Key Points
mAbs from aggressive CLL subset #8 display extreme antigen polyreactivity, in clear contrast with the mAbs from other aggressive CLL subsets.
Subset #8 CLL clones respond avidly to stimulation by multiple antigens and this may underlie their noted propensity to transform.
Abstract
Subset #8 is a distinctive subset of patients with chronic lymphocytic leukemia (CLL) defined by the expression of stereotyped IGHV4-39/IGKV1(D)-39 B-cell receptors. Subset #8 patients experience aggressive disease and exhibit the highest risk for Richter transformation among all CLL. In order to obtain biological insight into this behavior, we profiled the antigen reactivity and signaling capacity of subset #8 vs other clinically aggressive stereotyped subsets, namely subsets #1 and #2. Twenty-seven monoclonal antibodies (mAbs) from subsets #1, #2, and #8 CLL clones were prepared as recombinant human immunoglobulin G1 and used as primary antibodies in enzyme-linked immunosorbent assays against representatives of the major classes of established antigenic targets for CLL. Subset #8 CLL mAbs exhibited broad polyreactivity as they bound to all antigens tested, in clear contrast with the mAbs from the other subsets. Antigen challenge of primary CLL cells indicated that the promiscuous antigen-binding activity of subset #8 mAbs could lead to significant cell activation, again in contrast to the less responsive CLL cells from subsets #1 and #2. These features constitute a distinctive profile for CLL subset #8, supporting the existence of distinct mechanisms of aggressiveness in different immunogenetic subsets of CLL.
Introduction
Microenvironmental dependence has been established as relevant for the natural history of chronic lymphocytic leukemia (CLL).1,2 Supportive functional evidence is provided by the fact that CLL cells do not survive or proliferate autonomously in vitro, indicating that they are still dependent on external stimuli.3 Furthermore, they respond, though variably, to stimulation of both the B-cell receptor (BCR) immunoglobulin (Ig) with anti-Ig antibodies as well other receptors (eg, CD40, toll-like receptors [TLRs], and chemokine receptors).4-10 Finally, BCR signaling inhibitors have proven remarkably efficacious showing that interfering with immune signaling is clinically relevant.11-13
Molecular support for microenvironmental interactions is provided by the skewed repertoire of Ig heavy variable (IgHV) genes in CLL, suggesting that antigens and/or superantigens may be involved in CLL ontogeny.14,15 The case for antigen involvement was corroborated by the finding that patients carrying clonotypic IgHV genes with no or limited somatic hypermutation (SHM) (“unmutated CLL” [U-CLL]) experience aggressive disease, contrasting those with a heavier SHM load (“mutated CLL” [M-CLL]) who follow more indolent disease courses.16,17 The strongest molecular claim for antigen drive in CLL ontogeny stems from the fact that different patients can express highly similar if not altogether identical BCR Ig, a phenomenon at striking odds with serendipity, which is aptly coined BCR stereotypy.15,18-24 Remarkably, ∼30% of patients with CLL, both U-CLL and M-CLL, express stereotyped BCR Ig and can be classified into one of multiple, different subsets, each characterized by a distinct configuration of the BCR Ig.15
Mounting evidence suggests that the molecular classification of CLL based on BCR stereotypy is relevant. Indeed, patients belonging to the same stereotyped subset may display subset-biased clinicobiological profiles, distinct from those characterizing other subsets, even when the leukemic clones bear BCR Ig of similar SHM status.22,25-29 Therefore, one might argue that different processes initiated from the binding of distinct stereotyped BCR Ig to their cognate antigen(s) are implicated both in modulating the natural history of the disease, and, eventually, in determining clinical outcome.
A paradigmatic example concerns subset #8, which is defined by the expression of stereotyped, unmutated IgHV4-39/IgKV1(D)-39 BCR Ig of the γ isotype, a rarity for CLL,30 and a distinctive profile of genetic aberrations (ie, high frequency of trisomy 12 and NOTCH1 mutations).19,26,31,32 Remarkably, subset #8 displays a very high risk for Richter transformation19,26 that seems to be dissociated from other features of aggressive disease. Indeed, a much lower risk of transformation has been documented for other clinically aggressive subsets, including subsets #1 (clan I IgHV genes/IgKV1[D]-39, U-CLL) and #2 (IgHV3-21/IgVL3-21), which also exhibits a very different genetic background, alluding to different pathomechanisms of aggressiveness.19,21,22,26,29,33-36
In order to obtain biological insight into the noted propensity for transformation of CLL subset #8, we profiled the antigen reactivity and functional outcomes of immune stimulation in subset #8 vs subsets #1 and #2. We report that (auto)antigenic reactivity in subset #8 CLL is pronounced even in comparison with aggressive CLL subsets #1 and #2. Furthermore, we offer evidence that promiscuous antigen-binding activity can lead to significant CLL cell activation, potentially leading to progressive selection of the more aggressive clonal variants that may underlie the increased propensity for Richter transformation.
Materials and methods
Patient samples
Peripheral blood samples were collected from CLL patients belonging to clinically aggressive stereotyped subsets #1, #2, and #8 (see supplemental Tables 1 and 2 and supplemental Figure 1 available on the Blood Web site), as well as other cases not assigned to these subsets that were used for comparison in certain experiments (supplemental Tables 1 and 2). CLL diagnosis was established according to the revised guidelines of the International Workshop of Chronic Lymphocytic Leukemia/National Cancer Institute.37 All patients were either untreated or off therapy for at least 6 months before sampling. The study was approved by the local ethics committees and conducted in accordance with the Declaration of Helsinki.
Analysis of clonotypic Ig rearrangements and expression of recombinant CLL monoclonal antibodies (mAbs)
RNA from peripheral blood mononuclear cells was reverse transcribed into complementary DNA. Polymerase chain reaction amplification and sequence analysis of Ig heavy and light gene rearrangements were performed as described.22 Molecular characteristics of the clonotypic CLL Igs are provided in supplemental Table 3.
Cloning, expression, and purification of CLL mAbs were performed as reported.38
Enzyme-linked immunosorbent assays (ELISA)
Target antigens for ELISA and commercial ELISA kits are listed in supplemental Table 4. Target antigens were used at a concentration of 5 μg/mL. CLL mAbs were used as primary antibodies at a concentration of 20 μg/mL, or substituting patient serum at 50 μg/mL in commercial ELISA kits. The analysis was performed on the BioTek EL×800 ELISA reader.
Immunohistochemical studies
CLL recombinant mAbs were used for detecting autoantigen recognition in normal human tissues and in reactive lymphoid tissues. Frozen tissue sections were fixed in cold acetone at 100% for 10 minutes at 4°C. Sections were then left until dry and subsequently covered with TBS-Tween. The tissues were blocked with 3% bovine serum albumin for 10 minutes and with goat anti-human IgG Fab fragment (Jackson ImmunoResearch) at a final concentration of 100 μg/mL for 1 hour to block any internal tissue IgG. Thereafter, incubation with CLL IgG (20 μg/mL) was performed for 1 hour at room temperature; negative controls (ie, without primary antibody [Ab]) were included. The sections were then washed with TBS-Tween and incubated with horseradish peroxidase conjugated anti-human IgG1 Ab for 30 minutes at room temperature. The slides were developed with 3,3′-diaminobenzidine substrate and counterstained in hematoxylin. Visualization was performed using a Zeiss fluorescent microscope.
CLL cell isolation
CLL cells were negatively selected immediately after blood withdrawal using a B-lymphocyte enrichment kit (RosetteSep; Stemcell Technologies). The purity of all preparations exceeded 99%, and the cells coexpressed CD19 and CD5 (Beckman Coulter Inc. Brea, CA) as checked by flow cytometry.
Signaling studies
Purified CLL cells were cultured under standard conditions at a concentration of 3 × 106 cells/mL in the presence or the absence of specific ligands for BCR, TLRs, and nucleotide-binding oligomerization domain-containing protein 2 (NOD2) (supplemental Table 4) for a different duration of time appropriate for each experiment, as reported.8,39-41 For TLR2 blockage, cells were treated with anti-TLR2 mAb (Invivogen, San Diego, CA) for 1.5 hours prior to stimulation.
Flow cytometry.
Cells stimulated through the TLRs for 16 hours were analyzed for CD25 and CD86 expression by flow cytometry as reported.8,39 Cells triggered through the BCR for 10 minutes were examined for phosphorylated extracellular signal-regulated kinase (p-ERK) levels by flow cytometry as reported.41 Induction of apoptosis upon TLR stimulation for 3 and 6 days was evaluated by double-staining with fluorescein isothiocyanate-conjugated Annexin V and propidium iodide using the Annexin V–fluorescein isothiocyanate Apoptosis Detection Kit (eBioscience) according to the manufacturer’s instructions. For all flow cytometry studies, data were acquired on a FC500 flow cytometer (Beckman Coulter). All antibodies used for flow cytometry analysis are listed in supplemental Table 4.
Western blotting.
For western blotting experiments, whole protein lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. All antibodies used for immunoblotting are listed in supplemental Table 4. Immunoreactivity was revealed by enhanced chemiluminescence reaction (Pierce, Rockford, IL) and film exposures. Densitometric analysis of specific protein bands was performed with Image Studio Lite software (LI-COR Biosciences). Results were expressed as fold change relative to control cells and determined as the ratio of the optic density (OD) of p-PLCγ2 or p-ERK1/2 to the OD of total PLCγ2 or total ERK.
Identification and purification of CLL mAb-specific antigens from breast tissue
Specific recognition of breast tissue antigens was revealed by western blot analysis using CLL mAbs as primary antibodies at 10 μg/mL and goat horseradish peroxidase conjugated anti-human IgG (Bethyl Laboratories). Antigen(s) specifically recognized by CLL mAbs were isolated by immunoprecipitation using Immunoprecipitation Kit Dynabeads Protein G (Invitrogen, Paisley, United Kingdom) according to the manufacturer’s protocol. Purified proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. The Coomassie-stained proteins were excised from the gel, destained, trypsin-digested, and analyzed by nanoflow liquid chromatography electrospray ionization multistage tandem mass spectrometry (nLC-ESI-MS/MS) as described.42 Protein identification was performed with Mascot software; the database used was UniProtKB/Swiss-Prot.43
Vimentin specificity of CLL mAbs was tested by western blot analysis of immunoprecipitated proteins from breast tissue utilizing an anti-vimentin Ab (Santa Cruz, CA).
Statistical analysis
Data were compared with the use of either the paired Student t test or the nonparametric Mann-Whitney U test. Analyses were performed by GraphPad Prism 4 software (GraphPad Software, San Diego, CA). A P value < .05 was considered as statistically significant. Time to first treatment and overall survival curves were generated using the Kaplan–Meier method as described.29
Results
Subset #8 CLL mAbs display a remarkably broad antigen reactivity profile
We profiled the antigen reactivity of mAbs obtained from 27 CLL cases belonging to clinically aggressive subsets #1 (n = 11), #2 (n = 6), and #8 (n = 10) using ELISA, against antigens representing major classes of established antigenic targets for CLL, namely (1) autoantigens; (2) molecular structures on microbial pathogens; and (3) neo-epitopes created by chemical modifications during apoptosis.
Reactivity against autoantigens.
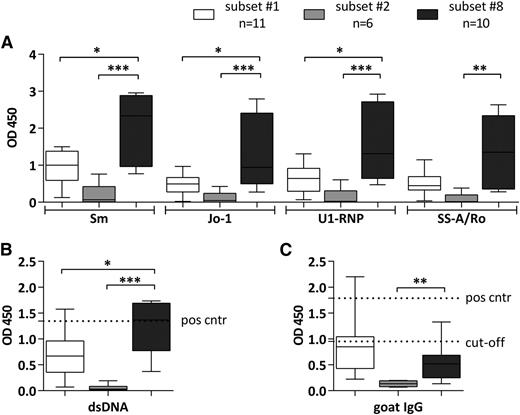
We first analyzed the binding capacity to common autoantigens associated with systemic autoimmunity (dsDNA, Sm, SS-A/Rro, U1RNP, Jo-1, and IgG-Fc) and observed that subsets #1 and #8 mAbs exhibited broad polyreactivity, as they bound to all autoantigens included in the panel. In sharp contrast, subset #2 mAbs were virtually nonreactive (Figure 1). Significantly stronger reactivity toward all tested autoantigens was found for subset #8 vs subset #1 mAbs (Figure 1A-B), except for IgG-Fc where an opposite pattern was noted (ie, subset #1 mAbs bound more strongly); however, the latter difference was not statistically significant (supplemental Table 5; Figure 1C).
Binding patterns of CLL mAbs assigned to stereotyped subsets #1, #2, and #8 to various autoantigens. Reactivity of CLL recombinant mAbs with common extractable nuclear antigens, including Sm, Jo-1, U1-RNP, and SS-A/Ro (A), goat IgG (B), and dsDNA (C). CLL mAbs were used as primary Ab for the ELISA. Samples were grouped on the basis of BCR stereotypy. The bars represent the mean OD value measured for CLL mAbs assigned to each of the stereotyped subsets #1, #2, or #8. The mAbs tested and the mean OD values of 3 replicates per sample are listed in supplemental Table 5. Rheumatoid factor activity was tested by commercial ELISA kit (C); positive and cutoff controls were provided by the kit. For anti-dsDNA activity (B), serum of a patient with systemic lupus erythematosus was used as positive control in a 1:200 dilution. *P < .05; **P < .005; ***P < .001. Cut-off, cutoff control; Pos cntr, positive control.
Binding patterns of CLL mAbs assigned to stereotyped subsets #1, #2, and #8 to various autoantigens. Reactivity of CLL recombinant mAbs with common extractable nuclear antigens, including Sm, Jo-1, U1-RNP, and SS-A/Ro (A), goat IgG (B), and dsDNA (C). CLL mAbs were used as primary Ab for the ELISA. Samples were grouped on the basis of BCR stereotypy. The bars represent the mean OD value measured for CLL mAbs assigned to each of the stereotyped subsets #1, #2, or #8. The mAbs tested and the mean OD values of 3 replicates per sample are listed in supplemental Table 5. Rheumatoid factor activity was tested by commercial ELISA kit (C); positive and cutoff controls were provided by the kit. For anti-dsDNA activity (B), serum of a patient with systemic lupus erythematosus was used as positive control in a 1:200 dilution. *P < .05; **P < .005; ***P < .001. Cut-off, cutoff control; Pos cntr, positive control.
Reactivity against microbial epitopes.
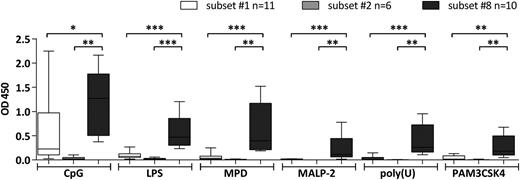
We screened subsets #1, #2, and #8 CLL mAbs against various molecular structures of microbial origin, typically also recognized by innate immunity receptors (TLRs and nod-like receptors). The antigen panel included lipopolysaccharides from Escherichia coli, lipopeptides (MALP-2 and Pam3CSK4), single stranded RNA (Poly[U]), DNA containing unmethylated cytidine phosphate guanosine (CpG) motifs commonly found in bacterial DNA, and bacterial peptidoglycan (muramyldipeptide). Overall, the subset #8 mAbs displayed a broad and strong reactivity to all tested antigens, thus contrasting both subset #1 mAbs (weakly reactive only to CpG) and subset #2 mAbs, again virtually nonreactive (Figure 2; supplemental Table 5).
Binding of CLL mAbs assigned to stereotyped subsets #1, #2, and #8 to TLR and nod-like receptor ligands. CLL mAbs were used as primary Ab for the colorimetric ELISA. Samples were grouped on the basis of BCR stereotypy. The bars represent the mean OD value as measured for CLL mAbs assigned to each of the stereotyped subsets #1, #2, or #8. The mAbs tested and the mean OD values of 3 replicates per sample are listed in supplemental Table 5. All determinations were performed in triplicate. *P < .05; **P < .005; ***P < .001. LPS, lipopolysaccharides; MPD, muramyldipeptide.
Binding of CLL mAbs assigned to stereotyped subsets #1, #2, and #8 to TLR and nod-like receptor ligands. CLL mAbs were used as primary Ab for the colorimetric ELISA. Samples were grouped on the basis of BCR stereotypy. The bars represent the mean OD value as measured for CLL mAbs assigned to each of the stereotyped subsets #1, #2, or #8. The mAbs tested and the mean OD values of 3 replicates per sample are listed in supplemental Table 5. All determinations were performed in triplicate. *P < .05; **P < .005; ***P < .001. LPS, lipopolysaccharides; MPD, muramyldipeptide.
Reactivity against neo-epitopes created by chemical modifications during apoptosis.
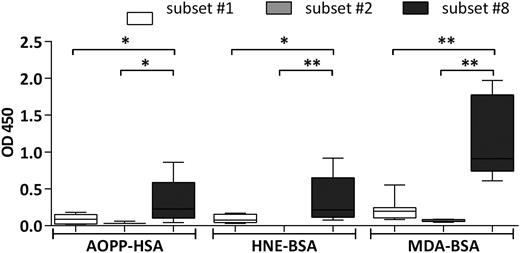
We also tested the reactivity profile of our mAb CLL library toward molecular adducts of lipid peroxidation, ie, bovine serum albumin modified by malondialdehyde (MDA) and 4-hydroxynonenal epitopes, as well as human serum albumin conjugated to Advanced Oxidation Protein Products. All these molecules were recognized by subset #8 mAbs and only to a significantly lesser degree by subset #1 mAbs; again, subset #2 mAbs were nonreactive (Figure 3; supplemental Table 5).
Binding of CLL mAbs to oxidation-induced molecular adducts and advanced oxidation protein products. CLL mAbs were used as primary Ab for the colorimetric ELISA. Samples were grouped on the basis of BCR stereotypy. The bars represent the mean OD value as measured for CLL mAbs assigned to each of stereotyped subsets #1, #2, or #8. The mAbs tested, as well as the mean OD values of 3 replicates per sample are listed in supplemental Table 5. All determinations were performed in triplicate. *P < .05, **P < .005. AOPP-HSA, human serum albumin conjugated to advanced oxidation protein products; HNE-BSA, BSA modified with 4-hydroxynonenal.
Binding of CLL mAbs to oxidation-induced molecular adducts and advanced oxidation protein products. CLL mAbs were used as primary Ab for the colorimetric ELISA. Samples were grouped on the basis of BCR stereotypy. The bars represent the mean OD value as measured for CLL mAbs assigned to each of stereotyped subsets #1, #2, or #8. The mAbs tested, as well as the mean OD values of 3 replicates per sample are listed in supplemental Table 5. All determinations were performed in triplicate. *P < .05, **P < .005. AOPP-HSA, human serum albumin conjugated to advanced oxidation protein products; HNE-BSA, BSA modified with 4-hydroxynonenal.
CLL mAbs bind normal human epithelia.
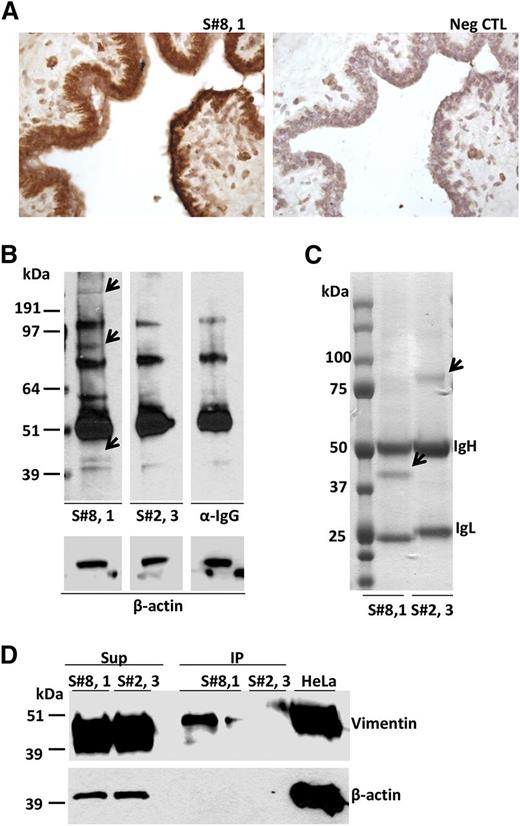
Immunohistochemical analysis revealed strong binding to normal breast and kidney tissues for 4/10 subset #8 mAbs and 2/5 subset #1 mAbs (Figure 4A). In contrast, none of the 4 tested subset #2 mAbs bound to antigenic elements on these tissues, although extracellular positivity in the lumen of some glands was observed (supplemental Table 6).
Subset #8 CLL BCR binds to autoantigens, including vimentin, on breast epithelial cells. (A) CLL 657 (S#8,1) IgG binding to breast epithelium. (B) SDS-PAGE electrophoresis of breast tissue protein extracts probed with CLL mAbs, CLL657 (S#8,1), and P326 (S#2,3), or with secondary Ab only (anti-human IgG). The arrows indicate breast tissue proteins specifically recognized by subset #8 CLL657 (S#8,1) mAb. The subset #2 P326 (S#2,3) mAb did not exhibit differential protein recognition relative to control (α-IgG). (C) CLL657 (S#8,1) and P326 (S#2,3) CLL mAbs were used to immunoprecipitate proteins from breast tissue. The immunoprecipitated proteins were resolved by SDS-PAGE electrophoresis, and thereafter the gel was subjected to Coomassie blue staining. The arrows indicate differentially immunoprecipitated proteins between CLL657 (S#8,1) and P326 (S#2,3) mAbs that were subjected to nLC-ESI-MS/MS analysis (supplemental Table 7). (D) IP and Sup samples from breast tissue extracts using CLL657 (S#8,1) and P326 (S#2,3) CLL mAbs were electrophoresed in 10% SDS-PAGE along with lysate of HeLa cells, and subsequently immunoprobed with anti-vimentin and anti–β-actin antibodies. Vimentin was recognized and immunoprecipitated by the subset #8 CLL657 (S#8,1) mAb. IP, immunoprecipitate; Sup, supernatant; Neg CTL, negative control.
Subset #8 CLL BCR binds to autoantigens, including vimentin, on breast epithelial cells. (A) CLL 657 (S#8,1) IgG binding to breast epithelium. (B) SDS-PAGE electrophoresis of breast tissue protein extracts probed with CLL mAbs, CLL657 (S#8,1), and P326 (S#2,3), or with secondary Ab only (anti-human IgG). The arrows indicate breast tissue proteins specifically recognized by subset #8 CLL657 (S#8,1) mAb. The subset #2 P326 (S#2,3) mAb did not exhibit differential protein recognition relative to control (α-IgG). (C) CLL657 (S#8,1) and P326 (S#2,3) CLL mAbs were used to immunoprecipitate proteins from breast tissue. The immunoprecipitated proteins were resolved by SDS-PAGE electrophoresis, and thereafter the gel was subjected to Coomassie blue staining. The arrows indicate differentially immunoprecipitated proteins between CLL657 (S#8,1) and P326 (S#2,3) mAbs that were subjected to nLC-ESI-MS/MS analysis (supplemental Table 7). (D) IP and Sup samples from breast tissue extracts using CLL657 (S#8,1) and P326 (S#2,3) CLL mAbs were electrophoresed in 10% SDS-PAGE along with lysate of HeLa cells, and subsequently immunoprobed with anti-vimentin and anti–β-actin antibodies. Vimentin was recognized and immunoprecipitated by the subset #8 CLL657 (S#8,1) mAb. IP, immunoprecipitate; Sup, supernatant; Neg CTL, negative control.
Interestingly, CLL mAbs of all 3 subsets did not recognize any antigen exposed on reactive human lymphoid tissues (supplemental Table 7; supplemental Figure 2).
Vimentin identified as autoantigenic target of a CLL subset #8 mAb
The mAb from subset #8 CLL case 657 that exhibited strong binding to epithelial breast cells was analyzed in detail by western blotting, immunoprecipitation, and nLC-ESI-MS/MS. Using this mAb for probing a protein lysate from the same breast tissue, 3 bands of approximately 45, 90, and 250 kDa MW were detected that were not evident when using either a subset #2 mAb or the secondary Ab (anti-human IgG) alone and, thus, deemed specific (Figure 4B). After immunoprecipitation of a breast tissue lysate with subset #8 CLL mAb CLL657, only the 45 kDa protein band was visible and its identity was investigated by nLC-ESI-MS/MS (Figure 4C). Besides Ig molecules of the expected mAb-specific heavy and light chain isotype (γ/κ), the major non-Ig protein in the 45 kDa band was vimentin. For comparison, using a subset #2 CLL mAb (P326) in immunoprecipitation material from breast tissue, only a 75 kDa band was visible (Figure 4C), generating peptide signals that matched only Ig molecules of the expected mAb-specific heavy and light chain isotype (γ/λ) (supplemental Table 8).
To confirm the nLC-ESI-MS/MS results, proteins immunoprecipitated using the same subsets #2 and #8 mAbs were immunoblotted and probed with antibodies specific for human vimentin. The immunoprecipitate by the subset #8 mAb was recognized by anti-vimentin antibodies, whereas the one obtained by subset #2 was not recognized, as expected. As control, vimentin was found to be abundant in the nonimmunoprecipitated material of all cases (Figure 4D). Altogether, these results strongly suggest that vimentin is the principal protein immunoprecipitated by the CLL subset #8 mAb.
CLL subset #8 cells respond avidly to triggering through adaptive and innate immunity receptors
BCR triggering.
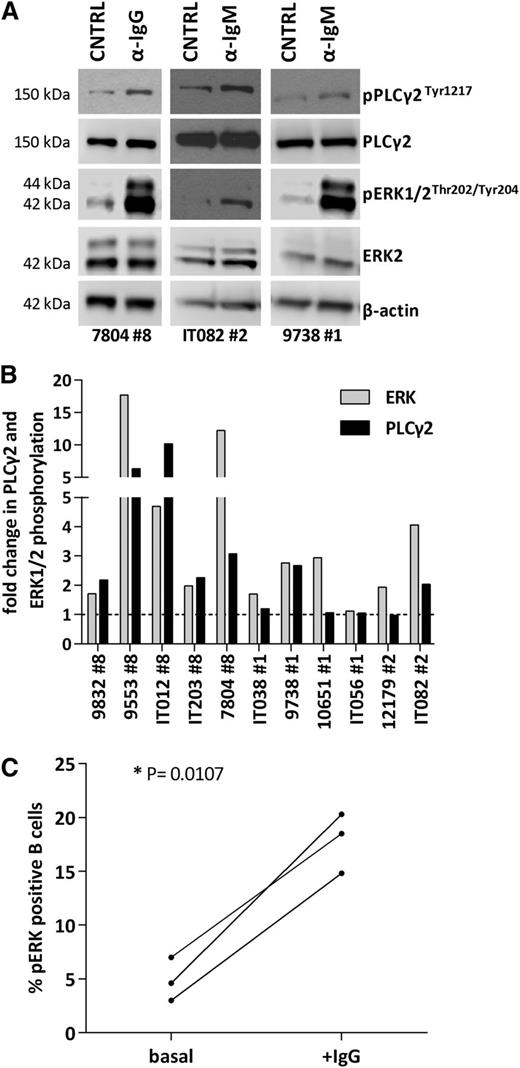
Primary CLL cells from stereotyped subsets #1 (n = 4), #2 (n = 2), and #8 (n = 5) cases were tested for their ability to transmit signals through the BCR. To this end, negatively isolated CD19+ CLL cells were stimulated with anti-Ig (anti-IgM for subsets #1 and #2 clones that express BCR Ig of the M/D isotype or anti-IgG for subset #8 clones that express BCR Ig of the G isotype). In all 5 subset #8 cases analyzed, Ig crosslinking consistently and strongly increased phosphorylation of PLCγ2 and ERK1/2. Τhe responses for subsets #1 and #2 were much more heterogeneous regarding p-PLCγ2 and p-ERK1/2 induction (p-PLCγ2: 1/4, 1/2; p-ERK1/2: 3/4, 2/2 in subsets #1 and #2 cases, respectively) (Figure 5A-B; supplemental Table 9). Furthermore, the responses obtained in subset #8 do not seem to be attributed to the expression of the G isotype as indicated by the finding that other IgG expressing CLL clones, unrelated to subset #8, did not display a similar signaling capacity (supplemental Figures 3 and 4).
Profiling of BCR signaling cascade in stereotyped subsets #1, #2, and #8. (A) Immunoblotting analysis of 3 representative responsive cases from stereotyped subsets #1, #2, and #8, in which BCR crosslinking increases p-PLCγ2 and p-ERK levels. (B) Densitometric analysis of p-ERK/ERK and p-PLCγ2/PLCγ2 levels in stereotyped subsets #1, #2, and #8 CLL samples after BCR crosslinking; the increase in p-PLCγ2 and p-ERK levels is more pronounced in subset #8 CLL cells. (C) Untreated subset #8 CLL cells exhibit low basal levels of p-ERK1/2 that increase after BCR engagement in vitro (+IgG), further documenting signaling capacity. *P < .05. CNTRL, unstimulated control.
Profiling of BCR signaling cascade in stereotyped subsets #1, #2, and #8. (A) Immunoblotting analysis of 3 representative responsive cases from stereotyped subsets #1, #2, and #8, in which BCR crosslinking increases p-PLCγ2 and p-ERK levels. (B) Densitometric analysis of p-ERK/ERK and p-PLCγ2/PLCγ2 levels in stereotyped subsets #1, #2, and #8 CLL samples after BCR crosslinking; the increase in p-PLCγ2 and p-ERK levels is more pronounced in subset #8 CLL cells. (C) Untreated subset #8 CLL cells exhibit low basal levels of p-ERK1/2 that increase after BCR engagement in vitro (+IgG), further documenting signaling capacity. *P < .05. CNTRL, unstimulated control.
In order to further probe the BCR signaling capacity of CLL subset #8, we determined by flow cytometry the levels of constitutive phosphorylation of pERK1/2, a biochemical feature of BCR-unresponsive (anergized) cells.40 All 3 tested cases displayed very low levels of basal phosphorylated ERK1/2 that increased upon BCR triggering with anti-Ig antibodies, indicating that they are well equipped and capable of transmitting signals through the BCR (Figure 5C).
TLR triggering.
Preliminary results from our group have suggested that subset #8 CLL cells respond intensely to TLR1/2, 2/6, 7, and 9 stimulation, often to a significantly greater extent than other U-CLL cases, including subset #1.8 Here, we confirm and extend these initial observations reporting upregulation of co-stimulatory molecules CD25 and CD86 upon TLR1/2, 2/6, 7, and 9 and NOD2 stimulation in all 4 subset #8 cases evaluated. In 2 of these cases, we also investigated the effects of TLR and NOD2 triggering on apoptosis and found that stimulation of TLR1/2, TLR2/6, TLR7, and NOD2 resulted in protection from apoptosis after 3 and 6 days, whereas stimulation of TLR9 had opposite effects (supplemental Figure 5). Altogether, these results support that TLR signaling is also functional in CLL subset #8 cells.
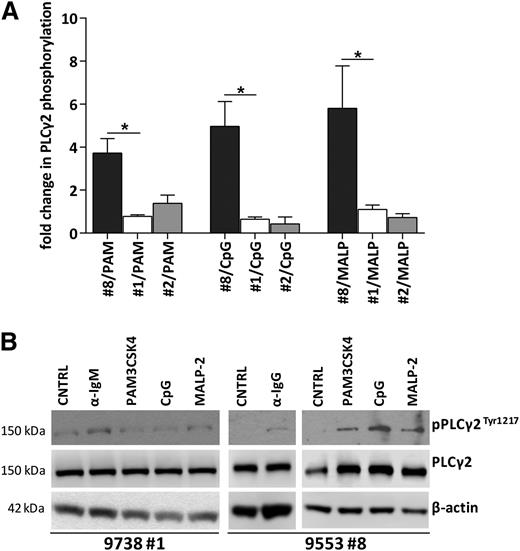
In order to further explore cell activation by TLR ligands in CLL cells, we set up short-term cultures of CLL cells stimulated by TLR ligands (PAM3CSK4, CpG ODN, and MALP-2). Western blot analysis revealed that CLL cells from subsets #1 (n = 4) and #2 (n = 2), as well as other CLL cases not assigned to subsets #1, #2, or #8 (n = 4), did not upregulate phospho-PLCγ2 upon TLR triggering. Intriguingly, however, CLL cells from all 5 subset #8 cases exposed to PAM3CSK4 (TLR1/2), CpG (TLR9), and MALP-2 (TLR2/6) displayed upregulation of phopsho-PLCγ2 levels compared with the unstimulated control (Figure 6A-B; supplemental Figures 6 and 7; supplemental Table 9).
TLR ligands increase PLCγ2 phosphorylation in CLL subset #8 CLL but not in subsets #1 or #2. (A) Densitometric analysis of p-PLCγ2/PLCγ2 normalized to the unstimulated control, in CLL samples after stimulation with TLR ligands: PAM3CSK4, CpG ODN 2006, and MALP-2. The graphs represent the mean values in subsets #8, #1, and #2 CLL cases (5, 4, and 2 cases, respectively).*P < .05. (B) Representative western blot of a subset #8 case displaying increased PLCγ2 phosphorylation after stimulation with anti-IgG Ab and TLR ligands PAM3CSK4, CpG ODN 2006, and MALP-2, contrasting a nonresponding subset #1 case.
TLR ligands increase PLCγ2 phosphorylation in CLL subset #8 CLL but not in subsets #1 or #2. (A) Densitometric analysis of p-PLCγ2/PLCγ2 normalized to the unstimulated control, in CLL samples after stimulation with TLR ligands: PAM3CSK4, CpG ODN 2006, and MALP-2. The graphs represent the mean values in subsets #8, #1, and #2 CLL cases (5, 4, and 2 cases, respectively).*P < .05. (B) Representative western blot of a subset #8 case displaying increased PLCγ2 phosphorylation after stimulation with anti-IgG Ab and TLR ligands PAM3CSK4, CpG ODN 2006, and MALP-2, contrasting a nonresponding subset #1 case.
Discussion
Independent studies have revealed that CLL BCR Igs may recognize and bind foreign antigens44-47 as well as self-antigens, in particular exposed in the context of apoptotic bodies produced during physiological cell turnover.46,48,49 Although monospecific, affinity-selected mAbs have been described, mainly deriving from M-CLL,44,50 poly-specificity is a common feature of CLL mAbs, especially from U-CLL cases, which exhibit binding reactivities similar to those of natural antibodies.51,52 Interestingly, poly/autoreactivity may constitute a determinant of clinical aggressiveness.53,54
In the current study, we investigated the antigen-reactivity profile and functional relevance of antigen binding in stereotyped subset #8, a paradigmatic CLL subgroup displaying aggressive clinical behavior and the highest reported risk for Richter syndrome among all CLL.19,26 Critically, subset #8 profiles were compared with those obtained in two other well characterized and also clinically aggressive CLL subsets, namely subsets #1 and #2, with the main aim of identifying potential causes into the unique propensity of subset #8 clones to transform that seems to be dissociated from other features of aggressive disease.
Overall, we demonstrate that subset #8 CLL mAbs exhibit promiscuous antigen reactivity. Indeed, they bind strongly to representatives of the major classes of established antigenic targets for CLL, namely molecular structures on microbial pathogens, autoantigens, and neo-epitopes created by chemical modifications during apoptosis, thus sharply contrasting subset #1 and, to an even greater extent, subset #2 mAbs. More importantly, subset #8 CLL cells responded to stimulation with these antigenic elements, alluding to the functional significance of the observed mAb-antigen interactions.
The recognition by CLL mAbs of antigenic particles exposed or created during apoptosis is well documented.46,52 However, not all CLL mAbs can bind such autoantigens and reactivity likely depends on the specific BCR configuration. This is corroborated by the results of the present study, showing that among subsets #1, #2, and #8 mAbs, only the latter displayed strong binding of autoantigens and oxidation-associated epitopes, whereas the other mAbs bound to a much lesser extent (subset #1) or if at all (subset #2). In particular, subset #8 mAbs showed prominent reactivity against MDA, which is the principal product of polyunsaturated fatty acid peroxidation. MDA, along with phosphorylcholine, another previously identified target of subset #8 CLL mAbs,52 are dominant determinants of in vivo murine immune responses to apoptotic cells.55 In this context, it is relevant to mention our previous findings that subset #8 mAbs (1) recognize apoptotic Ramos B cells and apoptotic Jurkat T cells; and (2) exhibit the strongest binding among all tested CLL mAbs, including those derived from subsets #1 and #2 to a class of apoptotic cells with exposed nonmuscle myosin heavy chain IIA (ie, myosin-exposed apoptotic cells).49,52
Antibodies recognizing autoantigens and oxidation-specific epitopes on self-antigens have been shown to cross-react with microbial antigens.56-64 With this in mind, we also screened our CLL mAb library against various microbial particles that are typically recognized by innate immunity receptors and found that subset #8 mAbs indeed bound to all such antigens, including lipopolysaccharides, lipopeptides, peptidoglycan, ssRNA, and unmethylated DNA.
Both apoptosis or infection can make epitopes accessible in peripheral tissues that may deliver activating stimuli through the BCR to subset #8 CLL cells. Notably, immunohistochemical analysis unveiled strong binding toward certain normal human epithelia for antibodies belonging to subsets #1 and #8. Immunoprecipitation experiments revealed that one of the principal antigens recognized by a subset #8 mAb (CLL657) was vimentin, that we and others have previously established as an antigenic target of CLL mAbs, including those from subsets #1 and #8.46,65,66 Interestingly, vimentin is also recognized by the BCR Igs of various other B-cell malignancies, including follicular lymphoma and mantle cell lymphoma, and can be expressed in lymphoma tissues, thus being available for binding to the lymphoma BCR Ig directly within the tumor microenvironment.67
To investigate the functional consequences of antigen binding, we assessed BCR signaling capacity in CLL cells from subsets #1, #2, and #8 by determining the phosphorylation status of PLCγ2 and ERK, both shown to have a critical role in BCR signal transduction.68,69 In particular, PLCγ2 and ERK phosphorylation is induced in CLL cells by in vitro crosslinking of the BCR with anti-IgM, especially in poor prognostic CLL.6,40,70 Furthermore, ERK phosphorylation has also been detected in vivo within the proliferative centers of CLL, sites of CLL cell proliferation, and encounter with antigens.71
We found significant qualitative differences between the 3 stereotyped subsets regarding the outcomes of BCR stimulation. In general, stronger responses were elicited by antigen challenge in subset #8 vs either subsets #1 or #2; this pattern was more pronounced for PLCγ2, where a significant increase in phosphorylation was seen exclusively in subset #8. These differences cannot be attributed to the different isotype expressed by subset #8 (IgG vs IgM in subsets #1 and #2), as other non-subset IgG+ cases did not display a similar signaling capacity. This finding suggests that aggressive CLL clones do not display equal BCR signaling capacity, even when the SHM status is similar, which is the case for subsets #1 and #8. In clear contrast, the indolent subset #4 is characterized by higher basal levels of PLCγ2 phosphorylation that cannot be further augmented upon stimulation, in line with the known biochemical signature of anergy, with constitutive activation of the pathways downstream the BCR.40 Furthermore, it underscores the relevance of BCR Ig stereotypy which offers a refined, more compartmentalized view into CLL biology, superseding distinction into the rather crude U-CLL and M-CLL categories and allowing an additional dissection at the clinical level, even among aggressive cases.
Subset #8 CLL cells exhibited strong functional responses to stimulation with TLR ligands, which was not the case with subsets #1 or #2 cells. Because most of the autoantigens recognized by subset #8 mAbs (eg, dsDNA and U-rich ribonucleoproteins) can also activate TLRs,72 synergism between BCRs and TLRs, a well documented mechanism both in normal and in autoreactive cells,73-78 may be relevant for the pathogenesis and evolution of subset #8 CLL as well, further supporting the parallelism between autoimmunity and CLL at least in a portion of cases. In the future, elucidation of the crystal structure of these Igs can shed light on the structural specificity that may underlie such functional behavior.
The role of classic (auto)antigen binding in CLL ontogeny was recently challenged by a report that CLL BCRs possess the unique capacity to activate antigen-independent signaling through BCR autorecogition.79 However, autonomous signaling was observed in all CLL cases, independently of SHM status, suggesting that BCR autorecognition alone cannot determine the clinical outcome, since in general, U-CLL cases have very different outcomes from M-CLL cases, while, even within U-CLL, the clinical course may differ depending on the expression of a particular BCR Ig, as exemplified by subsets #1 vs #8. Possibly, basal autonomous signaling could be generated in all CLL cases leading to enhanced survival, whereas signaling due to classical (auto)antigen recognition could lead to proliferation and expansion of the leukemic clone.80 The actual contribution of these two types of signaling should be further investigated, perhaps within the concept of stereotyped CLL subsets that allow to study more homogeneous groups of cases, thus overcoming the incapacitating heterogeneity characterizing more generic CLL cohorts.
Altogether, our results support the notion that the noted clinical aggressiveness of CLL subset #8 is linked to its promiscuous antigen reactivity and pronounced signaling capacity, indicating addiction to microenvironmental stimuli. That notwithstanding, genetic aberrations may also contribute significantly to the evolution and, in certain cases, transformation of subset #8 CLL clones to Richter syndrome. Indeed, subset #8 displays a distinctive profile of genomic aberrations, being enriched for trisomy 12, t(14;19) and NOTCH1 mutations.26,31,32,81 Therefore, it is plausible that an unlimited capacity to respond to an extensive range of microenvironmental stimuli, through the surface BCR Ig and the TLRs, may elicit unabated stimulation throughout the natural history of subset #8 patients resulting in enhanced survival and expansion of the malignant clone. How this relates to the distinctive genetic profile or conversely may facilitate the accumulation of genetic aberrations in subset #8 remains to be elucidated. This will ultimately help to understand how the progressive selection of the most aggressive clones occurs, likely influencing the clinical outcome of these patients, which is also relevant in view of BCR signaling inhibition therapy for CLL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported in part by grants from the Associazione Italiana per la Ricerca sul Cancro AIRC (Investigator Grant and Special Program Molecular Clinical Oncology, 5 per mille #9965), Milano, Italy; Ricerca Finalizzata 2010, Ministero della Salute, Roma; the ENosAI project (code 09SYN-13-880); co-funded by the EU and the General Secretariat for Research and Technology of Greece; the KRIPIS action; the General Secretariat for Research and Technology of Greece; the European Regional Development Fund of the EU under the O.P. Competitiveness and Entrepreneurship (NSRF 2007-2013); and the National Institutes of Health National Cancer Institute (RO1 CA081554).
Authorship
Contribution: M.G. performed research, analyzed the data, and wrote the manuscript; S.N., B.A., and N.P. performed research and analyzed the data; M.P. performed research, analyzed the data, supervised research, and assisted in writing the manuscript; C.C.C., D.R., G.G., and N.C. provided samples, supervised research, and assisted in writing the manuscript; and K.S. and P.G. designed the study, supervised research, and wrote the manuscript.
Conflict-of-interest disclosure: K.S. received research support from Roche SA and Janssen Pharmaceuticals. P.G. received research funding from Roche SA and GlaxoSmithKline. The remaining authors declare no competing financial interests.
Correspondence: Paolo Ghia, IRCCS Istituto Scientifico San Raffaele and Department of Onco-Hematology, Via Olgettina 58, Milan, MI 20132, Italy; e-mail: ghia.paolo@hsr.it; Kostas Stamatopoulos, Institute of Applied Biosciences, Center for Research and Technology Hellas, 57001 Thermi, Thessaloniki, Greece; e-mail: kostas.stamatopoulos@gmail.com.
References
Author notes
K.S. and P.G. contributed equally to this study as senior authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal