Key Points
Large-scale loss-of-function RNAi screens in patient-derived AML cells are feasible and able to pinpoint therapeutic targets.
ROCK1 inhibition exerts antileukemic effects in primary human AML cells in vitro and in vivo.
Abstract
Acute myeloid leukemia (AML) is characterized by a marked genetic heterogeneity, which complicates the development of novel therapeutics. The delineation of pathways essential within an individual patient’s mutational background might overcome this limitation and facilitate personalized treatment. We report the results of a large-scale lentiviral loss-of-function RNA interference (RNAi) screen in primary leukemic cells. Stringent validation identified 6 genes (BNIPL1, ROCK1, RPS13, STK3, SNX27, WDHD1) whose knockdown impaired growth and viability of the cells. Dependence on these genes was not caused by mutation or overexpression, and although some of the candidates seemed to be rather patient specific, others were essential in cells isolated from other AML patients. In addition to the phenotype observed after ROCK1 knockdown, treatment with the approved ROCK inhibitor fasudil resulted in increased apoptosis and decreased viability of primary AML cells. In contrast to observations in some other malignancies, ROCK1 inhibition did not foster growth of immature malignant progenitors but was toxic to this cell fraction in feeder coculture and xenotransplant experiments, indicating a distinct effect of ROCK1 inhibition on leukemic progenitors. We conclude that large-scale RNAi screens in primary patient–derived cells are feasible and can complement other methods for personalized cancer therapies, such as expression and mutation profiling.
Introduction
Adult acute myeloid leukemia (AML), unlike other hematologic malignancies, is still associated with a dismal prognosis.1 Although patients with chronic myeloid leukemia (CML) can be treated very efficiently in the era of targeted therapy,2 there have not been similar improvements in AML so far. Even though anthracycline dose escalation3 and optimization of stem cell transplant4 resulted in modest improvements for subsets of patients, efforts to change the classic “3+7” daunorubicin/cytarabine induction regimen have largely failed.5 Although mutations in receptor tyrosine kinases are frequently found in AML, kinase inhibition, at least with multikinase inhibitors, has not resulted in a CML-like success.6 Although more selective inhibitors such as quizartinib may represent a significant progress in this regard,7 the difficulties in substantially improving treatment probably relate to greater genetic heterogeneity inherent to leukemic blasts in AML. In contrast to CML, tyrosine kinase mutations in AML are usually not key leukemogenic drivers but rather one of several mutations contributing to the malignant phenotype.8 Recent whole-genome sequencing approaches have estimated that an AML sample contains, on average, 13 mutations in expressed regions of the genome, of which 3 to 5 are recurrent.9 Pharmacologic targeting of one of these aberrations might produce differential results, depending on the patient specific-mutational background. Therefore, novel personalized forms of treatment focusing on individual vulnerabilities of a given leukemia may be necessary to improve the prognosis of AML patients.
RNA interference (RNAi) screens may be a tool to delineate such individual cancer cell–specific susceptibilities.10,11 Lentiviral-packed short hairpin (sh)RNAs represent an effective format for RNAi screens because they allow for stable protein knockdown in a wide variety of dividing and nondividing cell types.12 In addition, pooling of different shRNAs in large-scale libraries facilitates the assessment of hundreds of genes in a single experiment. Whereas shRNAs beneficial to cell survival and proliferation enrich over time within the pool, shRNAs targeting vital pathways are negatively selected. Focusing on these depleted shRNAs allows for the identification of novel therapeutic targets.
Indeed, as exemplified by Brd4 and Wee1, the results of such screens have already been implemented in clinical trials in leukemia.13-15 However, most of the screens published thus far relied on established cell lines adapted for growth in culture. The generation of stable cell lines from each individual patient is not practical and, with the adaption to growth in culture, the cells are unlikely to closely reflect the original cells isolated from the patient. A personalized treatment approach would therefore necessitate the transfer of RNAi screening technology to primary leukemic cells derived directly from the patient.
Using primary leukemic cells from an AML patient, we established conditions for a pooled in vitro loss-of-function shRNA screen. We demonstrate that this approach can delineate patient-specific genes required for growth and viability of the AML cells that would not have been nominated by other methods. More specifically, we identify ROCK1 as a crucial gene for the leukemic bulk and stem/progenitor cells and demonstrate that pharmacologic inhibition of ROCK1 exerts antileukemic effects in vitro and in vivo. Our results show that large-scale RNAi screens in primary AML cells are feasible and able to pinpoint novel therapeutic targets. We propose that these results set the stage for further efforts to develop RNAi technology as a guide to personalized treatment in AML and possibly other cancers.
Materials and methods
Primary cells and cell lines
Primary cells from AML patients and CD34-selected (MACS MicroBeads; Miltenyi Biotec) hematopoietic stem/progenitor cells (HSPCs) derived from healthy donors underwent controlled-rate freezing and were subsequently stored in the vapor phase of liquid nitrogen. Freshly thawed B- and T-lymphocyte–depleted (MACS MircoBeads; Miltenyi Biotec) leukemic or normal HSPCs were used for experiments. All primary cells were grown in StemSpan SFEM with 2% fetal calf serum (both STEMCELL Technologies) supplemented with rhIL3, rhTPO, rhFLT3-ligand, and rhSCF (all R&D Systems). Leukemic cell preparations contained <0.5% lymphocytes, whereas normal HSCs were >95% CD34+.
All primary cells were obtained under protocols approved by the local institutional review board, and all donors gave written informed consent for the use of their cells for research purposes.
Lentiviral loss-of-function screen
For the lentiviral screen, we transduced 12 × 107 cells from the index patient (AML2) overnight with subpool 1 of The RNAi Consortium Library16 (Sigma-Aldrich) at a multiplicity of infection (MOI) of 1.5 using RetroNectin-coated plates (Takara Bio). Transduction rate was assessed as the percentage of puromycin-resistant cells using quantitative flow cytometry (MACSQuant; Miltenyi Biotec). After 9 days, cells were harvested for DNA extraction (DNeasy kit; Qiagen). The barcodes contained within the proviral inserts were amplified and appended with adapter sequences for next-generation sequencing (NGS) using a 2-step polymerase chain reaction (PCR) approach (for details, see http://www.broadinstitute.org/rnai/public/ and supplemental Table 1, which is available on the Blood Web site).
NGS
All NGS was done on an Illumina HiSequation 2000. For shRNA barcode analysis, purified PCR products were single-end sequenced in packages of 4 samples per lane. Transcriptome analysis was carried out by single-end direct sequencing using total RNA extracted from 10 × 106 cells (RNeasy Mini Kit; Qiagen). Exome sequencing was carried out with 50 ng of genomic DNA, enriched for exonic sequences (Nextera Rapid Capture Exome Kit; Illumina) and paired-end sequencing. Raw sequences will be deposited in the Gene Expression Omnibus database.
Validation of candidate genes
For validation experiments, 293T cells were cotransfected with pLKO.1-puro (MISSION TRC1 Human Glycerol Library; Sigma-Aldrich), psPAX2, and VSV-G plasmids (Addgene), and lentiviral particles were harvested 72 hours later. Primary cells were transduced at MOIs of 2.5 to 5, puromycin selected, and grown for 2 weeks in suspension culture. Validated shRNAs were required to reduce the count of transduced cells to <50% as assessed by quantitative flow cytometry, whereas nontarget shRNA–transduced cells grown in parallel had to increase to >100% of the initial count.
Knockdown efficacies were determined by quantitative reverse-transcription PCR and, for ROCK1, also by western blot analysis (Clone 46/ROCK-I; BD Biosciences).
Apoptosis was quantified with the help of the Annexin V Apoptosis Detection Kit (BD Biosciences), and cell cycle distribution was analyzed by staining ethanol-fixed cells with propidium iodide.
Pharmacologic ROCK1 inhibition and long-term culture–initiating cell assay
Fasudil hydrochloride and Y-27632 (Selleck Chemicals) were used to inhibit ROCK1 pharmacologically. Cell counts in suspension culture were assessed by quantitative flow cytometry. ROCK activity was quantified using an enzyme immunoassay (ROCK Activity Assay; EMDMillipore) according to the manufacturer’s instructions.
For long-term culture–initiating cell assay (LTC-IC), bulk leukemic cells were seeded in MyeloCult H5100 (STEMCELL Technologies) on irradiated M2-10B4 murine bone marrow stromal cells (MBSCs). After 4 to 5 weeks, cobblestone areas were manually counted by 2 independent investigators using a Celigo cytometer (Brooks). Fasudil (final concentration 40 μM) or water was added, and plates were reanalyzed 4 days later. Cobblestone areas were defined as any area of ≥5 hematopoietic cells growing immediately adjacent to each other and having a rectangular shape.
Xenotransplant experiments
A total of 2 × 106 primary AML cells were intravenously injected into 4- to 6-week-old female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory). Five or 10 weeks after transplant, animals were either treated with fasudil (50 μg/g/d subcutaneously via osmotic minipumps; Alzet Osmotic Pumps) or control treated. After 2 weeks, animals were euthanized, and murine bone marrow (BM) and or peripheral blood composition was analyzed by flow cytometry using antibodies against murine CD45 (eBiosciences) and human CD45, CD33, CD2 or CD3, and CD19 (all BD Biosciences). Human chimerism was calculated by dividing human CD45-positive cells by the total number of CD45-positive cells (murine and human). All animal work was approved by the relevant authorities (Landesdirektion Sachsen, AZ 24-9168.11-9/2010-1).
Bioinformatics and statistics
Reads obtained by NGS were trimmed on both ends to the final size of 46 bp and aligned to the library of all possible amplicons containing shRNA sequences using Bowtie (v.1.0).17 For each read, all potential alignments were reported and only the shRNA sequence with the least amount of mismatches was used for the final read annotation. Ambiguous reads (ie, mapping to >1 shRNA with the same number of mismatches) were discarded.
To normalize samples for differences in total read counts and possible amplification biases, we performed a loess regression of shRNAs fitting log2-transformed counts from day 9 to the day 0 sample. Residuals of the regression were considered estimates of shRNA abundance fold change and converted into modified z scores, where median and median absolute deviation were used as measures of a central tendency and an amount of variation in the sample. All calculations were performed using functions implemented in R, v.3.0.
Results
Large-scale shRNA screen in primary AML cells identifies essential candidate genes
To establish the conditions for a primary cell–based shRNA screen in AML, we obtained leukemic blasts (blast percentage 94%) from a 67-year-old female patient diagnosed with de-novo AML of French-American-British subtype M1 (see supplemental Materials and supplemental Table 2 for details). Cells from the initial diagnosis and prior to the initiation of any chemotherapy were used for all experiments outlined in Figure 1A.
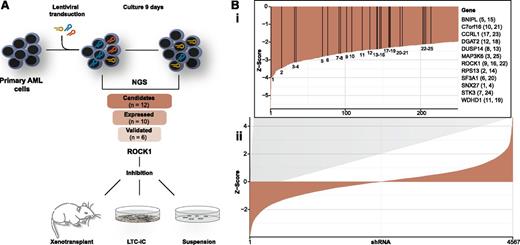
Experiment outline and results of the shRNA screen. (A) An overview of the experimental strategy. shRNAs targeting different genes are exemplified in brown, red, and blue. (B) Waterfall plot (ii) showing all shRNAs recovered with >100 reads in the day 0 sample. The y-axis depicts the z score for the comparison of the individual shRNA abundance between day 0 and day 9. Negative z scores refer to depleted genes; positive z scores refer to enriched genes. A zoomed-in version of the negative z scores (i) depicts the shRNAs targeting the 12 candidate genes. Vertical lines refer to the position of the individual shRNAs. Corresponding gene abbreviations are given to the right of the plot.
Experiment outline and results of the shRNA screen. (A) An overview of the experimental strategy. shRNAs targeting different genes are exemplified in brown, red, and blue. (B) Waterfall plot (ii) showing all shRNAs recovered with >100 reads in the day 0 sample. The y-axis depicts the z score for the comparison of the individual shRNA abundance between day 0 and day 9. Negative z scores refer to depleted genes; positive z scores refer to enriched genes. A zoomed-in version of the negative z scores (i) depicts the shRNAs targeting the 12 candidate genes. Vertical lines refer to the position of the individual shRNAs. Corresponding gene abbreviations are given to the right of the plot.
A total of 12 × 107 B cell– and T cell–depleted primary leukemic blasts were transduced with the library containing 7709 shRNAs (on average, 5 per gene) at an MOI of 1.5. The transduction rate was kept <25%, limiting the likelihood of multiple infections of a single cell. The posttransduction sample (day 0 sample) recovered 95.8% of the 7709 shRNAs, demonstrating near comprehensiveness of the screen. Cells were grown in suspension culture, and a sample for the final analysis of shRNA abundance was retrieved 9 days later (day 9 sample, corresponding to ∼4.5 population doublings). From this sample, we retrieved 7428 individual barcodes, indicating that most shRNAs were not lethal to the cells. However, the abundance of several shRNAs varied significantly between the 2 samples (supplemental Table 3), signifying effects on growth and viability for these shRNAs.
To identify candidate genes essential to leukemic cell survival, we compared the abundance of each individual shRNA in the day 0 and day 9 samples. Criteria for candidate gene definition were set with the aim of minimizing the likelihood of false-positive results. Therefore, we focused only on shRNAs present at high abundance in the day 0 sample and selected genes only when ≥2 specific shRNAs were depleted by a z score of ≤−2. Applying these criteria, we identified a total of 12 candidate genes targeted by 25 shRNAs (Figure 1B). Of special interest were enzyme-coding genes (eg, ROCK1, STK3, MAP3K6), because pharmacologic inhibition of the corresponding proteins could be envisioned as a straightforward approach for clinical translation.
Except for C7orf16 and CCRL1, RNA sequencing verified expression of all candidate genes. Because altered gene expression is a frequent phenomenon in AML and other malignancies and has successfully guided therapeutic innovations,18-20 we compared candidate gene expression levels in AML2 and in 3 additional primary AML patient samples to normal CD34+ HSPCs. Although we could recapitulate the previously reported overexpression of WT1, IL3RA, and HOMER320 in AML samples, none of the candidate genes identified by our screen showed a consistent misexpression when compared to normal controls (supplemental Figure 1). Hence, these genes would likely not have been nominated as candidate targets by expression profiling.
A classical way to define novel therapeutic strategies is the identification of oncogenic driver mutations.2 Exome and RNA sequencing of AML2 cells revealed several genetic aberrations, including NPM1 and TET2 mutations (supplemental Table 4). However, we were unable to detect any mutations within the coding DNA or expressed RNA of all candidate genes identified in our screen, indicating that these genes were generally essential, or that the effects were caused by nononcogene addiction.21
We conclude that RNAi screens are able to identify potentially essential target genes in primary AML cells, which would be missed by expression- and mutation-based approaches.
Validation of viability phenotype and knockdown efficacy
All 21 shRNAs targeting the 10 expressed candidate genes were subjected to single-shRNA validation experiments using AML2 cells. The number of nontarget shRNA–transduced cells included in all these experiments as controls increased over time (mean percentage increase, 142.3%; standard deviation, 25.5%). In contrast, for 14 of the 21 candidate shRNAs, we observed a reduction of the initial cell count to ≤50%, which we had prespecified as a condition for validation of the viability phenotype. For the following analyses, we focused on 6 candidate genes for which at least 2 shRNAs could be validated (Table 1). Compared to nontarget shRNAs, all 12 validated shRNAs targeting these candidate genes potently reduced the expression of their respective target genes (supplemental Figure 2).
Summary of the hit-validation results
| Expressed target gene . | No. of depleted shRNAs . | Mean z score of depleted shRNAs . | No. of validated shRNAs . |
|---|---|---|---|
| BNIPL | 2 | −2.6 | 2 |
| DGAT2 | 2 | −2.4 | 1 |
| DUSP14 | 2 | −2.5 | 0 |
| MAP3K6 | 2 | −2.6 | 2 |
| ROCK1 | 3 | −2.4 | 2 |
| RPS13 | 2 | −2.9 | 2 |
| SF3A1 | 2 | −2.5 | 1 |
| SNX27 | 2 | −3.5 | 2 |
| STK3 | 2 | −2.4 | 2 |
| WDHD1 | 2 | −2.4 | 0 |
| Expressed target gene . | No. of depleted shRNAs . | Mean z score of depleted shRNAs . | No. of validated shRNAs . |
|---|---|---|---|
| BNIPL | 2 | −2.6 | 2 |
| DGAT2 | 2 | −2.4 | 1 |
| DUSP14 | 2 | −2.5 | 0 |
| MAP3K6 | 2 | −2.6 | 2 |
| ROCK1 | 3 | −2.4 | 2 |
| RPS13 | 2 | −2.9 | 2 |
| SF3A1 | 2 | −2.5 | 1 |
| SNX27 | 2 | −3.5 | 2 |
| STK3 | 2 | −2.4 | 2 |
| WDHD1 | 2 | −2.4 | 0 |
All expressed target genes were reanalyzed in single-transduction hit-validation experiments. The numbers of shRNAs depleted by a z score of <−2 in the primary screen are given with their mean z score. The last column shows the number of shRNAs that could successfully be validated in single-transduction experiments.
Identification of general- and patient-specific vulnerabilities
Next, we set out to determine whether these genes were of equal importance in 2 other AML donors (AML24 and AML29) known to express them at levels detectable by RNA sequencing (supplemental Figure 1). Although knockdown of ROCK1 and BNIPL1 largely had comparable effects in AML2 and the 2 other donors, we observed differences in the individual sensitivity: AML2 cells were most sensitive to depletion of MAP3K6, and AML29 cells were most sensitive to depletion of STK3 (supplemental Figure 3). The most pronounced differences, however, were noted with respect to SNX27, which was essential to AML2 but seemed to be dispensable for AML24 and AML29 (supplemental Figure 3). These results indicate that pooled shRNA screens are able to identify both patient-specific leukemic vulnerabilities as well as dependencies of potential relevance for a larger subset of AML patients.
Identification of ROCK1 as a potential target for antileukemic therapy
Among the 6 validated candidate genes, 3 were marked as amenable to pharmacologic inhibition (MAP3K6, ROCK1, and STK3; www.sophicalliance.com). To further evaluate the possible therapeutic implications of our screen, we chose ROCK1 as an example because of its known involvement in cancer biology22,23 and because several inhibitors for this enzyme are already available.24
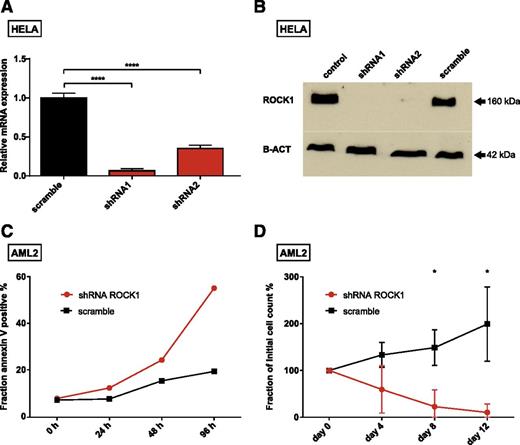
First, we confirmed knockdown efficacy of ROCK1-targeting shRNAs on messenger RNA and protein levels (Figure 2A-B; supplemental Figure 4). These experiments were partially done in HeLa cells because of the limited availability of primary patient material. Interestingly, knockdown of ROCK1 did not affect viability of HeLa cells, indicating that ROCK1 is not generally required for cell survival.
Consequences of ROCK1 knockdown. Knockdown levels of 2 shRNAs targeting ROCK1 are presented for messenger RNA (mRNA) (A) and protein (B). (C) Fraction of apoptotic cells after ROCK1 knockdown (red) and a nontarget control shRNA (scramble, black) at indicated time points. (D) Percentage of initial cell counts after transduction of cells with shRNAs targeting ROCK1 (red) and a nontarget control (black) at indicated time points. Due to the limited amount of primary cells available, the experiments shown in panels A and B were done with HeLa cells, whereas primary AML cells from our donor (AML2) were used in the experiments depicted in panels C and D. The cell source is indicated at the top of each panel. The means and standard deviations of experiments done in triplicates are given in panels A and D; significance was assessed by means of Student t test; *P < .05, ****P < .0001.
Consequences of ROCK1 knockdown. Knockdown levels of 2 shRNAs targeting ROCK1 are presented for messenger RNA (mRNA) (A) and protein (B). (C) Fraction of apoptotic cells after ROCK1 knockdown (red) and a nontarget control shRNA (scramble, black) at indicated time points. (D) Percentage of initial cell counts after transduction of cells with shRNAs targeting ROCK1 (red) and a nontarget control (black) at indicated time points. Due to the limited amount of primary cells available, the experiments shown in panels A and B were done with HeLa cells, whereas primary AML cells from our donor (AML2) were used in the experiments depicted in panels C and D. The cell source is indicated at the top of each panel. The means and standard deviations of experiments done in triplicates are given in panels A and D; significance was assessed by means of Student t test; *P < .05, ****P < .0001.
Next, we analyzed the effect of ROCK1 knockdown on cell cycle progression and proliferation of AML2 cells. Depletion of ROCK1 had modest effects on cell cycle distribution (supplemental Figure 5A). In contrast, we observed a marked increase of apoptotic cells during the first 4 days after transduction (Figure 2C), arguing for a proapoptotic effect of ROCK1 knockdown. After 12 days of suspension culture, almost all ROCK1 shRNA–transduced AML cells had perished, whereas nontarget shRNA–transduced cells grown in parallel increased by almost twofold (Figure 2D). Importantly, CD34+ HSPCs of 2 healthy donors appeared to be less sensitive to shRNA-mediated ROCK1 knockdown (supplemental Figure 6).
Taken together, these data demonstrate that the leukemic blasts of our donor were dependent on ROCK1 expression in vitro.
Assessment of the antileukemic potential of the ROCK inhibitor fasudil
Next, we set out to determine whether pharmacologic inhibition of ROCK1 results in a phenotype comparable to the results obtained by shRNA-mediated protein knockdown. To test this, we made use of fasudil, an isoquinoline derivative, which has already been approved for clinical use.25 To save on primary material, we confirmed inhibition of ROCK1 by fasudil in OCI-AML3, a leukemic cell line with comparable sensitivity toward ROCK1 knockdown (data not shown) and fasudil treatment (supplemental Figure 7).
In AML2 cells, fasudil treatment had modest effects on cell cycle distribution (supplemental Figure 5B) but led to a marked concentration- and time-dependent increase in apoptotic cells (supplemental Figure 8). For a 4-day exposure to the drug, the median lethal dose equaled 36 μM. A concentration-dependent effect on cell viability was also seen with Y-27632, another ROCK inhibitor (supplemental Figure 9), providing further support that inhibition of ROCK1 was causing the observed phenotype.
Because the antileukemic effects of ROCK1 knockdown were not limited to AML2, we were interested in knowing whether this was also applicable to pharmacologic inhibition of the enzyme. We therefore treated primary cells from 3 other AML patients (see supplemental Table 2 for details) with fasudil. Although the proliferation kinetics differed (Figure 3A), a cell count reduction comparable to the observations in AML2 was observed in all donors (Figure 3B). In contrast, although small numbers precluded valid statistical comparisons, normal HSPCs appeared to be less sensitive to fasudil treatment (Figure 3B).
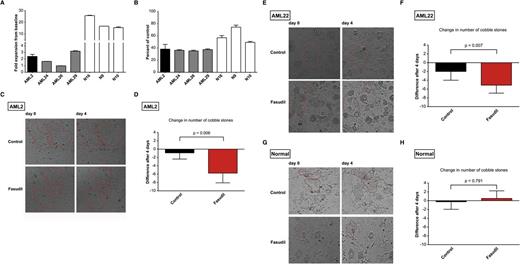
Effect of the ROCK inhibitor fasudil on malignant and nonmalignant hematopoietic cells. (A) Proliferation kinetics of untreated cells from the indicated donors over 96 hours of suspension culture (fold expansion from baseline to the end of the suspension-culture period is given). To allow for the cross-donor comparisons (B), all results obtained after 96 hours of fasudil treatment (40 μM), were normalized to wells containing vehicle-treated cells of each respective donor. The index patient AML2 (black) is shown in comparison with 3 other AML donors (AML24, AML26, and AML29; gray) and 3 normal donors (N16, N9, and N10; white). Means and standard deviations of experiments done in triplicate are presented. (C-H) Results of experiments done with leukemic and nonmalignant hematopoietic progenitors in LTC-IC. Representative microscopic images before (day 0) and after 4 days of treatment with carrier (control) or active drug (fasudil) at 40 μM concentration are shown for AML2 (C), AML22 (E), and normal CD34+ HSPCs isolated from volunteer donors (G). Red lines highlight cobblestone areas. Note the disappearance of cobblestone cells in the fasudil-treated sample in the AML cells but not in normal HSPCs. All images were taken using the Celigo cytometer according to the manufacturer’s instructions; native live cell imaging. Quantification of the changes of cobblestone numbers after 4 days of treatment with fasudil or carrier (control) in AML2 (D), AML22 (F), and normal CD34+ HSPCs (H). Means and standard deviations of 4 to 8 replicates are shown. All cobblestones present within each well were counted.
Effect of the ROCK inhibitor fasudil on malignant and nonmalignant hematopoietic cells. (A) Proliferation kinetics of untreated cells from the indicated donors over 96 hours of suspension culture (fold expansion from baseline to the end of the suspension-culture period is given). To allow for the cross-donor comparisons (B), all results obtained after 96 hours of fasudil treatment (40 μM), were normalized to wells containing vehicle-treated cells of each respective donor. The index patient AML2 (black) is shown in comparison with 3 other AML donors (AML24, AML26, and AML29; gray) and 3 normal donors (N16, N9, and N10; white). Means and standard deviations of experiments done in triplicate are presented. (C-H) Results of experiments done with leukemic and nonmalignant hematopoietic progenitors in LTC-IC. Representative microscopic images before (day 0) and after 4 days of treatment with carrier (control) or active drug (fasudil) at 40 μM concentration are shown for AML2 (C), AML22 (E), and normal CD34+ HSPCs isolated from volunteer donors (G). Red lines highlight cobblestone areas. Note the disappearance of cobblestone cells in the fasudil-treated sample in the AML cells but not in normal HSPCs. All images were taken using the Celigo cytometer according to the manufacturer’s instructions; native live cell imaging. Quantification of the changes of cobblestone numbers after 4 days of treatment with fasudil or carrier (control) in AML2 (D), AML22 (F), and normal CD34+ HSPCs (H). Means and standard deviations of 4 to 8 replicates are shown. All cobblestones present within each well were counted.
Taken together, these results raise the possibility that fasudil exerts antileukemic effects, which might not be limited to our index patient, whereas normal HSPCs seem less dependent on intact ROCK1 function. Although the consistency of shRNA-mediated knockdown and chemical inhibition with 2 independent inhibitors strongly implicates ROCK1 in the phenotype, we note that the drugs also inhibit other kinases26 that could contribute to the phenotype.
Efficacy of fasudil on leukemic progenitors
Because inhibition of ROCK1 facilitates induced pluripotent stem cell maintenance27 and promotes self-renewal in cancer stem cells,28 we wanted to rule out that targeting ROCK1 might foster leukemic progenitor survival. An in vitro approach to study immature progenitors is the LTC-IC,29 which relies on the fact that immature hematopoietic cells tend to form clusters of clonal growth (referred to as cobblestone areas) when seeded on a layer of MBSCs.
To this end, we seeded AML2 cells on a feeder layer of irradiated M2-10B4 MBSCs. After 5 weeks of coculture, cobblestone areas were readily identifiable (Figure 3C). The myeloid origin of these cells was demonstrated by flow cytometry, and the NPM1 mutation was readily detectable (supplemental Figure 10; data not shown), validating the authenticity of leukemic cells. Established cobblestones were treated with fasudil or carrier and monitored over time. Surprisingly, we observed a significant decrease in colony size and number and even the complete disappearance of smaller colonies in fasudil-treated, but not control-treated, wells (Figure 3C-D). We repeated these experiments with primary leukemic cells from a second donor (AML22; supplemental Table 2) and obtained comparable results (Figure 3E-F). Pretreatment of cells from this donor with fasudil for 2 days before seeding in the LTC-IC resulted in a significant decrease in the number of cobblestone areas after 4 weeks (supplemental Figure 11). In contrast to the results obtained with AML cells, cobblestone areas grown from healthy CD34+ HSPCs appeared to be less sensitive to fasudil treatment (Figure 3G-H). We conclude that fasudil did not spare leukemic progenitors in the LTC-IC, whereas healthy HSPCs again appeared less dependent on intact ROCK1 signaling.
AML2 cells are sensitive to fasudil treatment and ROCK1 knockdown in a xenograft model
The ability to engraft immunosuppressed mouse strains and to recapitulate key features of the human disease has been proposed as a defining characteristic of leukemic progenitors.30 Moreover, xenograft models represent a valuable tool to assess the efficacy of antileukemic treatments in an in vivo microenvironment.
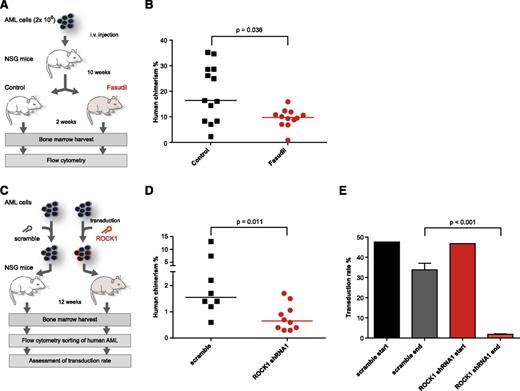
Encouraged by the results of our LTC-IC experiments, we therefore aimed at testing the potential antileukemic effect of fasudil in a xenograft model. To avoid xenogeneic graft-versus-host disease,31 we injected lymphocyte-depleted primary leukemic AML2 cells intravenously into 4- to 6-week-old female NSG mice. Animals were kept for 10 weeks to allow for establishment of leukemic disease. Fasudil was then subcutaneously administered for 2 weeks. Treated mice did not show any signs of weight loss, abnormal behavior, or decreased blood counts, indicating that the drug was well tolerated (supplemental Figure 12). After treatment, mice were euthanized and BM was analyzed for the presence of human cells by flow cytometry. Compared with untreated controls, fasudil-treated mice had a significantly lower fraction of human CD45-positive cells in the murine BM (median chimerism, 16.4% vs 9.8%; P = .036; Figure 4A-B), demonstrating an effect of the drug on leukemic proliferation in vivo. The myeloid origin of all leukemic cells in the mouse BM was verified by flow cytometry, as was the presence of the NPM1 type A mutation (supplemental Figure 13; data not shown). Transplant of AML2 cells transduced with shRNAs targeting ROCK1 was also associated with a significant decrease in human chimerism in the BM of NSG mice (Figure 4C-D). Of note, although the transduction rate in flow-sorted human cells isolated from murine BM 12 weeks after transplant approached zero in ROCK1 shRNA–transduced cells, it remained comparable to baseline in nontarget shRNA controls (Figure 4E), consistent with the observation that ROCK1 knockdown negatively affects the growth of the leukemic cells in vivo.
Fasudil is effective against transplanted AML cells in vivo. (A) Schematic outline of the treatment regime. After 10 weeks, animals were either fasudil treated (n = 12, 50 μg/g/d) or control treated (n = 13). After 2 weeks of treatment, mice were euthanized, and the percentage of human cells in the murine BM was assessed by flow cytometry. (B) Comparison of the percentage of human chimerism (percentage of human CD45-positive per total CD45-positive cells in the BM) between fasudil-treated (red) and control-treated animals (black). The difference between fasudil-treated and control-treated mice was statistically significant by Mann-Whitney U test (P = .036). (C) Overview of the experiments to address the effect of shRNA-mediated ROCK1 knockdown on the growth of primary cells from AML2 in NSG mice. At the end of the experiment, murine BM from each mouse was analyzed by flow cytometry, and human cells identified by positivity for human CD45 and human CD33 were flow sorted to purity. The sorted cells were spilt and grown in the presence or absence of puromycin to assess the transduction rate. (D) The human chimerism in murine bone marrow 12 weeks after transplant with AML2 cells containing nontarget control shRNAs (scramble, n = 8) or shRNAs targeting ROCK1 (n = 10). Compared with mice receiving scramble-transduced cells, those animals receiving ROCK1 shRNA cells had a significantly lower median chimerism (0.6% vs 1.6%; P = .011 by Mann-Whitney U test). Note that the chimerism in both groups is lower than in the experiments shown in panel B, probably due to the prolonged ex vivo culture period needed for transduction. (E) Comparison of the transduction rate in ROCK1 shRNA–transduced and scramble-transduced cells before (start) and 12 weeks after (end) transplant into NSG mice. Although the transduction rate at both time points was comparable in cells transduced with nontarget control shRNAs, it decreased to almost 0 in ROCK1 shRNA–transduced cells (median, 33.8% vs 1.85%; P < .001). In panels B and D, horizontal lines depict the median within each group.
Fasudil is effective against transplanted AML cells in vivo. (A) Schematic outline of the treatment regime. After 10 weeks, animals were either fasudil treated (n = 12, 50 μg/g/d) or control treated (n = 13). After 2 weeks of treatment, mice were euthanized, and the percentage of human cells in the murine BM was assessed by flow cytometry. (B) Comparison of the percentage of human chimerism (percentage of human CD45-positive per total CD45-positive cells in the BM) between fasudil-treated (red) and control-treated animals (black). The difference between fasudil-treated and control-treated mice was statistically significant by Mann-Whitney U test (P = .036). (C) Overview of the experiments to address the effect of shRNA-mediated ROCK1 knockdown on the growth of primary cells from AML2 in NSG mice. At the end of the experiment, murine BM from each mouse was analyzed by flow cytometry, and human cells identified by positivity for human CD45 and human CD33 were flow sorted to purity. The sorted cells were spilt and grown in the presence or absence of puromycin to assess the transduction rate. (D) The human chimerism in murine bone marrow 12 weeks after transplant with AML2 cells containing nontarget control shRNAs (scramble, n = 8) or shRNAs targeting ROCK1 (n = 10). Compared with mice receiving scramble-transduced cells, those animals receiving ROCK1 shRNA cells had a significantly lower median chimerism (0.6% vs 1.6%; P = .011 by Mann-Whitney U test). Note that the chimerism in both groups is lower than in the experiments shown in panel B, probably due to the prolonged ex vivo culture period needed for transduction. (E) Comparison of the transduction rate in ROCK1 shRNA–transduced and scramble-transduced cells before (start) and 12 weeks after (end) transplant into NSG mice. Although the transduction rate at both time points was comparable in cells transduced with nontarget control shRNAs, it decreased to almost 0 in ROCK1 shRNA–transduced cells (median, 33.8% vs 1.85%; P < .001). In panels B and D, horizontal lines depict the median within each group.
In summary, these results reveal that this RNAi screen was able to identify a potential therapeutic target, essential not only for bulk leukemic cells but also for more immature leukemic progenitors in the tested donors.
Discussion
We have shown that complex lentiviral shRNA-screens in primary cells derived directly from a patient are feasible and readily identify specific dependencies of the leukemic clone. Our experimental conditions and hit-calling criteria resulted in reasonable cell consumption and a high reproducibility of the primary screening results. The example of ROCK1, for which several specific inhibitors are already available, highlights the potential therapeutic implications of such screens.
ROCK1 is a serine/threonine kinase, which has long been known to be a key downstream effector of the Rho family of guanosine triphosphate hydrolase enzymes involved in various intracellular processes, including cytoskeleton assembly, cell contraction, and apoptosis.32,33 As a strategy to treat blood cancer, inhibition of the ROCK1 pathway might look surprising at first, because it is also known to be an essential part of caspase-3–induced apoptotic processes.34,35 In concordance, it has recently been demonstrated that ROCK1 is indispensable for the antileukemic effects of the apoptosis-inducing compound triptolide.36 ROCK1 inhibition has further been shown to prevent cell death of neurons37 and to induce proliferation of breast cancer cells.38
However, accumulating evidence suggests that therapeutic targeting of ROCK1 may also have a considerable antineoplastic potential. Application of ROCK1 inhibitors reduced the metastatic potential in cells from several kinds of solid tumors, presumably by decreasing cell motility.39,40 This mechanism is unlikely to explain the observed activity of ROCK1 inhibition in our feeder-free AML suspension-culture experiments. However, the destabilization of c-MYC, observed in ROCK inhibitor–treated prostate cancer cells,39 might have played a role because this oncogene is frequently deregulated in AML.41
Although the precise mode of action by which ROCK1 inhibition reduced leukemic burden in our in vitro and in vivo experiments was beyond the focus of our research, our results suggest that further exploration of ROCK1 inhibition as a potential antileukemic strategy is warranted. Even though our data in additional primary AML donors are limited in number, they are well in line with data generated by other groups in CML.23,42 Interestingly, both our data and 1 of the earlier reports in CML suggest that normal HSPCs are less sensitive to ROCK1 inhibition than leukemic blasts. These observations and the excellent tolerability of prolonged fasudil exposure in humans43,44 and in our mouse model argue for the feasibility of repetitive dosing to increase peak serum levels observed after a single application of this drug25 to levels needed for efficient ROCK1 inhibition in vivo. The absence of any signs of hematotoxicity in clinical trials on fasudil is further arguing in favor of this proposition.25,43
Of note, our shRNA screen delineated a potential therapeutic strategy without necessitating a comprehensive genome-wide mutation analysis. On the contrary, it identified putative therapeutic targets that were neither overexpressed nor mutated and might therefore have been missed using other approaches. Some of the candidate genes seem to be patient specific (eg, SNX27), whereas others, such as ROCK1, might be relevant in a broader population of AML patients.
Taken together, our results highlight the therapeutic potential of primary cells in combination with large-scale pooled shRNA screens to delineate patient-specific and general dependencies of leukemic blasts in AML. Given the improvement of cell culture conditions,45 lentiviral vectors,46 and transduction protocols,47,48 as well as the progress made in the field of RNAi-based therapeutics,49 we envision that RNAi screens could assist therapeutic decisions in AML in the future (supplemental Figure 14). We therefore believe that our results should encourage the further development of this technology.
The data reported in this article have been deposited in the Gene Expression Omnibus database.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Claudia Schönefeldt, Anja Liebkopf, and Robert Kuhnert for excellent technical assistance.
This work was supported by grants from the Sächsische Ministerium für Wissenschaft und Kunst and the Bundesexzellenzinitiative ZUK 64, and a Seed Grant from the Center for Regenerative Therapy Dresden (F.B. laboratory); a MeDDrive grant from the Medical Faculty of the Technische Universität Dresden (M.W.); the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 655 (S.T., A.D., G.E., S.B., and F.B.); and a Gerok Rotation Position from the Sonderforschungsbereich 655 (M.W. and M.v.B.).
Authorship
Contribution: M.W., A.C., M.v.B., and S.T. performed the experiments and analyzed the results; M.P.-R. developed the algorithms for and performed the biostatistics and bioinformatics analyses; A.D. was responsible for all deep-sequencing experiments; M.W. and A.C. made the figures; M.W. and F.B. designed the research and wrote the paper; and all authors interpreted the results and read and commented on the article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin Wermke, Universitätsklinikum C.-G.-Carus, Medizinische Klinik I, Fetscherstrasse 74, D-01307 Dresden, Germany; e-mail: martin.wermke@uniklinikum-dresden.de; and Frank Buchholz, Technische Universität Dresden, Medizinische Fakultät, UniversitätskrebsCentrum–Medizinische Systembiologie, Fetscherstrasse 74, D-01307 Dresden, Germany: e-mail: frank.buchholz@tu-dresden.de.
References
Author notes
M.W. and A.C. contributed equally to this study.