX). Moreover, this study was supported in part by the Marga and Walter Boll-Stiftung (210-05-13) and by the German Jose Carreras Leukemia Foundation (R12/26 [L.P.F.]).
Authorship
Contribution: L.P.F. and C.-M.W. conceived and designed the present work; V.B., M.R., N.V., C.J., A.S., and C.H. performed the research; V.B., C.J., A.S., N.V., and L.P.F. conducted statistical analysis; V.B., C.P.P., M.H., H.W., P.I.H.B., C.-M.W., and L.P.F. analyzed the data; and V.B. and L.P.F. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lukas P. Frenzel, Department I of Internal Medicine, University of Cologne, Kerpener Strasse 62, 50937 Cologne, Germany; e-mail: lukas.frenzel@uk-koeln.de.
References
Author notes
C.-M.W. and L.P.F. contributed equally to this work.

![Figure 1. APT1 and APT2 are regulated by miR-138 and miR-424. Quantitative PCR was used to compare expression of miRNAs potentially regulating APT1 and APT2 in CLL cells (n = 19 [miR-138] or n = 7 [miR-424]) and healthy B cells (n = 9 [miR-138] or n = 5 [miR-424]). miR-138 and miR-424 are significantly downregulated in primary CLL cells (A). Luciferase assays were performed to analyze the binding of both miRNAs to the respective 3′-UTR of APT1 or APT2. It could be shown that both miRNAs are in principle able to regulate APT1 (n = 4) or APT2 expression (n = 3) (B). By western blotting it could be demonstrated that miR-138 regulates APT1 (n = 3) and miR-424 regulates APT2 protein expression (n = 3) (C). MiR-138 and miR-424 regulate APT1- and APT2-mRNA levels by reducing mRNA levels (D). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SEM, standard error of the mean.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/19/10.1182_blood-2014-07-586511/4/m_2948f1.jpeg?Expires=1769354129&Signature=qWpt7Ma7iX6t61YsqhM4kXVqwu89Zz35hCWGHoW2YF1KiAdBXYdhAlESyoJKinZ8tIVz5EIlqdfbQAOuuPKYGt6ObMjkVnxTSA4WS5OOGcf6~~Jx58Jxm8kwco24ohx~jEca94iFn2w31AqsLXU5JUzHFfNmDhKJ8acE3HmDe80M-AerWfeQVOcXoUXiauwKp9-3O~5G6GGaIHLLcSFtZfAR-xf9oSPuliUB2RNyaX-qIE9vaR6zfHyHGlR7p59ODpP0CHYU2qEJo3VZCiQfuv7RLSoVyDFDzCc1JOfLfNA4OUS4R1afxqkRq~tYKYKE6LdJVxP8W8b3PJwtILm3iw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. APT1 and APT2 are overexpressed in primary CLL cells ex vivo. APT1 and APT2 mRNA expression was analyzed in healthy B cells (n = 4) and CLL cells (n = 7 [APT1] or n = 8 [APT2]) by quantitative PCR (A). Western blotting showed that APT1 and APT2 are overexpressed not only on mRNA, but also on protein level in CLL cells (n = 7 [APT1] or n = 9 [APT2]) compared with healthy B cells (n = 4 [APT1] or n = 5 [APT2]) (B-C). Global palmitoylation was analyzed by click chemistry. Comparison of CLL cells (n = 10), healthy PBMCs (n = 6), and healthy B cells (n = 6) revealed that the global palmitoylation level is significantly lower in CLL cells than in the healthy controls (D). Palmitoylation of proteins can be enhanced by the use of the depalmitoylation inhibitor PB (n = 5) (E). Means are given with their SEM. The statistical significance was determined using the Student t test. *P < .05, **P < .01, or ***P < .001 were considered significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/19/10.1182_blood-2014-07-586511/4/m_2948f2.jpeg?Expires=1769354129&Signature=Wb2zO31a2mRoC6~CAJoCEoduxACTsuHP6mj0r3cdLAN6yOjaTuBPY46WO2YWP-pmP6kIRypAm4t5H2qEuYkTavUfPKDw4WV0KHNxePDKgc7wWeuGB9YpmDKNJ8Np8TVcp7D0uofcmZ6kVZbXImsvM2T2Br6GYbAUFZcK9Px1EN7YTfNTX-SgGet9KOBR7Y6nC1jzSX-FtIRqHikEpiP3Ozv7JtXQvY4VstGzlkp3Is0b18KCVuU6wC~k-17h2vFn0jd8MaQ3k6BYNz7yko~Iapth70xXOzy53ij~zmDbfTWagcYM5-ou2EDQA4BFfXgueH272BnT6o1JVDIoz8rbHw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal