Key Points
Pulling of VWF A1 domain that is engaged to GPIb-IX induces unfolding of a hitherto unidentified mechanosensitive domain in GPIbα.
The spatial proximity of the mechanosensitive domain to GPIbβ and GPIX suggests a novel mechanism of platelet mechanosensing.
Abstract
How glycoprotein (GP)Ib-IX complex on the platelet surface senses the blood flow through its binding to the plasma protein von Willebrand factor (VWF) and transmits a signal into the platelet remains unclear. Here we show that optical tweezer-controlled pulling of the A1 domain of VWF (VWF-A1) on GPIb-IX captured by its cytoplasmic domain induced unfolding of a hitherto unidentified structural domain before the dissociation of VWF-A1 from GPIb-IX. Additional studies using recombinant proteins and mutant complexes confirmed its existence in GPIb-IX and enabled localization of this quasi-stable mechanosensitive domain of ∼60 residues between the macroglycopeptide region and the transmembrane helix of the GPIbα subunit. These results suggest that VWF-mediated pulling under fluid shear induces unfolding of the mechanosensitive domain in GPIb-IX, which may possibly contribute to platelet mechanosensing and/or shear resistance of VWF-platelet interaction. The identification of the mechanosensitive domain in GPIb-IX has significant implications for the pathogenesis and treatment of related blood diseases.
Introduction
The mechanical shear force generated by blood flow in the vasculature is an important factor that mediates physiologic hemostasis and pathologic thrombosis. The induction of platelet aggregation by the elevated shear stress requires von Willebrand factor (VWF) and its association with glycoprotein (GP)Ib-IX and GPIIb-IIIa, both of which are platelet-specific receptor complexes.1,2 VWF in flowing blood or immobilized at the damaged vessel wall responds to shear stress and exposes its A1-A2-A3 domains.3-5 Concurrently, ligation of VWF under flow with the N-terminal domain of GPIbα, the major subunit in GPIb-IX, transmits a signal into the platelet that eventually leads to activation of GPIIb-IIIa and aggregation of platelets.6-9 Although GPIb-IX has been recognized as the platelet mechanosensor for the past 20 years,10 how this receptor complex senses shear stress and converts this mechanical information into a protein-mediated signal that can be recognized and propagated has remained elusive.
The GPIb-IX complex consists of GPIbα, GPIbβ, and GPIX subunits in a 1:2:1 stoichiometry.11,12 GPIbα contains, starting from the N-terminus, a leucine-rich repeat domain that interacts with the A1 domain of VWF (VWF-A1), a highly glycosylated macroglycopeptide region, a stalk region of about 60 residues, a pair of cysteine residues that connect to GPIbβ via disulfide bonds, a single-span transmembrane helix, and a relatively short cytoplasmic domain that is connected to the cytoskeleton through filamin A (Figure 1A).13 GPIbβ and GPIX each contains an extracellular leucine-rich repeat domain that is much smaller than that of GPIbα, a transmembrane helix and a cytoplasmic tail.14 GPIb-IX is a highly integrated complex, with each subunit interacting with one another through its transmembrane helices and membrane-proximal extracellular domains (Figure 1A).15 Crystal structures of the GPIbα N-terminal domain in complex with VWF-A1 have been determined.16,17 Its association with VWF-A1 is classified as a catch-bond18,19 or flex-bond,20 which better facilitates the tethering of platelets to VWF under flow. However, numerous studies on the GPIbα N-terminal domain have not provided any clues about how it transmits the VWF-binding signal into the platelet. This is largely because the GPIbα N-terminal domain makes no contacts with the membrane-proximal parts of GPIb-IX except through the long and relatively flexible macroglycopeptide region (Figure 1A).21 There have been no reports of a highly glycosylated mucinlike region mediating a long-distance allosteric conformational change.
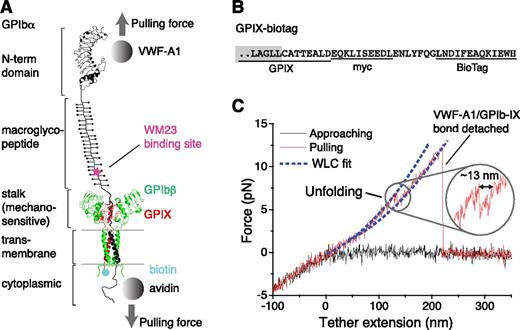
Pulling the engaged A1 domain of VWF induces unfolding of a domain in the full-length GPIb-IX complex. (A) A diagram of GPIb-IX illustrating the experimental setup for the optical tweezer single-molecule force measurement. The BioTag sequence that is specifically recognized and biotinylated by Escherichia coli biotin ligase was either appended to the C-terminus of GPIX cytoplasmic domain (as shown) or placed into the juxtamembrane region of the GPIbα cytoplasmic domain. The biotinylated GPIb-IX was expressed in transfected cells, solubilized in the Triton X-100–containing lysis buffer, and eventually immobilized on the streptavidin-coated bead. Recombinant VWF-A1 was linked through a DNA handle to a polystyrene bead that was placed in an optical trap as described before.3 Individual domains of GPIbα are marked on the left. (B) The cytoplasmic sequence of GPIX-biotag showing the appended site of biotinylation. A c-myc immunotag and a BioTag sequence (underlined) are attached to the C-terminal end of GPIX. Residues in the GPIX transmembrane domain are marked by a gray box. (C) A single-molecule force-distance trace illustrating the unfolding of MSD before the detachment of VWF-A1 from GPIb-IX. The inset highlights the observed unfolding event.
Pulling the engaged A1 domain of VWF induces unfolding of a domain in the full-length GPIb-IX complex. (A) A diagram of GPIb-IX illustrating the experimental setup for the optical tweezer single-molecule force measurement. The BioTag sequence that is specifically recognized and biotinylated by Escherichia coli biotin ligase was either appended to the C-terminus of GPIX cytoplasmic domain (as shown) or placed into the juxtamembrane region of the GPIbα cytoplasmic domain. The biotinylated GPIb-IX was expressed in transfected cells, solubilized in the Triton X-100–containing lysis buffer, and eventually immobilized on the streptavidin-coated bead. Recombinant VWF-A1 was linked through a DNA handle to a polystyrene bead that was placed in an optical trap as described before.3 Individual domains of GPIbα are marked on the left. (B) The cytoplasmic sequence of GPIX-biotag showing the appended site of biotinylation. A c-myc immunotag and a BioTag sequence (underlined) are attached to the C-terminal end of GPIX. Residues in the GPIX transmembrane domain are marked by a gray box. (C) A single-molecule force-distance trace illustrating the unfolding of MSD before the detachment of VWF-A1 from GPIb-IX. The inset highlights the observed unfolding event.
Related to the elusive platelet mechanosensing mechanism is another puzzling, and again unanswered, question about the function of GPIbβ and GPIX. Previous studies have demonstrated that expression of GPIb-IX in Chinese hamster ovary cells requires all 3 subunits; the surface expression level of GPIbα in the absence of GPIbβ and GPIX is drastically lower than that of GPIbα in GPIb-IX.22 This explains in principle why mutations causing Bernard-Soulier syndrome (BSS), a rare congenital bleeding disorder characterized by an abnormally low level of expression of functional GPIb-IX, are present in all 3 subunits.23-25 The phenomenon of interacting subunits necessary for coexpression has been documented in other receptor complexes such as TCR-CD3 and integrins.26-29 In those cases, all of the involved subunits take on a critical role in signaling in addition to coexpression. In comparison, no additional functions have been proposed for the extracellular domains of GPIbβ and GPIX, which can change conformation in response to environmental changes.30
We report here the first single-molecule force measurement on the full-length GPIb-IX complex. Pulling on the immobilized GPIb-IX with recombinant VWF-A1 induces unfolding of a domain in the juxtamembrane stalk region of GPIbα. This domain, hitherto unidentified and designated as the mechanosensitive domain (MSD) in this paper, is structured but relatively unstable. Identification of this MSD in GPIb-IX has potential implications for the mechanism of platelet mechanosensing, in which GPIbβ and GPIX extracellular domains play a critical role.
Methods
Materials
HEK293 Tet-on 3G cell line was obtained from Clontech (Mountain View, CA). Recombinant hexahistidine-tagged VWF-A1 and thiol-activated 802-bp DNA handles have been described before.3 Antibody WM23 was kindly shared by Dr. Michael Berndt. The monoclonal anti-GPIX antibody FMC25 was purchased from Millipore (Temecula, CA). Biotinylated antibody was prepared using sulfo-NHS-biotin (Thermo Scientific, Rockford, IL).
Cloning of mutant GPIb-IX constructs
To facilitate molecular cloning, a DNA fragment that encodes the same protein sequence of human GPIbβ but lacks the GC-rich nucleotide sequence was synthesized by Genscript (Piscataway, NJ). The encoded GPIbβ included an HA tag at its N-terminus as described before.31 Mammalian expression plasmid pBIG-5b (GenBank accession #KM042177) was generated by inserting the coding sequence of Escherichia coli biotin ligase (BirA) into multiple cloning site (MCS) II of pBI (Clontech) and a DNA cassette comprising, from 5′ to 3′-ends, a 13-residue biotin acceptor peptide (BioTag)–encoding sequence,32 an internal ribosome entry site, and an enhanced green fluorescent protein (EGFP)-encoding sequence into MCS I of pBI. The new GPIbβ gene was inserted into pBIG5b as an NheI/NsiI fragment to generate pBIG5b-BirA/Ibβ/EGFP.
To construct pBIG5b-Ibα/IX-biotag/mCherry, the gene fragment encoding GPIX was amplified, appended at the 5′-end with sequences encoding the myc tag and TEV protease cleavage sequence, and inserted into pBIG5b using the NheI/XhoI restriction sites. The resulting plasmid was digested by AgeI/BsrGI to replace the EGFP gene with mCherry. Subsequently, the pBIG5b-IX-biotag/mCherry plasmid was digested by EagI/SphI to replace the BirA gene with GPIbα. To enable ligation of the GPIbα fragment, the second SphI site in the MCSII was removed before ligation. Mutant GPIbα genes with altered MSD were subcloned into the pBIG5b-IX-biotag/mCherry plasmid in a similar fashion.
To construct the pcDNA-Ibα-biotag, the gene fragment encoding the BioTag sequence was inserted into the GPIbα cDNA to replace the sequence encoding residues Q519-A532. The mutated GPIbα gene was ligated into the pcDNA-3.1(–) vector (Invitrogen, Carlsbad, CA) using the NheI/XhoI sites. The wild-type GPIX gene was ligated into the pcDNA vector in a similar manner. All plasmids were confirmed by DNA sequencing.
Expression of biotinylated GPIb-IX and mutants
To establish a cell line stably expressing HA-GPIbβ, HEK293 Tet-on cells were cotransfected by plasmids pBIG5b-BirA/Ibβ/EGFP and pUC19-puro using Lipofectamine 2000 (Invitrogen). The pUC19-puro plasmid was generated by ligating the XhoI fragment of plasmid pTRE2pur (Clontech), which contains the puromycin N-acetyl-transferase expression cassette, into pUC19 vector that had been modified by insertion of an oligonucleotide cassette to contain a single XhoI restriction site in its MCS. Beginning 1 day posttransfection, cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 5 µg/mL blasticidin, and 2 μg/mL puromycin for 3 weeks. The surviving cells were treated with 3 μg/mL doxycycline for 1 day before being sorted for EGFP fluorescence and surface expression of HA-GPIbβ detected by anti-HA antibody (Sigma-Aldrich/Merck, Darmstadt, Germany). The cells stably expressing BirA/HA-GPIbβ/EGFP were further transfected with the pBIG5b-Ibα/IX-biotag/mCherry vector, cultured for 3 weeks, and sorted for stable surface expression of GPIbα detected by WM23, and EGFP and mCherry fluorescence. Alternatively, cells stably expressing HA-GPIbβ/BirA/EGFP were transiently transfected with pcDNA-Ibα-biotag and pcDNA-IX using Lipofectamine 2000.
To induce expression of biotinylated GPIb-IX, the cells were cultured in the FBS-free medium containing 3 µg/mL doxycycline and 100 µM d-biotin for 1 day. The cells were harvested and lysed in the lysis buffer (1% Triton X-100, 5 mM CaCl2, 58 mM sodium borate, 10% protease inhibitor cocktail, 5 mM N-ethylmaleimide, pH 8.0; at approx. 1-2 × 104 cells/µL). The supernatant containing biotinylated GPIb-IX was further analyzed by Western blot and flow cytometry largely as described previously,30,33,34 or it was stored at −80°C for the force measurement.
Expression and purification of recombinant GPIbα stalk region (Ibα-S)
The DNA fragment encoding GPIbα residues Ala417-Phe483 was amplified from the GPIbα cDNA and later ligated into the pHex vector12 as a BamHI/XhoI fragment. The DNA sequence was confirmed by sequencing. Ibα-S was expressed as a decahistidine-tagged glutathione S-transferase (GST) fusion protein from E coli BL21 cells, and its purification by nickel affinity chromatography followed published protocols.12,35 After the fusion protein was cleaved by thrombin (5 U/mg of fusion protein), Ibα-S was separated from GST by preparative reverse-phase high-performance liquid chromatography and stored at −80°C as lyophilized powder.12 Related proteins Ibα-cSc and Ibα-cS-biotag were prepared in a similar manner. The purity of each protein was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and analytical high-performance liquid chromatography, and its concentration estimated by the dry-weight method.
Laser optical tweezer measurement
Biotin and digoxygenin (Dig) DNA handles were prepared as described previously.3,20 Dithiobis-nitrilotriacetic acid (NTA) (Dojindo, Rockville, MA) was first reduced by Immobilized TCEP Disulfide Reducing Gel (Thermo Fisher Scientific) and then coupled to the activated biotin DNA handle via a disulfide bond. Recombinant Ibα-cSc was coupled to 2 pieces of DNA handles through disulfide bonds as described previously.3,20 Carboxyl-polystyrene beads of 2.0-μm diameter (Spherotech, Lake Forest, IL) were covalently coupled with streptavidin (Invitrogen), antidigoxigenin Fab (Roche), or WM23 as described previously.3,20 To couple VWF-A1 to the bead, streptavidin-coated beads were first incubated for 10 minutes with 1 nM biotin-DNA handle-NTA in Tris-buffered saline (150 mM NaCl, 10 mM Tris·HCl, 5 mM NiCl2, pH 7.5) and were then washed and incubated with 100 pM VWF-A1 for 15 minutes before the experiment. For capturing the biotinylated GPIb-IX, streptavidin-coated beads were incubated with 20 µL GPIb-IX–containing cell lysate for 10 minutes and washed with Tris-buffered saline containing 1% Triton X-100. Single-molecule pulling experiments were performed using an analytical minioptical tweezer apparatus that has been used in several single-molecule unfolding/unbinding studies previously.36-38 Force and bead-to-bead distance were recorded at 200 Hz. When appropriate, the force-extension data were fitted to the wormlike chain (WLC) model. The lifetime of bond as a function of force was estimated by the Dudko-Hummer-Szabo equation.39
Circular dichroism spectroscopy
Purified Ibα-S, or its variants, was weighed and dissolved in 50 mM Tris·HCl, 50 mM NaCl, 1 mM DTT, pH 7.4 buffer, or the same buffer containing various concentrations of urea, to a final concentration of 2.5 mg/mL. Far-UV circular dichroism (CD) spectra (190-260 nm) were collected on a JASCO J810 spectrometer using a 0.1-cm quartz cuvette at 20°C. The stepwise wavelength was set to 0.5 nm per step. Each spectrum was scanned 5 times and corrected for background signal.
Results
Force-induced unfolding of a domain in the full-length GPIb-IX
To enable single-molecule force measurement of full-length GPIb-IX, a new expression system was engineered to achieve site-specific biotinylation of GPIb-IX (supplemental Figure 1, available on the Blood Web site). Because the cytoplasmic domain of GPIX is not critical to the proper assembly of GPIb-IX,34 a 13-residue BioTag sequence in which a lysine residue is specifically recognized and biotinylated by E coli biotin ligase32 was appended to the C-terminus of GPIX (Figure 1 and supplemental Figure 1). Coexpression of the engineered GPIX (GPIX-biotag) with GPIbα, HA-tagged GPIbβ,31 and biotin ligase in human embryonic kidney (HEK)293 cells produced a well-assembled GPIb-IX complex that was uniformly biotinylated at the cytoplasmic end of GPIX (supplemental Figure 1). In our optical tweezer setup (Figure 1A), recombinant hexahistidine–tagged VWF-A1 was attached to an NTA-conjugated DNA handle. The other end of the DNA handle was immobilized to a 2-µm polystyrene bead via biotin-streptavidin linkage, which was controlled by the optical trap.3 The biotinylated GPIb-IX was captured by another streptavidin bead. This bead was held by a fixed micropipette. It should be noted that a small portion of GPIbα and GPIbβ in transfected cells form hmwGPIb complexes,30,34 in addition to the wild-type GPIb-IX (supplemental Figure 1C). Because hmwGPIb is formed in the absence of GPIX,34 specific biotinylation of the GPIX cytoplasmic domain ensured that only the native full-length GPIb-IX was immobilized to the fixed bead and analyzed here.
In each recorded contact-retraction cycle, the trapped VWF-A1 bead was brought into contact with the fixed GPIb-IX bead for ∼1 second at low contact force (∼5 pN) and then pulled away. Although no or very little (<1 pN) adhesion force was observed in ∼80% of the contact-retraction cycles, a tether that extended to >200 nm and ruptured at ∼10 to 30 pN was consistently observed in the other 20% of cycles (Figure 1C). The force-extension relationship of the tether fitted well with the WLC model40 in the low force (0-10 pN) regime (Figure 1C). It is noteworthy that the rupture force of 10 to 30 pN observed during the retraction step of pulling VWF-A1 from GPIb-IX was significantly smaller than those observed in the control experiment stretching DNA handle-NTA from VWF-A1 (supplemental Figure 2), but was comparable with those reported for the unbinding of VWF-A1 and the GPIbα N-terminal domain (Figure 2A).19,20 Thus, in our optical tweezer experiment we have observed the binding and unbinding of VWF-A1 and full-length GPIb-IX at the single-molecule level.
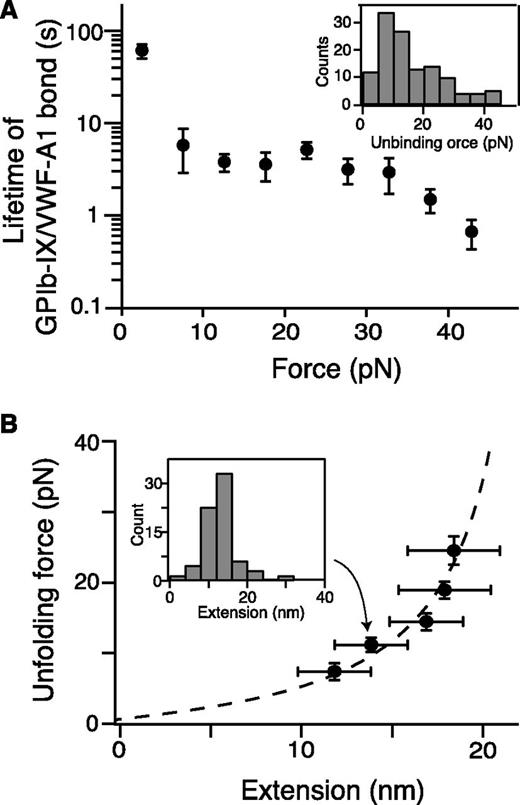
Quantitation of pulling VWF-A1 from biotinylated GPIb-IX. (A) Plot of lifetimes of the GPIb-IX/VWF-A1 bond vs force (mean ± SEM, n >3). Shown in the insert is a representative histogram of unbinding force (collected under a pulling speed of 100 nm/s) used to obtain bond lifetimes.39 (B) Fit of unfolding force vs extension data to the WLC model (dashed line), which yielded a contour length of 25.1 ± 0.3 nm. Extension distances were sorted by unfolding force into 4-pN bins. A histogram of extension (insert) was used to find peak extension. Unfolding forces were averaged for each bin (n = 23-51 per bin). Error bars are 1 SD for force and half-bin width for extension.
Quantitation of pulling VWF-A1 from biotinylated GPIb-IX. (A) Plot of lifetimes of the GPIb-IX/VWF-A1 bond vs force (mean ± SEM, n >3). Shown in the insert is a representative histogram of unbinding force (collected under a pulling speed of 100 nm/s) used to obtain bond lifetimes.39 (B) Fit of unfolding force vs extension data to the WLC model (dashed line), which yielded a contour length of 25.1 ± 0.3 nm. Extension distances were sorted by unfolding force into 4-pN bins. A histogram of extension (insert) was used to find peak extension. Unfolding forces were averaged for each bin (n = 23-51 per bin). Error bars are 1 SD for force and half-bin width for extension.
In the pulling curves of VWF-A1/GPIb-IX, an extension or unfolding event before the rupture, with forces ranging from 5 to 20 pN, was observed (Figures 1C and 2B). The WLC fit of the unfolding force and extension yielded a contour length of 25.1 ± 0.3 nm (Figure 2B) and a persistence length of 0.74 ± 0.04 nm. Assuming a contour length of 4 Å per residue, it indicated that a structural domain of ∼63 residues was stretched or unfolded by the pulling force while VWF-A1 was still bound to the GPIbα N-terminal domain. Intact domain structures are required for the interaction of VWF-A1 with the GPIbα N-terminal domain, both of which contain hundreds of residues.16 Furthermore, similar unfolding events were not observed in either the control experiment in which the NTA-DNA handle was pulled from VWF-A1 (supplemental Figure 2) or previous force measurements using VWF fragments and the GPIbα N-terminal domain.18-20 Therefore, the observed unfolding event in the pulling of VWF-A1 from GPIb-IX is not caused by unfolding of either of the interacting domains, but rather of a different domain in the GPIb-IX complex, which we designate here the MSD.
Localization of MSD to the stalk region of GPIbα
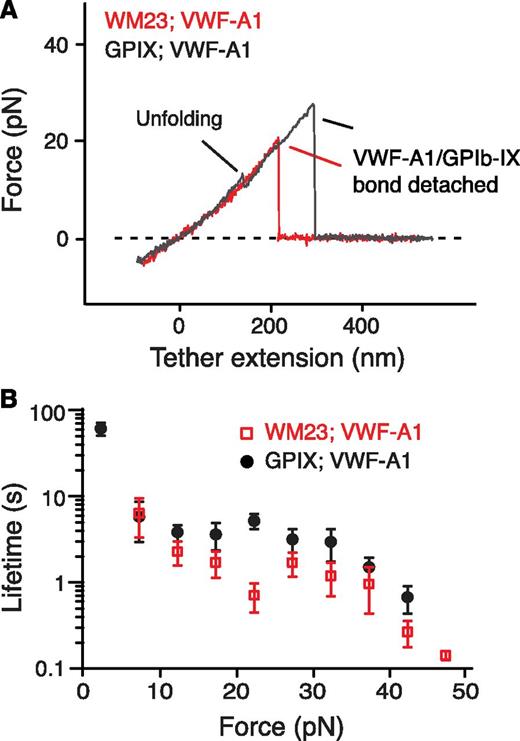
Next we sought to locate the MSD. First, monoclonal antibody WM23, which recognizes an epitope in the C-terminal portion of the macroglycopeptide region (Figure 1A),41 was biotinylated and attached to the fixed bead. No unfolding events were observed when pulling of VWF-A1 from GPIb-IX captured on the WM23 bead, whereas the lifetimes of the GPIb-IX/VWF-A1 bond remained unchanged (Figure 3). Moreover, a mutant GPIb-IX, in which a juxtamembrane residue in the GPIbα cytoplasmic domain was biotinylated, was expressed in HEK293 cells (supplemental Figure 3A-B). Pulling of DNA handle–conjugated WM23 on this GPIbα-biotinylated GPIb-IX produced similar unfolding events before the rupture (supplemental Figure 3C). Therefore, MSD should be located between the WM23 epitope in the macroglycopeptide region and the cytoplasmic domain of GPIbα. In other words, it should be in the stalk region or transmembrane helix of GPIbα. The force required to unfold a transmembrane helix is much stronger (>100 pN)42,43 than what was observed here (∼10 pN). In addition, unfolding of the 25-residue transmembrane helix of GPIbα would have produced an unfolding contour length of ∼10 nm, much shorter than >20 nm observed in the force curves (Figure 2C). Our results, therefore, suggest that the juxtamembrane stalk region of GPIbα contains MSD (Figure 1A).
Localization of MSD in the juxtamembrane stalk region of GPIbα. (A) Overlaid force-distance traces of pulling VWF-A1 on GPIb-IX captured by biotinylated WM23 (red trace) and on GPIb-IX captured by biotin at the GPIX cytoplasmic domain (gray). (B) Plots of lifetimes of the GPIb-IX/VWF-A1 bond vs force (mean ± SEM, n >3).
Localization of MSD in the juxtamembrane stalk region of GPIbα. (A) Overlaid force-distance traces of pulling VWF-A1 on GPIb-IX captured by biotinylated WM23 (red trace) and on GPIb-IX captured by biotin at the GPIX cytoplasmic domain (gray). (B) Plots of lifetimes of the GPIb-IX/VWF-A1 bond vs force (mean ± SEM, n >3).
The recombinant stalk region of GPIbα is structured but unstable
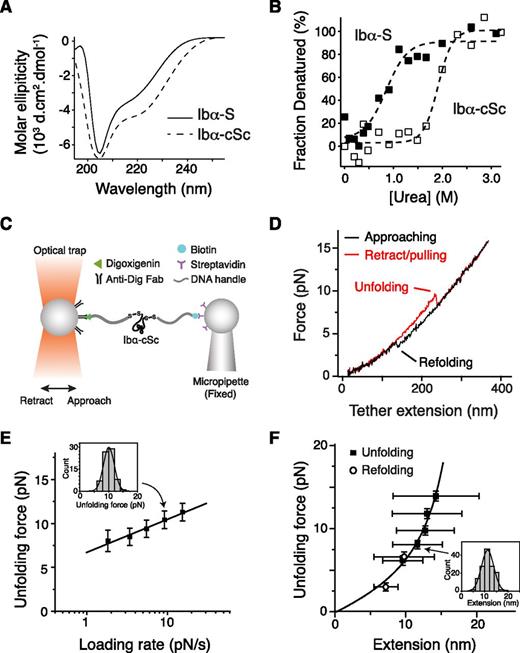
The stalk region of GPIbα has not been studied before, although it had been noted for its higher hydrophobic content than the neighboring macroglycopeptide region.44 A recombinant protein Ibα-S that contains the GPIbα stalk region (residues Ala417-Phe483) was produced (supplemental Figure 4). Deconvolution of its CD spectrum45 indicated that Ibα-S is a structured domain, with 24% of its residues taking α-helical conformation, 19% in β-strand and the rest in coils (Figure 4A). Ibα-S is relatively unstable; it was denatured in <1 M of urea (Figure 4B). Consistently, appending a biotinylated BioTag sequence to its C-terminus significantly altered its structure or stability (supplemental Figure 4). In comparison, a related protein called Ibα-cSc, in which the GPIbα stalk region was flanked by a cysteine on each side, displayed a similar CD spectrum as Ibα-S but was more stable as judged by urea denaturation (Figure 4A-B).
Unfolding of the recombinant stalk region of GPIbα. (A) Overlaid CD spectra of Ibα-S (solid trace) and Ibα-cSc (dashed) in 50 mM Tris, 50 mM NaCl, and 1 mM DTT; pH 7.4 buffer at 20°C. (B) Chemical denaturation plots of Ibα-S (filled squares) and Ibα-cSc (open squares). (C) Illustration of the optical tweezer setup to measure force-induced unfolding of Ibα-cSc. (D) Force-distance traces showing the force-induced unfolding and refolding of Ibα-cSc from pulling at 100 nm/s. (E) Plot of most probable unfolding force as a function of loading rate. Unfolding forces at 5 different loading rates were plotted as histograms (inset). Each histogram was fitted to a Gaussian curve (inset, solid line) to obtain the most probable force. Uncertainty in force is shown as half of the bin width. The solid line is a linear fit of the data to the Bell-Evans model.46 (F) Unfolding force-extension data (black squares) fitted to the WLC model, yielding a contour length of 22.3 ± 0.2 nm. Extension distances were sorted by unfolding force into 2-pN bins. A histogram of extension of each bin (inset) was fitted to a Gaussian curve (inset, solid line) to find peak extension. Unfolding forces were averaged for each bin (n = 43-149 per bin for unfolding, and 40-64 per bin for refolding). Error bars are 1 SD for force and half-width of the Gaussian fit for extension. Open circles represent refolding force-shortening data, which were treated the same way as the unfolding forces-extension.
Unfolding of the recombinant stalk region of GPIbα. (A) Overlaid CD spectra of Ibα-S (solid trace) and Ibα-cSc (dashed) in 50 mM Tris, 50 mM NaCl, and 1 mM DTT; pH 7.4 buffer at 20°C. (B) Chemical denaturation plots of Ibα-S (filled squares) and Ibα-cSc (open squares). (C) Illustration of the optical tweezer setup to measure force-induced unfolding of Ibα-cSc. (D) Force-distance traces showing the force-induced unfolding and refolding of Ibα-cSc from pulling at 100 nm/s. (E) Plot of most probable unfolding force as a function of loading rate. Unfolding forces at 5 different loading rates were plotted as histograms (inset). Each histogram was fitted to a Gaussian curve (inset, solid line) to obtain the most probable force. Uncertainty in force is shown as half of the bin width. The solid line is a linear fit of the data to the Bell-Evans model.46 (F) Unfolding force-extension data (black squares) fitted to the WLC model, yielding a contour length of 22.3 ± 0.2 nm. Extension distances were sorted by unfolding force into 2-pN bins. A histogram of extension of each bin (inset) was fitted to a Gaussian curve (inset, solid line) to find peak extension. Unfolding forces were averaged for each bin (n = 43-149 per bin for unfolding, and 40-64 per bin for refolding). Error bars are 1 SD for force and half-width of the Gaussian fit for extension. Open circles represent refolding force-shortening data, which were treated the same way as the unfolding forces-extension.
Optical tweezer measurement was performed on Ibα-cSc. Each of the flanking cysteine residues in Ibα-cSc was linked to a piece of DNA handle and a bead (Figure 4C). Upon pulling, Ibα-cSc unfolded at 5 to 20 pN and was stretched to 10 to 15 nm (Figure 4D-F), which is comparable with the unfolding force and extension observed in the full-length GPIb-IX. Fitting the most probable unfolding force as a function of loading rate to the single-barrier Bell-Evans model yielded an unfolding rate in the absence of force of 0.008 seconds−1, and a barrier width of 2.6 nm.46 WLC fit of the unfolding force as a function of extension yielded a contour length of 22.3 ± 0.2 nm. The shortening/contraction events during refolding also fell on the same curve (Figure 4F). Because Ibα-cSc contains 67 residues between the 2 Cys residues, when fully stretched it should extend to 26.8 nm, assuming a contour length of 4 Å per residue. Thus, our measurement indicated that the end-to-end distance of folded Ibα-cSc is about 4.5 nm. Overall, these results demonstrated that the GPIbα stalk residues form a structured MSD that has similar unfolding force and extension to that in GPIb-IX.
Lack of unfolding in GPIb-IX that lacks MSD of GPIbα
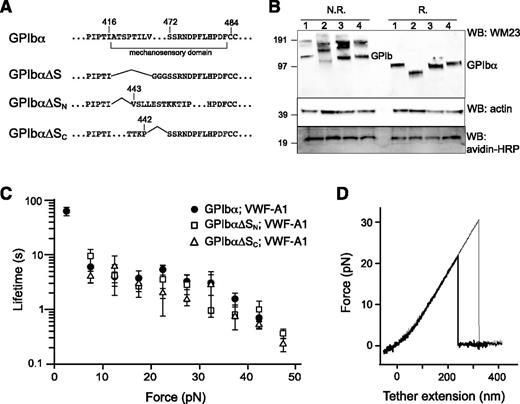
To confirm the location of MSD in GPIb-IX, GPIbα mutants that lack the entire MSD (GPIbα∆S), the N-terminal (GPIbα∆SN) and C-terminal (GPIbα∆SC) halves of the domain were constructed (Figure 5A). Considering the relative instability of MSD, we reasoned that deletion of either half of the domain should effectively leave the remaining sequence without a stable structure and as unfolded. Each GPIbα mutant was coexpressed with HA-GPIbβ and GPIX-biotag to produce the corresponding mutant GPIb-IX complex in the transfected HEK293 cells. Western blot analysis revealed the defective complex assembly by GPIbα∆S, because it was present mostly in the hmwGPIb complex30,34 instead of in the native GPIb complex (Figure 5B). In comparison, both GPIbα∆SN and GPIbα∆SC formed native GPIb and were therefore analyzed further by optical tweezer. VWF-A1 was used in the pulling experiments on mutant GPIb-IX complexes containing either GPIbα∆SN or GPIbα∆SC, yielding similar bond lifetimes as those observed for the wild-type GPIb-IX (Figure 5C). In contrast to the wild-type, no unfolding events were observed in the pulling of either mutant complex (Figure 5D). Overall, these results demonstrated that disrupting the structure in the GPIbα stalk region eliminates the force-induced unfolding event in GPIb-IX, thus confirming the presence of MSD in this region.
Lack of force-induced unfolding in GPIb-IX complexes with altered MSD. (A) Sequences illustrating various deletion mutations in MSD. (B) Expression and assembly of the mutant complexes, as shown by Western blots under nonreducing (N.R.) or reducing (R.) conditions. Lane 1, cells expressing wild-type complex GPIbα/GPIbβ/GPIX; lane 2, GPIbα∆S/GPIbβ/GPIX; lane 3, GPIbα∆SN/GPIbβ/GPIX; lane 4, GPIbα∆SC/GPIbβ/GPIX. (C) Lifetimes (mean ± SEM of >3 experiments) of the bonds between VWF-A1 and the 3 constructs as a function of force. Each plot is identified by the identity of GPIbα subunit in the complex and the pulling ligand. (D) Representative force-distance traces of pulling VWF-A1 on GPIbα∆SN/GPIbβ/GPIX (thick trace) or GPIbα∆SC/GPIbβ/GPIX (thin trace) complex that was captured by biotin at the GPIX cytoplasmic, showing the lack of force-induced unfolding.
Lack of force-induced unfolding in GPIb-IX complexes with altered MSD. (A) Sequences illustrating various deletion mutations in MSD. (B) Expression and assembly of the mutant complexes, as shown by Western blots under nonreducing (N.R.) or reducing (R.) conditions. Lane 1, cells expressing wild-type complex GPIbα/GPIbβ/GPIX; lane 2, GPIbα∆S/GPIbβ/GPIX; lane 3, GPIbα∆SN/GPIbβ/GPIX; lane 4, GPIbα∆SC/GPIbβ/GPIX. (C) Lifetimes (mean ± SEM of >3 experiments) of the bonds between VWF-A1 and the 3 constructs as a function of force. Each plot is identified by the identity of GPIbα subunit in the complex and the pulling ligand. (D) Representative force-distance traces of pulling VWF-A1 on GPIbα∆SN/GPIbβ/GPIX (thick trace) or GPIbα∆SC/GPIbβ/GPIX (thin trace) complex that was captured by biotin at the GPIX cytoplasmic, showing the lack of force-induced unfolding.
Discussion
In this paper we first present evidence for a juxtamembrane MSD in the GPIb-IX complex. VWF-A1–mediated mechanical pulling on the N-terminal domain of GPIbα induced unfolding of an MSD in GPIb-IX (Figures 1 and 2). Follow-up studies using different pulling ligands and immobilization sites localized MSD to the stalk region of GPIbα (Figure 3). Direct characterization of the recombinant stalk region revealed that it contains a structured but relatively unstable domain and that its sensitivity to tensile force is similar to that of the full-length complex (Figure 4). Finally, disrupting the domain structure by deleting a significant portion of the domain eliminated the VWF-A1–induced unfolding event in GPIb-IX (Figure 5), confirming the location of MSD in the juxtamembrane stalk region of GPIbα. Before this study, there have been no reports about the existence of a structured domain in the stalk region of GPIbα. Little information is currently available about this MSD. The CD spectrum of Ibα-S indicates the presence of some secondary structure (Figure 4A), but it is likely that the conformation of MSD in GPIb-IX may be somewhat different from that in recombinant Ibα-S, considering that MSD is relatively unstable and sensitive to its surroundings.
Numerous studies have been carried out to characterize the interaction of VWF or its A1 domain with the N-terminal domain of GPIbα, such as the complex structure, the binding kinetics, and the influence of force.16-20,47,48 In comparison, only a few biophysical studies have focused on the purified GPIb-IX,21,49 primarily because of the difficulty in obtaining this heterotetrameric membrane protein complex. A previous study has used the optical tweezer to measure the unbinding forces between VWF-coated beads and GPIb-IX–expressing Chinese hamster ovary cells,50 but it was not conducted under single-molecule experimental conditions and did not report the unfolding event. Here, site-specific biotinylation of the cytoplasmic domain enabled a systematic dissection of GPIb-IX under a condition that simulated the effect of shear flow on the VWF/GPIb-IX pair. Only through the single-molecule force measurement on the full-length receptor complex was the MSD in GPIb-IX uncovered. This novel setup will be useful in future investigation of GPIb-IX structure-function and could be applied to other mechanosensing receptors. Moreover, we found that the MSD unfolds at forces ranging from 5 to 20 pN, which is similar to the level of forces reported to induce catch- or flex-bond formation in the complex of VWF-A1 with the N-terminal domain of GPIbα.18-20 The unfolding and extension of the MSD could lower the force applied to the VWF/GPIb-IX complex and, therefore, may function together with the VWF-A1/GPIbα interaction to stabilize the tethering and adhesion of platelets to VWF under flow.
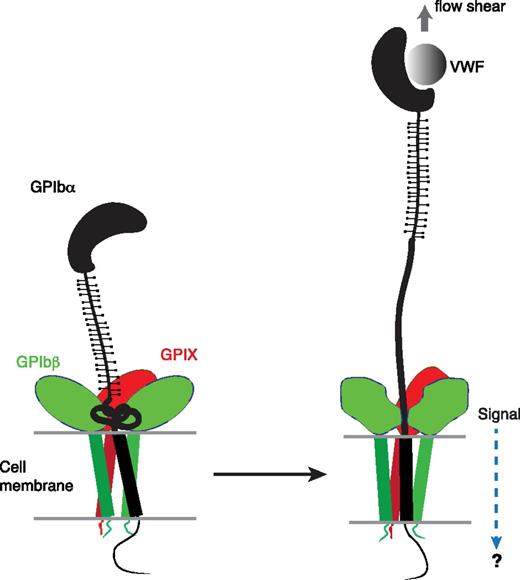
The identification of a juxtamembrane MSD in GPIb-IX has potential implications for the mechanism of platelet mechanosensing. We propose that force-induced unfolding of MSD is the step by which GPIb-IX converts a mechanical signal into a change in protein conformation (Figure 6), a type of signal that could be recognized and transduced further. Under the assumption that platelets behave as spherical beads with the radius of 1 µm, it can be estimated that at 20 dyn/cm2, a typical shear stress found in the microvasculature, the drag force exerted on a single platelet is 64 pN.51,52 Because it takes only 5 to 20 pN to unfold MSD in GPIb-IX, it is plausible that VWF, under physiologic or pathologic shear stress, could exert sufficient force on the tethered GPIb-IX, and the platelet, to induce unfolding of MSD therein. Moreover, the long and unstructured macroglycopeptide region of GPIbα contains 1 to 4 copies of tandem nucleotide repeat sequence (VNTR), and the VNTR polymorphism contributes to the variation in the molecular weight and length of GPIbα (by as much as 15 nm).53 The number of VNTR repeat is not correlated with the occurrence of coronary heart disease, in which arterial thrombosis and the interaction of VWF/GPIb-IX is thought to play an important role.54 This lack of correlation has not been explained in the previously proposed receptor clustering model of GPIb-IX,55 because one would expect that the VNTR polymorphism, with its significant impact on GPIbα length, should affect the extent of GPIbα clustering on the platelet surface and downstream signaling. By comparison, in our model of mechanotransduction, the macroglycopeptide region is proposed to transduce only the tensile force, which should be minimally affected by its length and is therefore consistent with the lack of correlation between VNTR polymorphism and the occurrence of coronary heart disease. Finally, MSD is located in direct contact with the extracellular domains of GPIbβ and GPIX (Figure 1A), which can change conformation in response to an alteration in intersubunit contacts.14,30 Therefore, we propose that unfolding of MSD induces a conformational change in the neighboring GPIbβ and GPIX, which transmits a signal into the cell (Figure 6). This mechanotransducing model, instead of the receptor clustering model, can explain that anti-GPIbβ monoclonal antibody RAM.1 blocks VWF-initiated GPIb-IX–mediated signaling into the platelet without affecting VWF binding to GPIb-IX.49,56 It may also help to explain the critical role of GPIbβ in mediating the procoagulant activity of platelets.57
A model of the proposed mechanosensing mechanism of GPIb-IX. We found in this study that the juxtamembrane MSD in GPIbα is folded, and it unfolds upon VWF-mediated pulling. Our results suggest that on the cell surface, the juxtamembrane MSD in GPIbα is folded in the absence of shear flow (left panel). VWF binding under shear to the N-terminal domain of GPIbα induces unfolding of MSD, and subsequently a conformational change in the adjacent extracellular domains of GPIbβ and GPIX, which sends in a signal across the platelet membrane (right).
A model of the proposed mechanosensing mechanism of GPIb-IX. We found in this study that the juxtamembrane MSD in GPIbα is folded, and it unfolds upon VWF-mediated pulling. Our results suggest that on the cell surface, the juxtamembrane MSD in GPIbα is folded in the absence of shear flow (left panel). VWF binding under shear to the N-terminal domain of GPIbα induces unfolding of MSD, and subsequently a conformational change in the adjacent extracellular domains of GPIbβ and GPIX, which sends in a signal across the platelet membrane (right).
The identification of MSD in GPIb-IX also provides new insights into the pathogenesis of BSS. GPIbα is quickly degraded in transfected cells when it is not expressed with GPIbβ and GPIX,58 but the underlying molecular basis has remained unclear. Here we showed that Ibα-S in isolation is not stable (Figure 4 and supplemental Figure 4). It is therefore conceivable that the instability of MSD in GPIbα could contribute to the rapid degradation of GPIbα when it is expressed in the absence of GPIbβ and GPIX.58 GPIbβ and GPIX, being close to MSD in the complex, may either stabilize the domain or prevent its induction of degradation. Furthermore, the potential involvement of GPIbβ and GPIX in detecting the unfolding of MSD and further propagating the signal offers a plausible functional justification for their elaborative complexation with GPIbα.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Michael C. Berndt for sharing of the WM23 antibody, and the Emory Children’s Pediatric Research Center Flow Cytometry Core for technical support.
This work was supported in part by National Institutes of Health, National Heart, Lung, and Blood Institute grant HL082808 (R.L.) and American Heart Association grant 11SDG5420008 (X.F.Z.).
Authorship
Contribution: W.Z., W.D., L.Z., Y.X., W.Y., X.L., and Y.W. performed research and analyzed results; W.Z., W.D., and L.Z. prepared the figures and wrote the paper; J.D.K. provided critical reagents; and X.F.Z. and R.L. designed research, analyzed results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Renhao Li, Department of Pediatrics, Emory University, 2015 Uppergate Dr NE, Room 440, Atlanta, GA 30322; e-mail: renhao.li@emory.edu; and X. Frank Zhang, Department of Mechanical Engineering and Mechanics, Bioengineering Program, Lehigh University, 19 Memorial Dr West, Bethlehem, PA 18015; e-mail: frank.zhang@lehigh.edu.
References
Author notes
W.Z. and W.D. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal