Abstract
Posttranslational modifications of histone proteins represent a fundamental means to define distinctive epigenetic states and regulate gene expression during development and differentiation. Aberrations in various chromatin-modulation pathways are commonly used by tumors to initiate and maintain oncogenesis, including lymphomagenesis. Recently, increasing evidence has demonstrated that polycomb group (PcG) proteins, a subset of histone-modifying enzymes known to be crucial for B-cell maturation and differentiation, play a central role in malignant transformation of B cells. PcG hyperactivity in B-cell lymphomas is caused by overexpression or recurrent mutations of PcG genes and deregulation of microRNAs (miRNAs) or transcription factors such as c-MYC, which regulate PcG expression. Interplays of PcG and miRNA deregulations often establish a vicious signal-amplification loop in lymphoma associated with adverse clinical outcomes. Importantly, aberrant enzymatic activities associated with polycomb deregulation, notably those caused by EZH2 gain-of-function mutations, have provided a rationale for developing small-molecule inhibitors as novel therapies. In this review, we summarize our current understanding of PcG-mediated gene silencing, interplays of PcG with other epigenetic regulators such as miRNAs during B-cell differentiation and lymphomagenesis, and recent advancements in targeted strategies against PcG as promising therapeutics for B-cell malignancies.
Introduction
Histone posttranslational modifications represent a fundamental mechanism for regulating DNA accessibility in various DNA-templated processes such as gene transcription.1 Dysregulation of chromatin-modifying mechanisms is one of the central oncogenic pathways in human cancer,1-3 including B-cell malignancies.4-6
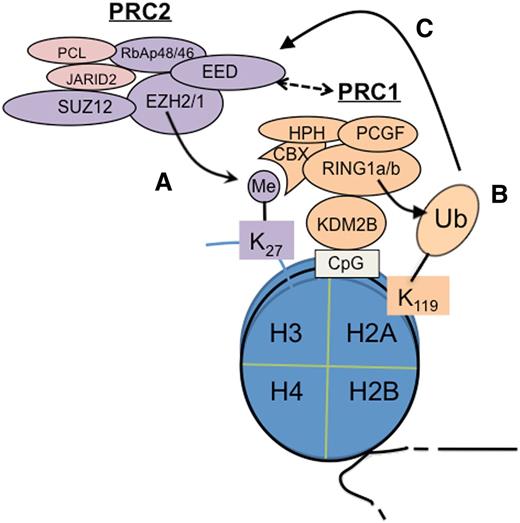
Among various chromatin-modifying factors, polycomb group (PcG) proteins are critical for controlling gene expression, maintaining repressive chromatin states, and defining cellular identities during development.7,8 PcG proteins act in multimeric complexes known as polycomb repressive complexes (PRCs). Two major PcG complexes exist in mammalian cells: PRC1 and PRC2. Biochemically, PRC1 employs an E3 ligase, RING1A or RING1B, to induce monoubiquitination of histone H2A, lysine 119 (H2AK119ub1) (Figure 1), a reaction that requires essential cofactors such as BMI1.8 PRC2 utilizes an enzymatic subunit, enhancer of zeste homolog 2 (EZH2) or related EZH1, to methylate histone H3, lysine 27 (H3K27; Figure 1)7 ; other PRC2 subunits (EED and SUZ12) and accessory cofactors such as JARID2 and polycomb-like harbor either DNA- or histone-binding activities to modulate PRC2 activity and mediate its targeting or spreading on chromatin.7-9 H2AK119ub1 and H3K27 trimethylation (H3K27me3) are prominent histone markers associated with gene silencing, indicating a causal role of PcG-mediated enzymatic activity in transcriptional regulation.7,8 H3K27me3 also coexists with the gene-activation–associated trimethylation of histone H3, lysine 4 (H3K4me3) at “bivalent domain genes” to maintain genes in a repressed but poised conformation, which can be subsequently activated or stably repressed according to lineage-specific differentiation programs.1
Cooperation of PRC2 and PRC1 in epigenetic silencing of genes. PRC2 catalyzes trimethylation of histone H3 at lysine 27 (H3K27me3) (A), which is recognized and bound by CBX proteins such as CBX7, a PRC1 subunit, to subsequently recruit PRC1 for induction of monoubiquitination of histone H2A at lysine 119 (H2AK119ub1)7,8 (B).Conversely, recent studies show that a variant form of PRC1 can act upstream of PRC2 to initiate formation of the polycomb domain; in this case, H2AK119ub1 serves as a PRC2 recruitment mechanism (C).12-14 In addition, EED is also shown to interact to PRC1 physically.15 CpG, cytosine guanine dinucleotide; Me, trimethylation; Ub, ubiquitination.
Cooperation of PRC2 and PRC1 in epigenetic silencing of genes. PRC2 catalyzes trimethylation of histone H3 at lysine 27 (H3K27me3) (A), which is recognized and bound by CBX proteins such as CBX7, a PRC1 subunit, to subsequently recruit PRC1 for induction of monoubiquitination of histone H2A at lysine 119 (H2AK119ub1)7,8 (B).Conversely, recent studies show that a variant form of PRC1 can act upstream of PRC2 to initiate formation of the polycomb domain; in this case, H2AK119ub1 serves as a PRC2 recruitment mechanism (C).12-14 In addition, EED is also shown to interact to PRC1 physically.15 CpG, cytosine guanine dinucleotide; Me, trimethylation; Ub, ubiquitination.
In a simplistic hierarchical model, PRC2 acts upstream of PRC1 as H3K27me3 serves as a “docking” site for CBX, a chromodomain-containing protein (Figure 1A), which then recruits PRC1 to induce H2AK119ub17,8 (Figure 1B). However, more recently, data have demonstrated that PRC1 recruitment is both PRC2 dependent and PRC2 independent.10,11 Furthermore, recent studies show that PRC1 can act upstream of PRC2. In this case, a PRC1 variant utilizes KDM2B, a CxxC-domain protein, to bind to the nonmethylated cytosine guanine dinucleotide sequence where PRC1-induced H2AK119ub1 recruits PRC2 via an unknown mechanism12-14 (Figure 1C). EED, a PRC2 subunit, also physically interacts with PRC1, thus linking PRC2 to PRC1.15 Overall, PRC2 and PRC1 cooperate and enforce gene silencing via positive-feedback loops.
Increasing evidence has revealed crucial roles of PcG proteins in myriad biological processes, including self-renewal, differentiation, cell-cycle control, senescence, and gene expression and imprinting,7,8,16,17 all of which have been linked to oncogenesis when deregulated. Notably, PcG genes were found mutated in B-cell malignancies. B lymphoma Mo-MLV insertion region 1 homolog (BMI1, also known as polycomb group ring finger 4 or PCGF4 [Figure 1]) was originally isolated as a gene upregulated in murine B-cell lymphomas18 ; recurrent gain-of-function mutations of EZH2 were identified in germinal center (GC) B-cell lymphomas.4,19,20 Here, we focus on deregulations of PcG and cofactors during the initiation and development of B-cell malignancies. We also discuss the interplays between PcG and other epigenetic regulators such as microRNAs (miRNAs), histone deacetylases (HDACs), and DNA methytransferases (DNMTs). Finally, we summarize recent progress in development of PcG-specific inhibitors as novel therapies of B-cell malignancies.
Biological function of PcG proteins in B-cell development and lymphomagenesis
The development and differentiation of B-cell lineages initially occur with progenitor B-cell expansion and V(D)J gene rearrangement, a DNA recombination process that produces clonally unique, immunoglobulin variable regions for antigen recognition.21 Upon antigen stimulation, B cells undergo activation through proliferation, somatic hypermutation, and antibody class switching, which occur in the GCs of secondary lymphoid tissues. A proliferative feature of GC B lymphocytes, with concomitant attenuation of their DNA damage repair function and ongoing somatic hypermutation, increases the likelihood of oncogenic mutation, genomic instability, and subsequent lymphomas. B-cell development is tightly controlled by genetic and epigenetic mechanisms, including DNA methylation, histone modification, chromatin remodeling,22 and noncoding RNAs.23 During normal B-lymphocyte differentiation, expression of PRC1 and PRC2 genes shows a restricted, stage-specific pattern. BMI1 and its PRC1 partners are primarily detected among resting B cells in the GC mantle zone and in nondividing centrocytes of the GC follicles; these PRC1 genes are silenced in proliferating follicular centroblasts, which then express the PRC2 genes instead.24-26 In contrast, lymphomas generally lose such a mutually exclusive expression pattern, and altered expression of PRC1 and PRC2 genes is a general theme in lymphomas, including diffuse large B-cell lymphomas (DLBCL),27 follicular lymphomas (FLs), and mantle cell lymphomas (MCLs).28 These findings suggest essential regulatory roles of PRC1 and PRC2 in both normal B-lymphocyte development and lymphoma pathogenesis.
PRC1 in B-cell development and lymphomagenesis
BMI1
Bmi1 (also known as PCGF4 [Figure 1]) was initially discovered from a locus activated by viral integration in murine lymphomas.18 BMI1 controls a range of B-cell developmental genes, including lineage master regulators Ebf1 and Pax5.29 Bmi1 deficiency causes conversion of the “bivalent domain” states associated with Ebf1 and Pax5 to a monovalent active state, resulting in their premature expression and accelerated lymphoid differentiation.29 BMI1 also directly represses expression of the tumor suppressors p16Ink4a/p19Arf and p15Ink4b; therefore, BMI1 overexpression prevents c-MYC–mediated apoptosis and was sufficient to induce lymphoma, a process further accelerated by c-MYC.18,30 Furthermore, BMI1 represses the proapoptotic genes Noxa and Bim, supporting its prosurvival role in B-cell development and lymphomas.31,32 In human B-cell lymphomas, BMI1 overexpression is common in almost all subtypes.33 Expression of BMI1 alone or in combination with EZH2 characterizes aggressive B-cell lymphomas with unfavorable prognosis.27,33,34 Recently, a novel t(10;14)(p12;q32) translocation was identified in chronic lymphocytic leukemia and MCLs leading to IgH-BMI1 rearrangement and BMI1 overexpression35 ; IgH-BMI1 rearrangement was acquired during tumor high-grade transformation and correlated with chemoresistance.35 Transcriptome analyses of multiple cancers found that BMI1-driven gene signatures define a phenotype of cancer stem cells,36 suggesting that BMI1 confers malignant cells with features of cancer stem cells, the rare cancerous subpopulations that confer drug resistance and regeneration abilities.16,37 Indeed, BMI1-mediated repression of p16Ink4a/p19Arf was shown to be essential for self-renewal of hematopoietic stem cells.38 Overall, these studies support critical roles of BMI1 in promoting lymphoma progression and conferring therapy resistance.
Other PRC1 factors
Evidence exists showing direct involvement of other PRC1 components in B-cell lymphomas. CBX7 (Figure 1) was found highly expressed in GC lymphocytes and GC-derived FLs, where its elevated expression was correlated with c-MYC expression and an aggressive feature.39 Lymphoid-specific overexpression of Cbx7 in mice initiated lymphomagenesis and cooperated with c-MYC to produce aggressive B-cell lymphomas.39 Similar to BMI1, Cbx7 overexpression was linked to repression of p16Ink4a/p19Arf.39 RING1A is associated with the risk of non-Hodgkin lymphomas40 and high expression of RING1B detected in lymphomas such as DLBCLs and Burkitt lymphoma.41 However, in the absence of p16Ink4a, RING1B deficiency accelerated lymphomagenesis through upregulation of cyclin D2 and Cdc6.42 Thus, PRC1 harbors both oncogenic and tumor-suppressive roles in different contexts, which is reminiscent of PRC2’s dual functions described among different hematopoietic malignancies.43-45
PRC2 in B-cell development and lymphomagenesis
EZH2 is highly expressed in lymphoid progenitors and required for efficient V(D)J recombination in pro–B cells.46 EZH2 is silenced in resting GC B cells but massively upregulated when GC B cells get activated and undergo rapid proliferation and immunoglobulin affinity maturation26,47 ; EZH2 blocks the DNA damage response pathways, allowing cells to survive the somatic hypermutation during antibody maturation.26 Expression of EZH2 strongly associates with B-cell malignancies, with its high levels correlated with the Ki67 labeling index, lymphoma aggressiveness, and unfavorable prognosis.33,34 The highest percentage of EZH2 positivity was found in 100% of Burkitt lymphomas, 87.5% of grade-3 FLs, and 85.7% of DLBCLs. Multivariate survival analysis identified EZH2 as the strongest prognostic predictor of inferior outcomes of MCLs.27 SUZ12 expression was also found to be restricted to proliferating lymphoid cells during development and at high levels in MCLs, in comparison with its general absence in nontumorous mantle zone cells.48
The importance of PRC2 in lymphomagenesis is further strengthened by recent identification of recurrent missense mutations in EZH2, with the most prevalent ones altering a single residue in the catalytic domain, Y641 (the numeration of EZH2 amino acids based on a short isoform of EZH2 [National Center for Biotechnology Information accession Q15910.2]), among ∼10% to 20% of GC-derived B-cell lymphomas such as DLBCLs and FLs.4,5,49 These EZH2 mutations are likely to be early lesions during lymphomagenesis.6,49 Biochemically, EZH2Y641 mutations alter substrate specificity of EZH2.19,20,50 Being a catalytic subunit of PRC2, EZH2 induces sequential mono-, di-, and trimethylation of H3K27, with the highest methylation status most strongly associated with gene silencing.51 Wild-type EZH2 has a greater catalytic efficiency for conducting monomethylation of H3K27 (H3K27me1) and a diminished efficiency for subsequent reactions (mono- to di- and di- to trimethylations).19,20,50 In contrast, lymphoma-associated EZH2Y641X mutations (X means Asn/Phe/Ser/His/Cys) show the exactly opposite substrate specificity, displaying limited ability to induce H3K27 monomethylation yet extremely high efficiency catalyzing the H3K27 di- to trimethylation reaction.19,20,50 Such enzymatic differences between wild-type and EZH2Y641X mutant protein suggest that EZH2Y641X mutations must occur heterozygously in lymphomas, which is indeed the case in human patients,4,5,49 allowing for EZH2Y641X to cooperate with wild-type EZH2 to induce a global increase in H3K27me319,20,50 and aberrant transcriptional alteration. Later on, 2 additional somatic mutations, EZH2A677G and EZH2A687V, were identified at a lower frequency (∼1% to 3%) among GC B-cell lymphomas,5,49,52,53 and they demonstrate enzymatic properties distinct from EZH2Y641X mutants.50,52-54 EZH2A677G and EZH2A687V possess the almost equally enhanced catalytic activity towards all the H3K27 substrates with different methylation status.50,52-54 Thus, EZH2A677G and EZH2A687V mutants are able to induce a global increase in H3K27me3 without the need for wild-type EZH2.50 Besides kinetics, EZH2Y641X also affects protein stability.55 Phosphorylation of Y641, a known phosphorylation site of JAK2 kinases, leads to interaction of EZH2 with β-TrCP, a SCF E3 ubiquitin ligase, and promotes EZH2 degradation. Loss of this phosphorylation site due to somatic Y641 mutations reduces EZH2 turnover, which has been postulated to contribute to the hyper-H3K27me3 phenotype.55 Taken together, different gain-of-function mutations induce EZH2 hyperactivity through distinct molecular mechanisms, and despite the fact that lymphomas carrying different EZH2 mutations may possess different levels of the lowly methylated H3K27, they all have a consistently higher level of H3K27me3.50,52-54
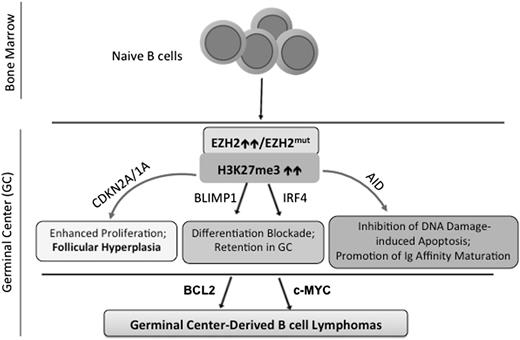
Recent studies have provided a better understanding of the in vivo function of EZH2 and mutation in normal B-cell development and lymphomagenesis.47,56-58 Using Ezh2 knockout mice, Beguelin et al56 and Caganova et al47 have independently shown that EZH2 is crucial for the formation of GCs and GC B-cell development. Mechanistically, EZH2 represses myriad downstream genes including the negative cell-cycle regulators Cdkn2a (p16Ink4a/p19Arf) and Cdkn1a/p21 and crucial transcription factor genes IRF4 and BLIMP1/PRDM1, which are known to be essential for post GC B-cell development47,56,57 (Figure 2). Indeed, depletion of EZH2 from lymphomas suppressed their proliferation and attenuated tumor formation.57 These studies support the notion that EZH2 hyperactivation promotes malignant transformation by repressing both antiproliferative and differentiation-inducing programs. Furthermore, Ezh2-deficient GC B cells had profound impairments in GC responses and memory B-cell formation and failed to protect themselves from the genotoxic damages induced by activation-induced cytidinedeaminase,47 an enzyme critical for somatic hypermutation and antibody affinity maturation,21 demonstrating an essential role of EZH2 in the GC B-cell development (Figure 2). B-cell–specific expression of the EZH2Y641N or EZH2Y641F mutant in transgenic mice elevated the global H3K27me3, promoted a high proliferation of GC B cells, and resulted in follicular hyperplasia.56,58 However, additional oncogenic events are required for neoplastic transformation, although GC-derived lymphomas remain addicted to EZH2 mutations. It has been shown that EZH2Y641N/F mutants cooperate with BCL2 to generate malignant GC B-cell lymphomas56 ; similarly, genetic interaction of EZH2Y641F and MYC in transgenic mice gave rise to high-grade lymphomas with a mature B-cell phenotype.58 Collectively, these findings have shown that dynamic expression of EZH2 allows expansion and development of GC B cells, which undergo terminal differentiation and develop into antibody-secreting cells and plasma cells as EZH2 expression declines (Figure 2). EZH2 hyperactivity perturbs the fine balance of GC B-cell proliferation and differentiation, permanently locking GC B cells in an immature and proliferative state, a prelude to full-blown lymphoma. Although not sufficient on its own to cause lymphoma, EZH2 gain-of-function mutations serve as a driver of lymphomagenesis and collaborate with additional lesions to generate and/or accelerate GC B-cell lymphomas (Figure 2).
Biological functions of EZH2 in normal B-cell development and lymphomagenesis. During B-cell differentiation, naive B cells enter the GC and EZH2 is transcriptionally upregulated during GC B-cell maturation.26,47 Via induction of H3K27me3, EZH2 then transcriptionally represses a myriad of downstream effector genes, which at least include the negative cell-cycle regulators (CDKN2A and CDKN1A) and B-cell differentiation-promoting transcription factors (IRF4 and BLIMP1/PRDM1), hence allowing for rapid expansion of immature B cells47,56,57 ; in addition, EZH2 protects GC B cells from the genotoxic damages induced by activation-induced cytidinedeaminase (AID),47 an enzyme critical for immunoglobulin affinity maturation via a mechanism of somatic hypermutation that modifies the immunoglobulin variable region of the rearranged antibody genes in GC B cells.21 EZH2 levels decrease as B cells exit the GC, enabling derepression of EZH2-targeted genes and hence terminal differentiation.47,56,57 However, EZH2 hyperactivity (either somatic mutation or overexpression) disrupts such fine equilibrium, continuously enhances H3K27me3, and results in exaggerated silencing of EZH2 targeted genes, which then block GC B-cell differentiation and promote their proliferation and survival. EZH2 mutations alone lead to follicular hyperplasia, and, with acquisition of additional oncogenic events such as upregulation of BCL2 or c-MYC, EZH2 mutations cooperatively enable or accelerate malignant transformation of GC B cells.56,58
Biological functions of EZH2 in normal B-cell development and lymphomagenesis. During B-cell differentiation, naive B cells enter the GC and EZH2 is transcriptionally upregulated during GC B-cell maturation.26,47 Via induction of H3K27me3, EZH2 then transcriptionally represses a myriad of downstream effector genes, which at least include the negative cell-cycle regulators (CDKN2A and CDKN1A) and B-cell differentiation-promoting transcription factors (IRF4 and BLIMP1/PRDM1), hence allowing for rapid expansion of immature B cells47,56,57 ; in addition, EZH2 protects GC B cells from the genotoxic damages induced by activation-induced cytidinedeaminase (AID),47 an enzyme critical for immunoglobulin affinity maturation via a mechanism of somatic hypermutation that modifies the immunoglobulin variable region of the rearranged antibody genes in GC B cells.21 EZH2 levels decrease as B cells exit the GC, enabling derepression of EZH2-targeted genes and hence terminal differentiation.47,56,57 However, EZH2 hyperactivity (either somatic mutation or overexpression) disrupts such fine equilibrium, continuously enhances H3K27me3, and results in exaggerated silencing of EZH2 targeted genes, which then block GC B-cell differentiation and promote their proliferation and survival. EZH2 mutations alone lead to follicular hyperplasia, and, with acquisition of additional oncogenic events such as upregulation of BCL2 or c-MYC, EZH2 mutations cooperatively enable or accelerate malignant transformation of GC B cells.56,58
In contrast to the oncogenic role of EZH2 in B-cell lineages, the tumor-suppressive roles of PRC2 in T-cell acute lymphoblastic leukemia44,45 and myeloid malignancies43 were identified due to a range of missense, nonsense, and frameshift mutations in EZH2, SUZ12, or EED (Figure 1). These lesions can be homozygous, are found throughout the gene, and are generally predicted to disable PRC2 activity, implying its disease-dependent functions. These observations emulate those obtained in Eµ-Myc lymphoma models showing that PRC2 can be a tumor suppressor in Eµ-Myc–induced lymphomagenesis, wherein the lymphoma onset was accelerated by knockdown of Suz12 or Ezh2.59 Such an effect is likely due to the enhanced self-renewal of B-lymphoid progenitors upon PRC2 loss,59 which is in contrast to cooperation between EZH2Y641F and Eµ-Myc reported by Berg et al in GC B-cell compartments.58 These studies emphasize the complicated, context-dependent role of PRC2 in oncogenesis, which might be due to a tightly controlled expression pattern of EZH2 throughout B-cell lineage differentiation. As a result, PRC2 activity at various developmental stages may either suppress or facilitate lymphomagenesis. Indeed, EZH2 expression is high in pro–B cells and decreased in pre–B cells and becomes nearly undetectable in immature naive cells46,57 ; EZH2 is then upregulated again during affinity maturation in GC B cells.26,47 Therefore, it could be the case that whereas PRC2 restricts the proliferative and self-renewal potential of immature B-lymphoid progenitors, its gain-of-function mutations stimulate proliferation specifically in maturing GC B cells. Further work with mouse models engineered to overexpress or delete EZH2 at each specific stage of B-cell differentiation shall provide insight into the role of PRC2 in various B-cell malignancies.
Interplay of PcG with other epigenetic enzymes
In addition to intrinsic enzymatic functions, PcG complexes also recruit other chromatin-modifying factors such as HDACs and DNMTs to re-enforce transcriptional repression. PRC2 recruits HDAC1-3, linking 2 distinctive repressive machineries together.60 Several broad-spectrum HDAC inhibitors, including sodium butyrate, decrease the messenger RNA and protein levels of BMI1 and EZH2 in cancer cells.61 These findings implicate that HDACs positively regulate cellular PcG levels and that epigenetic control of gene expression is governed by cooperation of PRC2 and HDACs. In addition, histone methylation influences DNA methylation and, in turn, DNA methylation serves as an instructive template for histone modification. In cancer, PcG-suppressed genes are likely to be associated with DNA hypermethylation, and hypermethylated promoters more frequently premarked with PcG.8,62,63 Indeed, EZH2 directly associates with DNMTs.64 This mechanism also appears to be operative in B-cell lymphoma because DNA methylation profiling of lymphomas revealed a significant enrichment of PcG targets at the de novo methylated genes,65 indicating that crosstalk between histone and DNA methylation may form a double “locking” mechanism of an undifferentiated cell state during malignant transformation. Perturbation of cellular factors that antagonize PcG, such as trithorax group (TrxG) proteins,17 may equally influence the regulatory roles and biological outputs of PcG complexes. Indeed, direct sequencing of patients with B-cell malignancies has recently led to identification of recurrent damaging mutations of several TrxG genes such as MLL2, p300, and CBP.5,6,66 Loss-of-function mutations of TrxG and gain-of-function mutations of PcG genes may equally perturb a fine equilibrium of histone methylation dynamics during B-cell lymphomagenesis.
Interplay of PcG with miRNAs
miRNAs are 22-nucleotide, noncoding single-stranded RNAs that can repress gene expression at a posttranscriptional level. miRNAs are increasingly recognized as one of the major players in numerous biological processes, and their downderegulation is often seen in tumors, suggesting their tumor-suppressive roles. It can be anticipated that miRNA deregulation can contribute to PcG deregulation. Indeed, EZH2 was the first PcG gene shown to be regulated by miRNA.67,68 By targeting the 3′ untranslated region of EZH2 messenger RNA, miR-101 and miR-26 repress cellular EZH2 levels.67,68 miRNAs that repress PRC1 genes were also identified.69-71 Downregulation and deletion of these miRNAs are frequent in various tumors, including prostate cancer and lymphomas.67,68 Conversely, PcG proteins also contribute to miRNA expression and deregulation during malignant development, given their frequent alterations found in tumors. Indeed, many miRNA genes are repressed by PRC2 and demarcated with H3K27me3.72 PRC2 represses miR-31 in adult T-cell lymphoma, leading to activation of nuclear factor κB oncogenic signaling.73 Thus, these findings have shown an intriguing interplay between miRNAs and PcG. Below, we summarize recent advances in understanding their interactions in cancers, especially B-cell malignancies, which reveal the hitherto-unappreciated regulatory circuits involving miRNA and epigenetic factors.
PRC2–miRNA–PRC1 circuitry
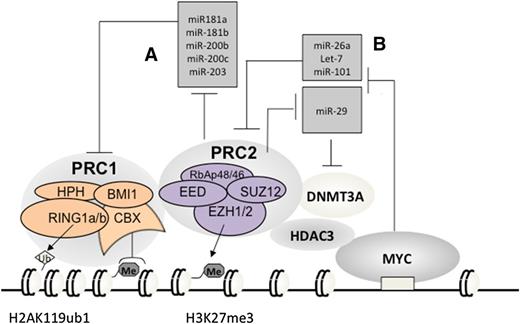
A subset of miRNAs, including miR-181a, miR-181b, miR-200b, miR-200c, and miR-203 (Figure 3A), are transcriptionally silenced by PRC2 in cancer.69 Interestingly, these miRNAs repress PRC1 genes such as BMI1 and RING1B69-71 (Figure 3A). It has been shown that downregulation of these miRNAs such as miR-200c ensures the cellular level and functionality of PRC1 in stem cells70,71 and cancers including lymphoma,69-71,74 promoting cell “stemness” properties. These data demonstrate that expression of PRC1 and PRC2 is integrated through a network of regulatory miRNAs wherein epigenetic repression of PRC1-targeting miRNAs by PRC2 establishes a positive feedback loop, ensuring coexpression and cooperation of 2 major PcG complexes.
Vicious amplification loops involving a myriad of PcG proteins and miRNAs. Repression of PRC1-repressing miRNAs by PRC2 (A) establishes a positive-feedback loop ensuring coexpression and cooperation of 2 main PcG repressor complexes in stem and cancer cells; c-MYC, which is frequently translocated or overexpressed in Burkitt lymphoma and other B-cell lymphoma types, assembles a gene-silencing complex with PRC2 and HDACs to downregulate a list of tumor-suppressive miRNAs that can repress EZH2 and DNMT3A (B), hence establishing positive-feedback loops to enforce expression and functionality of PRC2 in B-cell lymphomas. Me3, trimethylation. Ub1, mono-ubiquitination.
Vicious amplification loops involving a myriad of PcG proteins and miRNAs. Repression of PRC1-repressing miRNAs by PRC2 (A) establishes a positive-feedback loop ensuring coexpression and cooperation of 2 main PcG repressor complexes in stem and cancer cells; c-MYC, which is frequently translocated or overexpressed in Burkitt lymphoma and other B-cell lymphoma types, assembles a gene-silencing complex with PRC2 and HDACs to downregulate a list of tumor-suppressive miRNAs that can repress EZH2 and DNMT3A (B), hence establishing positive-feedback loops to enforce expression and functionality of PRC2 in B-cell lymphomas. Me3, trimethylation. Ub1, mono-ubiquitination.
EZH2/c-MYC–miRNA–EZH2 circuitry
Recent studies of B-cell malignancies also unveiled a second circuitry involving EZH2, miRNAs, and c-MYC, an oncogenic transcription factor almost invariably translocated in Burkitt lymphoma. c-MYC assembles a repressive complex with PRC2 and HDACs to downregulate a broad spectrum of tumor-suppressive miRNAs, including miR-15a/16-1, miR-26, miR-27, miR-29, let-7, miR-494, and miR-548m67,75-79 (Figure 3B). A similar c-MYC–PRC2 complex also represses miR-101 in hepatocellular carcinoma.80 Among these repressed miRNAs, miR-101 and miR-26 were recurrently deleted in tumors including lymphoma67,68 ; miR-15a/16-1 targets BCL2 and acts as tumor suppressor in chronic lymphocytic leukemia.81 Interestingly, several of these c-MYC–repressed miRNAs, including miR-26a, miR-101, and let-7, actually repress EZH2 directly67,75,78 (Figure 3B); miR-29, a family of miRNAs known to be involved in B-cell lymphomagenesis,75,82 was shown to downregulate DNMT3A, a PRC2-interacting factor, in chronic lymphocytic leukemia83 (Figure 3B). Thus, via recruitment of PRC2 and HDACs, c-MYC, a prominent lymphoma-promoting factor, represses miRNAs that negatively regulate EZH2 and its cofactors, establishing a positive-feedback loop for enforcing polycomb genes expression and functionality in B-cell lymphomas. As knocking down EZH2 and HDACs led to re-expression of the MYC-repressed miRNAs,75,78,84,85 the existing pharmacologic agents for inhibition of these c-MYC–associated corepressors shall represent a promising way to disrupt such a vicious amplification loop associated with lymphomagenesis.
Epigenetic therapy and perspective
Epigenetic deregulation of chromatin structure and function leads to aberrant gene expression and oncogenesis. Consequently, epigenetic therapies aim to restore normal chromatin-modification patterns through inhibition of the deregulated epigenetic machinery. HDAC and DNMT inhibitors are among the first promising agents for epigenetic therapies,2 and, more recently, specific inhibitors for PcG proteins have been developed.
Targeting PRC1
A recent high-throughput screen discovered a small-molecule compound PTC-209 as an inhibitor of BMI1.86 PTC-209 inhibited expression of BMI1 and induced a dose-dependent reduction of global H2AK119ub1.86 BMI1 knockdown conferred PTC-209 insensitivity, indicating its specificity.86 Similar to BMI1 knockdown, PTC-209 treatment inhibited self-renewal of cancer-initiating stem cells.86 This study implicates that BMI1 has the potential to be developed as a drug target for treating B-cell lymphomas with BMI1 overexpression.
Targeting PRC2
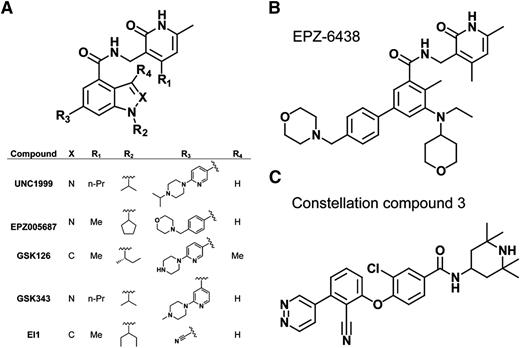
Several highly selective small-molecule inhibitors of PRC2 (with Ki values within the low-nanomolar range) have recently been discovered,87-93 many of which possess a common pyridone-containing motif that confers EZH2 or EZH1 inhibition (Figure 4A). Among them, EPZ00568788 and GSK12687 show high selectivity for EZH2 vs other methyltransferases, with >50- to 150-fold selectivity for EZH2 over EZH1 (Figure 4A). Early success was seen in treating B-cell lymphomas bearing EZH2 gain-of-function mutation with these inhibitors in DLBCL xenografts in mice.56,87-93 GSK126,87 EPZ005687, and EPZ-643888,94 (Figure 4A-B) show their particular effectiveness in suppressing growth of the EZH2-mutant lymphomas vs those with wild-type EZH2. At the transcriptome level, and in contrast to minimal transcriptional responses in wild-type EZH2, drug-insensitive lines, reactivation of the formerly H3K27me3-demarcated genes was generally seen in drug-sensitive GC B-cell (GCB) type of DLBCL lines.87,93 However, only a limited number of upregulated genes were found common across different inhibitor-sensitive lines, although upregulated genes in each individual line are enriched in those related to cell-cycle and apoptotic regulation,87 thus highlighting a challenge to define relevant targets presumably due to variations of genetic backgrounds. The biological responses such as growth suppression after treatment with EZH2 inhibitors show a delayed pattern in comparison with the biochemical responses; the diminution of H3K27me3 was apparent within 24 to 48 hours posttreatment, and yet the cellular response is usually not fully presented until days 4 to 7 posttreatment and beyond.87 Such a delayed effect with the PRC2 inhibitors was seen in acute myeloid leukemia95 and for inhibitors of the histone methyltransferase DOT1L,96 which is in contrast to quick responses associated with HDAC inhibitors. It has been speculated that additional time and/or chromatin factors may be required to reverse the histone methylation-regulated events, and, alternatively, catalytic inhibition induces compensatory recruitment of more enzymatic complexes at crucial gene targets, thus delaying demethylation. It is worth noting that PRC2 possesses noncanonical functions such as methylation of nonhistone substrates97 or acting as transcriptional activator,98 but it remains unclear if these mechanisms exist in lymphomas. Treatment of EZH2-mutant, GCB-DLBCL xenograft models with GSK126 or GSK343 (Figure 4A) resulted in tumor regression,56,87 and the inhibitor was well tolerated. 87 Currently, various PRC2 inhibitors are under clinical evaluation and it would be exciting to see whether PRC2 inhibition provides clinical benefits for lymphoma patients.
Highly selective, small-molecule inhibitors of PRC2. (A) Scaffold demonstrating that several of the recently developed EZH2 or EZH2/1 inhibitors all possess a pyridone motif as well as an indole or indazole core. The inserted table details the identity of each designated substituent of the described inhibitors. (B) Chemical structure of EPZ-6438. (C) Chemical structure of Constellation Pharmaceuticals compound 3, the first non–pyridone-containing EZH2 inhibitor.
Highly selective, small-molecule inhibitors of PRC2. (A) Scaffold demonstrating that several of the recently developed EZH2 or EZH2/1 inhibitors all possess a pyridone motif as well as an indole or indazole core. The inserted table details the identity of each designated substituent of the described inhibitors. (B) Chemical structure of EPZ-6438. (C) Chemical structure of Constellation Pharmaceuticals compound 3, the first non–pyridone-containing EZH2 inhibitor.
EZH2 mutation (EZH2Y641X, EZH2A677G, or EZH2A687V) is a known predictor of EZH2 inhibitor sensitivity; however, later studies showed that GCB-DLBCL is addicted to EZH2 and shows general sensitivity to the EZH2 inhibitor independent of its mutational state,56 and such an EZH2 addiction was not seen in the activated B-cell type of DLBCLs, a more differentiated lymphoma subtype with EZH2 repressed.56 These findings are consistent with the clinical observation that EZH2 mutations exclusively occur in GCB-DLBCLs and not activated B-cell DLBCL,4 providing a rationale for a personalized medicine for lymphoma therapy. Furthermore, the efficacy of EZH2 inhibitors has been established in various B-cell lymphomas with wild-type EZH2, including MCLs and Burkitt lymphoma.75,93 Therefore, other molecular determinants for EZH2 inhibitor sensitivity remain to be defined for B-cell lymphomas in order to improve personalized therapy. Such genetic determinants were defined in other cancers, including SNF inactivation in malignant rhabdoidtumors,91 MMSET/NSD2 translocation in multiple myeloma,99 and MLL rearrangement in acute leukemia,95,100 and all of these affected pathways have been connected to PRC2 genetically. Thus, it is likely that lymphomas carrying TrxG gene mutations,5,6,66 such as MLL2 mutations, may render sensitivity to PRC2 inhibition.
Furthermore, it remains to be examined if EZH1, a less-studied EZH2-related enzyme, is overexpressed in B-cell malignancies and if EZH1 inhibition improves the therapeutic potential. Given that EZH1 compensates the function of EZH2, inhibitors that target both EZH2 and EZH1 such as UNC199990 (Figure 4A) and Constellation Pharmaceuticals compound 392 (Figure 4C), are expected to have benefits for treating a broader spectrum of B-cell malignancies with overexpression of EZH2 or EZH1. Indeed, we have recently shown that UNC1999 offers advantages over EZH2-selective inhibitors and represents a novel efficient therapeutic for MLL-rearranged leukemias that coexpress EZH2 and EZH1.95
In line with therapeutic advances, combination therapy shall be explored, because EZH2 inhibitors can be used together with inhibitors against other oncogenic pathways that act in parallel. Indeed, our recent studies demonstrated synergy of EZH2 and HDAC inhibitors to inhibit lymphoma clonogenic cell growth, induce apoptosis, and suppress growth of MCLs or aggressive c-MYC–associated lymphomas.75,77 The effects are at least partially due to disruption of c-MYC/EZH2-mediated miRNA silencing and vicious amplification loops (Figure 3B). In addition, combination treatment with EZH2 and BCL2 inhibitors outperformed the single-drug therapies in lymphoma models.56 Lastly, given cooperation between DNA methylation and histone modification in transcriptional regulation, it would be of great interest to test if lymphoma cases with a higher degree of epigenetic silencing and hence reduced reversibility are less sensitive to PRC2 inhibitor single treatment but more responsive to a combined treatment with DNA demethylating agents.
Conclusion
Gene regulation by PcG complexes is critical for regulation of various biological programs related to normal development and oncogenesis. PcG aberration, caused by its deregulated expression, somatic mutation, and chromosomal translocation, is common in various B-cell malignancies, demonstrating PcG as a central mechanism in lymphoma initiation and development. PcG complexes interact with other epigenetic machineries such as HDACs, DNMTs, and miRNAs in a context-dependent manner to control gene expression and promote lymphomagenesis. Recent advances in developing targeted strategies against PcG have demonstrated early success and display great potential in treating incurable B-cell malignancies.
Acknowledgments
G.G.W. is supported by a National Institutes of Health National Cancer Institute “Pathway to Independence” Award in Cancer Research (CA151683), a Department of Defense Career Development Award (CA130247), and grants from Gabrielle’s Angel Foundation and Concern Foundation. G.G.W. is a Kimmel Scholar of Sidney Kimmel Foundation for Cancer Research and an American Society of Hematology Scholar in Basic Science. K.D.K. is supported by an American Chemical Society Medicinal Chemistry Predoctoral Fellowship. J.T. is supported by grants from the National Institutes of Health National Cancer Institute (R01 CA137123), Maher Fund, Lymphoma Research Foundation, National Functional Genomics Center Programmatic Research grant, and an American Society of Hematology Bridge Grant.
Authorship
Contribution: All authors participated in the preparation of the manuscript and illustrations and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jianguo Tao, Departments of Hematopathology and Laboratory Medicine, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612; e-mail: jianguo.tao@moffitt.org; and G. Greg Wang, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, 450 West Dr, CB 7295, Chapel Hill, NC 27599; e-mail: greg_wang@med.unc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal