Key Points
Analysis of CSF-1R pTyr-regulated messenger RNAs identifies novel signaling nodes and networks that can be targeted to modulate macrophage functions.
miR-21 is a novel CSF-1R pTyr-721–induced molecule that suppresses the macrophage M1 phenotype and enhances the M2 phenotype.
Abstract
Macrophage polarization between the M2 (repair, protumorigenic) and M1 (inflammatory) phenotypes is seen as a continuum of states. The detailed transcriptional events and signals downstream of colony-stimulating factor 1 receptor (CSF-1R) that contributes to amplification of the M2 phenotype and suppression of the M1 phenotype are largely unknown. Macrophage CSF-1R pTyr-721 signaling promotes cell motility and enhancement of tumor cell invasion in vitro. Combining analysis of cellular systems for CSF-1R gain of function and loss of function with bioinformatic analysis of the macrophage CSF-1R pTyr-721–regulated transcriptome, we uncovered microRNA-21 (miR-21) as a downstream molecular switch controlling macrophage activation and identified extracellular signal-regulated kinase1/2 and nuclear factor-κB as CSF-1R pTyr-721–regulated signaling nodes. We show that CSF-1R pTyr-721 signaling suppresses the inflammatory phenotype, predominantly by induction of miR-21. Profiling of the miR-21–regulated messenger RNAs revealed that 80% of the CSF-1–regulated canonical miR-21 targets are proinflammatory molecules. Additionally, miR-21 positively regulates M2 marker expression. Moreover, miR-21 feeds back to positively regulate its own expression and to limit CSF-1R–mediated activation of extracellular signal-regulated kinase1/2 and nuclear factor-κB. Consistent with an anti-inflammatory role of miRNA-21, intraperitoneal injection of mice with a miRNA-21 inhibitor increases the recruitment of inflammatory monocytes and enhances the peritoneal monocyte/macrophage response to lipopolysaccharide. These results identify the CSF-1R–regulated miR-21 network that modulates macrophage polarization.
Introduction
Macrophages protect the host against infection and injury and facilitate tissue remodeling.1 However, they frequently accumulate in pathological settings, including cancers,2 atherosclerosis,3 metabolic disease,4 and sepsis,5 where they respond to microenvironmental cues that can be detrimental to the host. Two distinct extreme states of polarized activation have been described in macrophages:6,7 the classically activated (M1) and the alternatively activated (M2) macrophage phenotypes, each characterized by well-described markers.5,6,8-11 M1 macrophages produce proinflammatory cytokines, elevate the expression of inducible nitric oxide synthase 2 (iNOS) and major histocompatibility complex class II (MHC II),12 and can play antitumorigenic roles.5,9 In contrast, the M2 macrophages have increased expression of scavenger receptors, increased activation of the arginase pathway, low expression of interleukin-12 (IL-12), high expression of IL-10 and IL-1RA, and increased anti-inflammatory responses and protumorigenic functions.5 Despite these observations, the detailed molecular networks controlling macrophage activation are not fully understood.
In the cellular response to growth factor stimulation, there are several, transient waves of gene transcription, including immediate early genes (IEG), delayed early genes (DEG), and secondary response genes.13-15 In addition, studies of epidermal growth factor (EGF) receptor tyrosine kinase signaling have shown that there are 2 major negative feedback mechanisms: immediate and delayed.16 The immediate wave of feedback regulation occurs within the first 20 minutes of ligand stimulation16-18 and relies exclusively on preexisting signaling components. It involves rapid enzyme-mediated posttranslational modifications such as phosphorylation,17 dephosphorylation,19 and ubiquitination.20 The delayed wave of feedback regulation that suppresses both ligand-mediated signaling and the expression of the IEGs involves newly synthesized molecules encoded by DEGs, including microRNAs (miRNAs), transcriptional repressors, proteases, and phosphatases.21 However, the precise feed-forward and feedback signaling and transcriptional events regulating macrophage activation are unknown.
The colony-stimulating factor 1 receptor (CSF-1R), regulated by its cognate growth factor ligands CSF-1 and IL-34,22,23 plays a major role in the regulation of tissue macrophage differentiation, growth, and survival.24,25 Macrophage CSF-1R signaling also favors the generation of immunosuppressive, protumorigenic, M2-polarized macrophages.10,24,26 The CSF-1R possesses 8 cytoplasmic domain tyrosines that are phosphorylated in the activated receptor (reviewed in Stanley and Chitu25 ). The conditional CSF-1R–deficient MacCsf1r−/− (M−/−) macrophage cell line has been used to probe the functions of these CSF-1R tyrosines.20,27-30 M−/− macrophages retrovirally transduced with the wild-type (WT) receptor behave like primary macrophages,27 whereas those reconstituted with a CSF-1R in which all 8 intracellular tyrosines phosphorylated upon activation are mutated to phenylalanine (YEF) and lack CSF-1R kinase activity, fail to support CSF-1–mediated survival, proliferation, or differentiation.27 By a CSF-1R Tyr deletion/replacement strategy, we have shown that Tyr-559 and Tyr-807 together are necessary and sufficient for CSF-1 responsiveness and that with the further “addition back” (AB) of Tyr-544 for full restoration of kinase activity, the resulting M−/−.YEF.Y544,559,807AB macrophages exhibit normal survival and proliferation responses.20,27,28 In the activated CSF-1R, phospho-Y721 (pTyr-721) creates the site for the binding and activation of phosphatidylinositol 3-kinase (PI3K),29,31 and PI3K pathways have been shown to regulate M1 and M2 activation programs in macrophages.32-35 Using the Y721F mutation in WT CSF-1R and by “adding back” Tyr-721 to the M−/−.YEF.Y544,559,807AB CSF-1R backbone, we have shown that CSF-1R pTyr-721 signaling promotes macrophage motility, spreading, and macrophage enhancement of tumor cell invasion.29,30 Although pan-PI3K inhibitors can be useful for suppressing macrophage M2 polarization because of their effects on other critical cellular functions, it is important to identify new molecular targets acting downstream of CSF-1R pTyr-721.
In the present study, we have used the M−/− cell line system coupled with transcriptomic, bioinformatic, and cell biological approaches to identify macrophage CSF-1R pTyr-721 signaling pathways. We show that CSF-1R pTyr-721 signaling suppresses the proinflammatory M1 phenotype and enhances the M2 macrophage phenotype via a miR-21 regulated network in which elevated miR-21 mediates the suppression of M1 and the enhancement of M2 gene expression. Consistent with these observations, we show that miR-21 attenuates the peritoneal monocyte/macrophage inflammatory response in vivo.
Materials and methods
Reagents
The anti-phospho-extracellular signal-regulated kinase1/2 (ERK1/2) (T202/Y204), anti-phospho-NF-κB p65 (S536), anti-ERK1/2, anti-nuclear factor-κB (NF-κB) p65, and anti-β-actin antibodies were from Cell Signaling Technology (Beverly, MA). Human recombinant CSF-1 was a gift from Chiron Corp. (Emeryville, CA). LY294002 was from EMD Millipore (Darmstadt, Germany), PD98058 from BioMol (Plymouth Meeting, PA), and PS1145 from Sigma-Aldrich (St. Louis, MO). The Locked Nucleic Acid (LNA)-antisense miR-21 inhibitors and mismatch control oligonucleotides, certified endotoxin-free, used previously for in vitro and in vivo inhibition of miR-21 function,36 were from Exiqon (Woburn, MA). The derivation of the cloned, granulocyte macrophage colony-stimulating factor–dependent MacCsf1r−/−(M−/−) cell line and its retrovirally transduced CSF-1R derivatives has been described elsewhere.27-29
Cell culture
M−/−.WT, M−/−.Y721F, M−/−.3ABY721, and M−/−.3AB cell lines, which had been frozen in multiple aliquots at the time of first characterization,27-29 were thawed and cultured in α modified Eagle medium containing 10% newborn calf serum (Invitrogen, Chicago, IL), 100 U/mL penicillin, 100 µg/mL streptomycin, and 120 ng/mL human recombinant CSF-1 for no longer than 6 passages to ensure phenotypic stability. For the kinase inhibition experiments, CSF-1–starved M−/−.WT macrophages were treated with PI3K inhibitor LY2940002 (100 μM) or vehicle (1% dimethylsulfoxide) for 2 hours. For combined inhibitor treatment, LY2940002-treated cells were subsequently treated with either vehicle, ERK1/2 inhibitor, PD98059 (50 μM), or IKK inhibitor PS1145 (2 μM) for 2 additional hours. Immediately following these treatments, cells were stimulated with CSF-1 for the indicated times, then harvested for total RNA extraction. For polarization assays, cells were plated at 106 cells/well in 24-well plates in culture medium and stimulated with either mouse recombinant IL-4 (20 ng/mL; Stemcell Technologies, Vancouver, BC, Canada), or with lipopolysaccharide (LPS) from Escherichia coli (100 ng/mL; Sigma-Aldrich) plus mouse recombinant interferon-γ (IFN-γ; 200 U/mL; Stemcell Technologies) for 18 hours. Cells were harvested for quantitative real-time polymerase chain reaction (qRT-PCR) analysis or arginase assays, whereas cell-free supernatants were collected for analysis of nitrite levels and cytokine measurements. In vitro inhibition of miR-21 function was performed as previously described.36
RNA extraction and qRT-PCR
Cell lysis was performed using the TRIzol reagent (Invitrogen). The miRNeasy kit (Qiagen, Germantown, MD) was used for total RNA and miRNA extractions from cell lysates, according to the manufacturer’s instructions. RNA amounts were quantitated by Nanodrop 1000 (Wilmington, DE) and RNA quality assessed using an Agilent 2100 Bioanalyzer (Santa Clara, CA). Total RNA preparations with ribosomal integrity numbers >9.5 were used for microarray and qRT-PCR assays. qRT-PCR for miR-21, miR-155, and endogenous control miR-234 was performed on total RNA. Reverse transcription using the TaqMan MicroRNA Reverse Transcription Kit and qRT-PCR using the TaqMan MicroRNA Assays (Life Technologies, Carlsbad, CA) were performed according to the manufacturer’s protocols.
Validation of the bioinformatically predicted miR-21–regulated transcripts was based on the combination of 2 previously published methods.37,38 The messenger RNA (mRNA) targets bound to their miRNAs were captured and subjected to qRT-PCR analysis (supplemental Figure 3 on the Blood Web site). To control for the specificity of the capture, the unrelated control Caenorhabditis elegans miR-67 was used.
For gene expression analysis, total RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System (Life Technologies). Mouse oligonucleotide primers were commercially synthesized (Fisher Scientific) and the amplification products were verified by sequencing. Real-time PCR was performed with SYBR (Qiagen) using previously described29 amplification conditions on a Real Time Thermal Cycler (Eppendorf, Hamburg, Germany). Results were normalized to HPRT gene expression (stable in CSF-1–starved and CSF-1–treated cells; data not shown) and analyzed using the ΔΔCT method.
Microarrays, bioinformatics analysis, and statistics
Gene microarrays were performed on Affymetrix Mouse Gene 1.0 ST Array (Affymetrix, Santa Clara, CA) at the Genomics Core at the Albert Einstein College of Medicine, according to the manufacturer’s instructions. Each array permitted detection of 28 361 coding transcripts. Differential expression analysis was performed using R/Bioconductor’s limma package to identify significantly differentially expressed mRNAs over time in response to CSF-1 treatment and to the genotype. The CSF-1–regulated genes for each cell line were obtained from the data as described in Figure 1. The expression of each coding transcript in cells grown with CSF-1 was normalized with respect to its expression in CSF-1–starved cells, and a relative expression ratio was assigned to each gene. The log2 relative expression ratios were calculated and cutoff values of fold change >1.5 and P < .05 were used to identify genes that were significantly regulated by CSF-1 in each of the 4 cell lines. CSF1-regulated genes were identified according to the cutoffs of fold change >1.5 and P < .05. Extensive qRT-PCR validations indicated a good correlation with microarray analysis based on several randomly selected differentially regulated genes (n = 60, data not shown).
The CSF-1R pTyr721-regulated macrophage transcriptome. (A) Schematic depiction of the CSF-1Rs in the macrophage cell lines used to assess the necessity (loss of function, upper panels) and sufficiency (gain of function, lower panels) of pTyr-721 signaling in macrophages. The CSF-1R domains (extracellular [ECD]; transmembrane [TM], and tyrosine kinase [TK]) and the intracellular CSF-1R Tyr (Y, black) residues mutated to Phe (F, red) are indicated. (B) Pairwise comparisons of gene expression data for all 28 361 transcripts detected on microarrays, showing high Pearson correlation coefficients between biological replicates and the expected hierarchical clustering of samples, as a function of genotype and CSF-1 treatment. Results for M−/−.WT and M−/−.Y721F (upper panel) and M−/−.3ABY721 and M−/−.3AB (lower panel) are shown as heat maps illustrating the significant differences in gene expression between Tyr-721–expressing and Tyr-721–nonexpressing cells, each with an associated color key and histogram (−, without CSF-1; +, with 120 ng/mL of CSF-1; biological replicates are indicated by the numerals 1 and 2). (C-E) Venn diagrams, representing the size of the pTyr-721–regulated transcriptome, identified by each approach. To obtain the pTyr-721–regulated genes, the CSF-1–regulated transcripts expressed in either M−/−.WT or M−/−.3ABY721 cells were compared with those of the corresponding Y721F mutant cells. (C) The Y721F mutation (1904 CSF-1–regulated mRNAs) reduced the number of CSF-1–regulated mRNAs of M−/−.WT macrophages (2174) by 270 transcripts. However, of the CSF-1–regulated genes in both cell types, 1713 (∼9% of 19 000 total mouse genes) were differentially regulated in both data sets. (D) The addition of CSF1R Y721 to the 3AB background increased the number of CSF-1–regulated transcripts by 250 (from 870 to 1120). Of the CSF-1–regulated genes in both cell types, 244 (∼1.3% of 19 000 total mouse genes) were differentially regulated in both data sets. (E) Only 34 genes are common between the 2 datasets.
The CSF-1R pTyr721-regulated macrophage transcriptome. (A) Schematic depiction of the CSF-1Rs in the macrophage cell lines used to assess the necessity (loss of function, upper panels) and sufficiency (gain of function, lower panels) of pTyr-721 signaling in macrophages. The CSF-1R domains (extracellular [ECD]; transmembrane [TM], and tyrosine kinase [TK]) and the intracellular CSF-1R Tyr (Y, black) residues mutated to Phe (F, red) are indicated. (B) Pairwise comparisons of gene expression data for all 28 361 transcripts detected on microarrays, showing high Pearson correlation coefficients between biological replicates and the expected hierarchical clustering of samples, as a function of genotype and CSF-1 treatment. Results for M−/−.WT and M−/−.Y721F (upper panel) and M−/−.3ABY721 and M−/−.3AB (lower panel) are shown as heat maps illustrating the significant differences in gene expression between Tyr-721–expressing and Tyr-721–nonexpressing cells, each with an associated color key and histogram (−, without CSF-1; +, with 120 ng/mL of CSF-1; biological replicates are indicated by the numerals 1 and 2). (C-E) Venn diagrams, representing the size of the pTyr-721–regulated transcriptome, identified by each approach. To obtain the pTyr-721–regulated genes, the CSF-1–regulated transcripts expressed in either M−/−.WT or M−/−.3ABY721 cells were compared with those of the corresponding Y721F mutant cells. (C) The Y721F mutation (1904 CSF-1–regulated mRNAs) reduced the number of CSF-1–regulated mRNAs of M−/−.WT macrophages (2174) by 270 transcripts. However, of the CSF-1–regulated genes in both cell types, 1713 (∼9% of 19 000 total mouse genes) were differentially regulated in both data sets. (D) The addition of CSF1R Y721 to the 3AB background increased the number of CSF-1–regulated transcripts by 250 (from 870 to 1120). Of the CSF-1–regulated genes in both cell types, 244 (∼1.3% of 19 000 total mouse genes) were differentially regulated in both data sets. (E) Only 34 genes are common between the 2 datasets.
The core analysis function in the Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com/) (Ingenuity System Inc, licensed to the Albert Einstein College of Medicine) was performed to identify the enriched biological processes, pathways, upstream regulators (including predicted miRNAs and their represented targets), and networks for those significantly differentially regulated genes. The Affymetrix gene identifiers were used for IPA, and both up- and downregulated genes were defined as value parameters for the analysis. We used 2 metrics to identify the most significantly enriched downstream effects (ie, biofunctions, networks, pathways, and predicted regulatory molecules) of the differentially expressed genes: the activation z score and P value.39 A positive z score indicates greater enrichment of functional activity for a process or molecules in 1 set relative to another. The P value, calculated by Fisher’s exact test, indicates the likelihood that the association between a set of genes in our dataset and a predicted function, pathway, or molecule in the IPA database is significant. In the end, we considered only functions, networks, and molecules with P < .05 and, if applicable, a z score ≥2 (activated) or ≤−2 (inhibited). This analysis tool generated networks and pathways in which the differentially regulated genes could be related according to previously known associations between genes or proteins, but independently of established canonical pathways.40 The graphic representation of the miR-21 target network was generated using the network visualization tool embedded in Cytoscape.41 Functional classification of CSF-1R–regulated miR-21 targets based on Gene Ontology terms was performed using PANTHER gene list analysis tool (version 9.0, January 201442 ). Significance was tested using the unpaired Student t test.
CSF-1 stimulation, preparation of cell lysates, and western blotting
Subconfluent (∼70%) 100-mm tissue culture dishes of macrophages were CSF-1–starved for 16 hours to upregulate CSF-1R expression. Starved cells were incubated with recombinant CSF-1 (240 ng/mL) at 37°C for the indicated time points. Cell lysate preparation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, western blotting, and the quantitation of chemiluminescent signals were performed as described previously.27-29
Flow-cytometric analysis of macrophages and peritoneal cells
Peritoneal cells were collected from mice subjected to the indicated treatments. Red blood cells were lysed using 3.5 mL of red blood cell lysis solution containing buffered 0.83% NH4Cl, at 37°C, for 5 minutes, followed by centrifugation at 300 g. Cultured macrophages were detached from culture dishes by brief incubation with cold, endotoxin-free, 2 mM EDTA in phosphate-buffered saline (PBS). To selectively label live cells, all cells were incubated with LIVE/DEAD fixable Aqua Dead Cell Stain (Life Technologies), according to the manufacturer’s instructions. The stained cells were washed with 3 mL of PBS containing 2% fetal calf serum. After Fc receptor blocking, cells were labeled with anti-CD11b-APC-Cy7, anti-CD115-PE, anti-Ly6C Pacific Blue, anti-Ly6G-PerCP, anti-F4/80-Pacific Blue, anti-B220-PE, anti-CD8α-PE-Cy5.5, anti-CD19-APC, anti-MHC II–fluorescein isothiocyanate, anti-CD206-APC, anti-IL-4Rα-PE, and anti-CD11c-fluorescein isothiocyanate (eBioscience). Macrophages43,44 and peritoneal cells45 were analyzed as described and sorted using a BD FACSAria II cell sorter (Becton Dickinson). Spectral overlaps between fluorophores were corrected by electronic compensation. FACS data were analyzed using the FACSDiva 6.3.1 software (BD Biosciences, San Diego, CA).
Enzyme-linked immunosorbent assay and cytokine arrays
Cell-free supernatants were collected from cell cultures at the indicated times and screened for cytokine and growth factor production using the RayBio Mouse Cytokine Antibody Array G-Series 2000 (RayBiotech, Norcross, GA) or the mouse IL-1β enzyme-linked immunosorbent assay Ready-SET-Go kit (eBioscience).
Enzyme assays
Macrophage arginase activity was determined in 100 μg of NP-40 cell lysate using the QuantiChrom Arginase Assay Kit (DARG-200) from BioAssay Systems (Hayward, CA), according to the manufacturer’s instruction. One unit of arginase activity was defined as the amount of enzyme necessary to convert 1 μmole of l-arginine to ornithine and urea per minute at pH 9.5 and 37°C. Using the Griess reaction, iNOS activity was measured as the amount of NO2− and NO3− detected in cell supernatants from cells subjected to indicated treatments, using the Nitrate/Nitrite Colorimetric Assay kit (Cayman Chemical, Ann Arbor, MI). For normalization purposes, adherent cells were stained with 4′,6 diamidino-2-phenylindole and fluorescence was recorded. Results were expressed as μMol of NO2− + NO3− per fluorescence unit.
Mouse experiments
Four- to 6 week-old C57BL/6 male mice (Charles River Breeding Laboratories), were injected intraperitoneally with either LNA miR-21 inhibitor (25 mg/kg), mismatch control oligonucleotide (25 mg/kg), LPS (100 μg/mouse), or LNA miR-21 inhibitor plus LPS, or vehicle (PBS). LNA miR-21 inhibitor or mismatch control were injected on days 1 and 3,36 and LPS (100 μg/mouse) at 18 or 48 hours before euthanasia and peritoneal lavage on day 5. Mouse peritoneal lavages were performed as previously described.46
Results
Transcriptional analysis indicates a distinct role for macrophage CSF-1R pTyr-721 signaling
To discover the regulatory molecules, networks, and phenotypes associated with macrophage CSF-1R pTyr-721 signaling, we used 4 previously described27-29 cell lines, 2 CSF-1R Tyr-721–deficient lines (M−/−.Y721F and M−/−.YEF.Y544,Y559,807AB [M−/−.3AB]), and 2 CSF-1R Tyr-721–expressing lines (M−/−.WT and M−/−.YEF.Y544,559,721,807AB [M−/−.3ABY721]) (Figure 1A). We initially analyzed the transcriptional response to CSF-1 in the steady state. We performed a microarray-based analysis of the gene expression profiles in cells with or without Tyr-721, cultured in the presence or absence of CSF-1 (Figure 1B). In macrophages expressing the WT CSF-1R (M−/−.WT cells), we identified 2174 genes that were differentially expressed (±CSF-1), indicating that approximately 10% of the macrophage transcriptome is CSF-1–dependent, whereas in macrophages expressing the Y721F receptor (M−/−.Y721F cells), only 1904 CSF-1–regulated genes were identified (Figure 1B-C; supplemental Table 1). A similar analysis identified 1120 CSF-1–regulated genes in M−/−.3ABY721 cells and 870 CSF-1–regulated genes in M−/−.3AB (Figure 1B-D; supplemental Table 2). These results indicate that, in the steady state, CSF-1R pTyr-721 signaling regulates approximately 1% of the macrophage transcriptome. However, although the estimated size of the CSF-1R pTyr-721–regulated transcriptome is approximately the same in both approaches (ie, 2174 − 1904 = 270; 1120 − 870 = 250 transcripts, respectively), only 34 genes (∼13% of the pTyr-721–regulated genes) are common to both datasets (Figure 1E), indicating that, despite pTyr-721–specific cellular morphology and functions,29 the identity of most pTyr-721–regulated transcripts is strongly influenced by the CSF-1R pTyr background. Thus, analysis of the pTyr-721 networks that are common to both cellular systems is more likely to reveal novel pTyr-721–specific cellular functions.
To delineate how CSF-1R pTyr-721 signaling regulates the early kinetics of gene expression, we stimulated M−/−.WT and M−/−.Y721F cells with CSF-1 for times ranging between 20 and 180 minutes and measured the changes in mRNA abundance (supplemental Figure 1). We observed a pTyr-721–dependent hierarchical clustering of samples as a function of genotype and CSF-1 treatment (supplemental Figure 1A) and a correlation matrix revealing well-defined waves of pTyr-721–regulated IEGs and DEGs per cell line and over time (supplemental Figure 1B; supplemental Table 3). The numbers of differentially pTyr-721–regulated genes at 20, 60, and 180 minutes of stimulation are shown in supplemental Figure 1C. Because most of the typical growth factor–induced IEGs or DEGs47 that are CSF-1R–dependent are not pTyr-721–regulated (supplemental Figure 1D), these results suggested novel roles for CSF-1R pTyr-721–regulated genes in macrophages.
Macrophage CSF-1R pTyr-721 signaling suppresses the expression of proinflammatory (M1) and stimulates the expression of repair (M2) genes
To identify the gene signatures predictive of cellular functions, molecular pathways, and network regulators altered downstream of pTyr-721, a bioinformatics analysis was performed on the genes differentially expressed in each system and for each time point using IPA software, followed by validation in both cellular systems. This unbiased analysis revealed that pTyr-721 signaling kinetically downregulates the abundance of a set of proinflammatory genes, including IL1α, IL1β, CXCL2, CCL4, LIF, and TNFSF15, while upregulating the expression of the M2 genes Arginase (Arg1) and IL10 (Figure 2A). Furthermore, pTyr-721 signaling suppressed the expression of 8 of 14 genes encoding proinflammatory molecules in the steady state (Figure 2B). These data suggest that pTyr-721 signaling inhibits inflammatory pathways and induces macrophage polarization toward an M2 phenotype. Further support for this conclusion was obtained by quantitation of the differential secretion of 144 cytokines, chemokines, growth factors, and proteases secreted by M−/−.WT and M−/−.Y721F cells in response to CSF-1 stimulation (Figure 2C-D). Consistent with pTyr-721 regulation of macrophage polarization, a pTyr-721–mediated increase in macrophage production of tumorigenic EGF, matrix metalloproteinase-3, and insulin-like growth factor-1 was observed (Figure 2C), whereas absence of pTyr-721 signaling was associated with an increase in the secreted amounts of inflammatory cytokines MIP-1α, MIP-2α, MIP-3β, and IL-12p70 (Figure 2D).
CSF-1R pTyr-721 controls expression of a specific subset of IEGs and DEGs associated with suppression of inflammation and promotion of an M2 phenotype. (A) Bar charts representing the relative abundance of a subset of pTyr-721–induced IEGs (left panel) and DEGs (center and right panels), illustrating pTyr-721–mediated decreased abundance of transcripts encoding proinflammatory (M1, *) molecules in conjunction with increased abundance of anti-inflammatory (M2, **) molecules. Known IEGs or DEGs are shown in bold. CSF-1–regulated transcript abundance in M−/−.WT cells (ie, log2 fold change [FC] between the expression of the indicated RNA at the indicated time point and its measured expression at time 0) was normalized to CSF-1–regulated mRNA abundance in M−/−.Y721F macrophages calculated in the same manner. For a full set of pTyr-721–regulated genes, see supplemental Table 3. (B) Relative abundance of a subset of pTyr-721–induced genes in cells growing in CSF-1 (see supplemental Table 1) that encode secreted cytokines, illustrating the pTyr-721–mediated change in abundance of transcripts encoding M1 (*) and M2 (**) molecules (P < .05, FC > 1.5). (C-D) pTyr-721 suppresses secretion of proinflammatory molecules (*) and induces secretion of tumorigenic EGF, insulin-like growth factor-1 (IGF-1), and matrix metalloproteinase-3 (MMP-3). Conditioned medium from CSF-1–treated (120 ng/mL) and CSF-1–starved M−/−.WT and M−/−.Y721F macrophages was assayed for production of proinflammatory and proangiogenic factors using a mouse cytokine array permitting simultaneous measurements of 144 cytokines and soluble factors. For each cell line, signals detected for each cytokine produced by CSF-1–treated cells were integrated and normalized to the corresponding signal obtained from CSF-1–starved cells (duplicates ± range). (C) Molecules exhibiting pTyr-721–dependent increased secretion. (D) Molecules exhibiting increased secretion in the absence of pTyr-721 signaling. Error bars indicate the range of levels of biological duplicates.
CSF-1R pTyr-721 controls expression of a specific subset of IEGs and DEGs associated with suppression of inflammation and promotion of an M2 phenotype. (A) Bar charts representing the relative abundance of a subset of pTyr-721–induced IEGs (left panel) and DEGs (center and right panels), illustrating pTyr-721–mediated decreased abundance of transcripts encoding proinflammatory (M1, *) molecules in conjunction with increased abundance of anti-inflammatory (M2, **) molecules. Known IEGs or DEGs are shown in bold. CSF-1–regulated transcript abundance in M−/−.WT cells (ie, log2 fold change [FC] between the expression of the indicated RNA at the indicated time point and its measured expression at time 0) was normalized to CSF-1–regulated mRNA abundance in M−/−.Y721F macrophages calculated in the same manner. For a full set of pTyr-721–regulated genes, see supplemental Table 3. (B) Relative abundance of a subset of pTyr-721–induced genes in cells growing in CSF-1 (see supplemental Table 1) that encode secreted cytokines, illustrating the pTyr-721–mediated change in abundance of transcripts encoding M1 (*) and M2 (**) molecules (P < .05, FC > 1.5). (C-D) pTyr-721 suppresses secretion of proinflammatory molecules (*) and induces secretion of tumorigenic EGF, insulin-like growth factor-1 (IGF-1), and matrix metalloproteinase-3 (MMP-3). Conditioned medium from CSF-1–treated (120 ng/mL) and CSF-1–starved M−/−.WT and M−/−.Y721F macrophages was assayed for production of proinflammatory and proangiogenic factors using a mouse cytokine array permitting simultaneous measurements of 144 cytokines and soluble factors. For each cell line, signals detected for each cytokine produced by CSF-1–treated cells were integrated and normalized to the corresponding signal obtained from CSF-1–starved cells (duplicates ± range). (C) Molecules exhibiting pTyr-721–dependent increased secretion. (D) Molecules exhibiting increased secretion in the absence of pTyr-721 signaling. Error bars indicate the range of levels of biological duplicates.
Further functional analysis performed on pTyr-721–modulated gene sets (from supplemental Tables 1 and 2) inferred a pTyr-721–mediated role in leukocyte activation, leukocyte infiltration, angiogenesis, cell invasion and motility (supplemental Figure 2). The gene networks generated from M−/−.WT cells indicated ERK1/2 as the main regulatory hub (P value = 10−40, 34 molecules), whereas the top predicted network generated from cells lacking pTyr-721 was centered on NF-κB (P value = 10−36, 30 molecules; supplemental Table 4), both of which have been linked to regulation of inflammation48-52 and macrophage polarization.51-53 Additional data mining on gene expression signatures previously associated with the CSF-1–mediated macrophage shift toward an M2 phenotype in steady-state conditions54,55 identified a significant overlap in gene identity and direction of regulation between the 96 genes differentially expressed in M1 vs M2 polarized macrophages54 and 19 of our pTyr-721–regulated transcripts (supplemental Figure 2D). Consistent with the increased lipid metabolism and glycolysis in M1 polarized macrophages,55 we observed a significant enrichment in cholesterol synthesis–, glycolysis-, and gluconeogenesis-related genes in pTyr-721–deficient macrophages (supplemental Figure 2E). Together, these results suggest that pTyr-721 signaling suppresses the M1 phenotype and activates the M2 phenotype; we next addressed how pTyr-721 participates in the regulation of these phenotypes by inducing either the M1 or the M2 phenotype in the presence and absence of CSF-1.
CSF-1R pTyr-721 signaling suppresses the proinflammatory (M1) phenotype
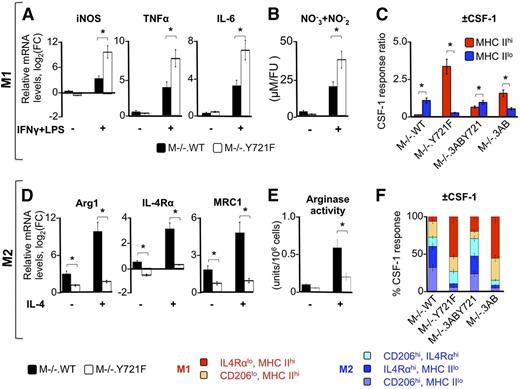
The combination of IFN-γ and LPS is a potent activator of the M1 phenotype.56,57 To examine the role of pTyr-721 signaling in the development of M1 polarization, we compared the response of M−/−.WT and M−/−.Y721F macrophages with IFN-γ and LPS, both in the presence and absence of CSF-1. To quantitate the effect of IFN-γ and LPS, we used the M1 macrophage polarization markers iNOS, tumor necrosis factor-α (TNF-α), and IL-6. CSF-1–stimulated M−/−.WT macrophages were less responsive to IFN-γ and LPS than M−/−.Y721F macrophages, as shown by the attenuated induction of M1-associated genes, the IFN-γ target gene iNOS, the LPS-target genes TNFα and Il6 (Figure 3A), and iNOS activity (Figure 3B). The expression of the mRNAs of the cognate receptors for both LPS (ie, TLR4) and IFN-γ (ie, IFNGR1 and IFNGR2) were similar in all gene sets (supplemental Tables 1 and 2), suggesting that suppression of M1 gene expression is independent of regulation of these receptors.
CSF-1R pTyr-721 signaling suppresses M1 responses and enhances M2 responses. (A) qRT-PCR results showing the enhancement by pTyr-721 signaling of the IFN-γ and LPS stimulation of iNOS, TNF-α, and IL-6 gene transcripts in cells stimulated with CSF-1. Cells were either starved for CSF-1 overnight, or constitutively grown in CSF-1 (120 ng/mL), then treated with either IFN-γ and LPS (+) or vehicle (−) for 18 hours. HPRT gene expression levels were used as endogenous control. mRNA levels are expressed as the log2 fold-change relative to CSF-1–starved cells. (B) Nitrate/nitrite levels in cell lysates from equivalent cultures. (C) Relative CSF-1–induced surface expression of the M1 polarization marker MHC II on CD11b+ cells, determined by flow cytometry in CSF-1–treated (18 hours, +CSF-1) vs CSF-1–starved (−CSF-1) cells and expressed as a ratio of the +CSF-1 to −CSF-1 responses. (D) qRT-PCR results showing the enhancement by pTyr-721 signaling of the IL-4–stimulated expression of arginase-1, IL-4Rα, and mannose receptor-1 (MRC1) transcripts. Experiments were carried out as described in panel A. (E) Total arginase 1 activity of supernatants of equivalent cultures. (F) Stacked bar chart showing relative contribution of combined sets of macrophage M2 (CD206 and IL-4Rα) and M1 (MHC II) cell surface–specific polarization markers in CSF-1–treated cells. The relative CSF-1 response ratio for each set of cell surface markers was calculated as in panel C and expressed as a percentage for each cell line. Data are representative of 3 independent experiments (error bars, ±standard deviation (SD); *P ≤ .05).
CSF-1R pTyr-721 signaling suppresses M1 responses and enhances M2 responses. (A) qRT-PCR results showing the enhancement by pTyr-721 signaling of the IFN-γ and LPS stimulation of iNOS, TNF-α, and IL-6 gene transcripts in cells stimulated with CSF-1. Cells were either starved for CSF-1 overnight, or constitutively grown in CSF-1 (120 ng/mL), then treated with either IFN-γ and LPS (+) or vehicle (−) for 18 hours. HPRT gene expression levels were used as endogenous control. mRNA levels are expressed as the log2 fold-change relative to CSF-1–starved cells. (B) Nitrate/nitrite levels in cell lysates from equivalent cultures. (C) Relative CSF-1–induced surface expression of the M1 polarization marker MHC II on CD11b+ cells, determined by flow cytometry in CSF-1–treated (18 hours, +CSF-1) vs CSF-1–starved (−CSF-1) cells and expressed as a ratio of the +CSF-1 to −CSF-1 responses. (D) qRT-PCR results showing the enhancement by pTyr-721 signaling of the IL-4–stimulated expression of arginase-1, IL-4Rα, and mannose receptor-1 (MRC1) transcripts. Experiments were carried out as described in panel A. (E) Total arginase 1 activity of supernatants of equivalent cultures. (F) Stacked bar chart showing relative contribution of combined sets of macrophage M2 (CD206 and IL-4Rα) and M1 (MHC II) cell surface–specific polarization markers in CSF-1–treated cells. The relative CSF-1 response ratio for each set of cell surface markers was calculated as in panel C and expressed as a percentage for each cell line. Data are representative of 3 independent experiments (error bars, ±standard deviation (SD); *P ≤ .05).
Another marker of M1 activation is cell surface MHC II,58,59 on which the efficiency of antigen presentation depends.60 Flow cytometric determination of cell-surface MHC II showed a CSF-1–stimulated increase in the frequency of MHC IIhi cells in Y721F-expressing cells and a decrease in their frequency in Tyr-721–expressing cells (Figure 3C). Also, in response to IFN-γ plus LPS, relative CSF-1–induced expression of cell surface MHC II was increased in Tyr-721–deficient cells compared with their Tyr-721–expressing counterparts (data not shown). These results demonstrate that pTyr-721 signaling suppresses the macrophage proinflammatory phenotype and significantly contributes to the known lower sensitivity of alternatively activated macrophages to proinflammatory factors.61
CSF-1R pTyr-721 signaling promotes the M2 phenotype and primes the IL-4–elicited M2 response
IL-4 is an activator of the M2 phenotype.57,62 To assess the contribution of pTyr-721–signaling to M2 activation, we used the M2 macrophage polarization markers, arginase, IL-4Rα, and CD206/MRC1. CSF-1–stimulated M−/−.WT macrophages exhibited a stronger M2 response and were more responsive to IL-4 than Tyr-721F cells, as shown by significantly higher induction of mRNAs encoding M2-associated genes Arg1, IL-4Rα, and MRC1 (Figure 3D) and of arginase activity (Figure 3E). Consistent with pTyr-721 signaling enhancement of the M2 phenotype, Tyr-721–expressing cells exhibited a higher proportion of MHC IIlo cells (Figure 3C) and other M2 cell surface markers (Figure 3F) than Y721-deficient cells. Because the gene (Figure 3D) and cell surface (Figure 3F) expression levels of the cognate receptor for IL-4 are positively regulated by pTyr-721, the increased responsiveness to IL-4 is possibly due, in part, to the pTyr-721–mediated increase in IL-4Rα abundance, consistent with a priming effect of pTyr-721 signaling on the M2 response. These results indicate that the previously described CSF-1–mediated promotion of the M2 phenotype61 requires pTyr-721 signaling and that pTyr-721 signaling increases the sensitivity to alternative activation by IL-4.
miR-21 directly mediates CSF-1R pTyr-721 suppression of proinflammatory (M1) genes
MiRNA activity can modulate monocyte and macrophages responses to environmental signals by fine-tuning gene expression networks in both homeostasis and disease.63-65 To identify the master regulators of CSF-1R–directed macrophage polarization, we performed downstream effector analysis in IPA using the set of CSF-1R–regulated transcripts identified in each cell line. This identified significant (P value < 10−6, z score >2) correlations between the set of pTyr-721–regulated genes and downstream activation of miR-21 in M−/−.WT and M−/−.3ABY721 cells (Figure 4A), predicting 31 (M−/−.WT) and 25 (M−/−.3ABY721) mRNAs as direct miR-21 targets. Additionally, miR-21 was the top predicted pTyr-721–dependent regulator (supplemental Table 5) and, in Y721F cells, the same approach identified only 8 (M−/−.Y721F) and 7 (M−/−.3AB) miR-21–modulated transcripts with higher P values and lower z scores (Figure 4A), suggesting lower CSF-1R–induced miR-21 activity in cells lacking pTyr-721. To demonstrate the correlation between the predicted miR-21 activity and the increased miR-21 abundance with pTyr-721 signaling, we used qRT-PCR and specific probes for mature miR-21. CSF-1 stimulation increased miR-21 expression with time in pTyr-721–expressing, but not Y721-deficient cells, relative to the endogenous control miR-234 (Figure 4B).
miR-21 expression is positively regulated downstream of CSF-1R pTyr-721 to decrease the abundance of proinflammatory molecules. (A) Downstream effector analysis performed by IPA on differentially regulated CSF-1 transcripts (FC > 1.5, P < .05), predicts miR-21 as the top statistically significant miRNA (supplemental Table 5) that is activated in a pTyr-721–dependent manner (z score >2, gray dotted line). The number of coexpressed mRNAs predicted as miR-21 targets (bold) and the statistical P values (italics) are indicated for each cell line at the top of each bar. (B) Validation of miR-21 as a pTyr-721–regulated molecule and of a miR-21 inhibitor. For miR-21 validation, qRT-PCR measurements were performed on complementary DNA prepared from M−/−.WT and M−/−.Y721F macrophages and from the M−/−.3ABY721 and M−/−.3AB cells that were either CSF-1–starved overnight, then treated with CSF-1 (120 ng/mL) for the indicated time points, or constitutively grown in CSF-1 (C). For inhibitor validation, cells were constitutively grown in CSF-1. The miR-21 inhibitor (C+I), or mismatch inhibitor control (C+M), were added 48 hours before harvesting the cells and determining the miR-21 levels by qRT-PCR. (C) Cytoscape representation of the IPA-predicted miR-21 targets that are regulated in a pTyr-721–dependent manner. Note that 80% of these molecules are associated with macrophage polarization toward an inflammatory phenotype (red circles), whereas only 10% are associated with the M2 phenotype (blue circles). (D-E) CSF-1 negatively regulates IL-1β mRNA levels and IL-1β secretion in a pTyr-721– and miR-21–dependent manner. (D) qRT-PCR measurements of IL-1β mRNA in Tyr-721– and Tyr-721F–expressing macrophages. Cells were treated as described in panel B. (E) Conditioned media from Tyr-721– and Tyr-721F–expressing macrophages treated as described in panel B were used to measure the amount of soluble mature IL-1β released in the medium, by enzyme-linked immunosorbent assay. (B,D,E) Key for cell lines as in panel A. Five biological replicates; error bars, ±SD; *P ≤ .05.
miR-21 expression is positively regulated downstream of CSF-1R pTyr-721 to decrease the abundance of proinflammatory molecules. (A) Downstream effector analysis performed by IPA on differentially regulated CSF-1 transcripts (FC > 1.5, P < .05), predicts miR-21 as the top statistically significant miRNA (supplemental Table 5) that is activated in a pTyr-721–dependent manner (z score >2, gray dotted line). The number of coexpressed mRNAs predicted as miR-21 targets (bold) and the statistical P values (italics) are indicated for each cell line at the top of each bar. (B) Validation of miR-21 as a pTyr-721–regulated molecule and of a miR-21 inhibitor. For miR-21 validation, qRT-PCR measurements were performed on complementary DNA prepared from M−/−.WT and M−/−.Y721F macrophages and from the M−/−.3ABY721 and M−/−.3AB cells that were either CSF-1–starved overnight, then treated with CSF-1 (120 ng/mL) for the indicated time points, or constitutively grown in CSF-1 (C). For inhibitor validation, cells were constitutively grown in CSF-1. The miR-21 inhibitor (C+I), or mismatch inhibitor control (C+M), were added 48 hours before harvesting the cells and determining the miR-21 levels by qRT-PCR. (C) Cytoscape representation of the IPA-predicted miR-21 targets that are regulated in a pTyr-721–dependent manner. Note that 80% of these molecules are associated with macrophage polarization toward an inflammatory phenotype (red circles), whereas only 10% are associated with the M2 phenotype (blue circles). (D-E) CSF-1 negatively regulates IL-1β mRNA levels and IL-1β secretion in a pTyr-721– and miR-21–dependent manner. (D) qRT-PCR measurements of IL-1β mRNA in Tyr-721– and Tyr-721F–expressing macrophages. Cells were treated as described in panel B. (E) Conditioned media from Tyr-721– and Tyr-721F–expressing macrophages treated as described in panel B were used to measure the amount of soluble mature IL-1β released in the medium, by enzyme-linked immunosorbent assay. (B,D,E) Key for cell lines as in panel A. Five biological replicates; error bars, ±SD; *P ≤ .05.
To better characterize the role of CSF-1R–modulated miR-21 in macrophages, we used the list of differentially expressed genes (supplemental Tables 1-3), TargetScan, TarBase, miRecords, and the Ingenuity Knowledge–based algorithms embedded into IPA’s target filter tool to identify a set of 63 CSF-1–regulated, miR-21 canonical mRNA targets (Figure 4C). The expression of 47 of these mRNAs was significantly and negatively correlated with miR-21 abundance in our macrophage cell lines (supplemental Table 6). Subsequent classification of these gene targets according to Gene Ontology (supplemental Table 7) revealed a significant association with known macrophage-mediated or CSF-1–mediated functions (supplemental Table 7). Other significantly enriched processes, including regulation of apoptosis and cell death, the M1-specific enhancement of nitric oxide biosynthesis, and the regulation of NF-κB functions (supplemental Table 7) have previously been associated with altered miR-21 expression.66-68 These results suggest that the CSF-1R pTyr-721/miR-21/miR-21 substrate network negatively regulates macrophage M1 polarization.
To validate the bioinformatically predicted miR-21–regulated transcripts, we used a technique based on 2 previously published methods37,38 to capture mRNA targets that bound their miRNAs in a format amenable to downstream quantitative analysis (supplemental Figure 3). For each cell line and growth condition, qRT-PCR analysis of the affinity-purified mRNA targets showed a significant (2.5- to fivefold) enrichment of the 47 predicted miRNA targets (supplemental Table 8) compared with miR-21 target levels detected in preparations that did not undergo the capture procedure, whereas no significant enrichment (<twofold) was detected when using the unrelated control C. elegans miR-67.
To further confirm the suppressive role of miR-21 in inflammation, we used an LNA-miR-21 inhibitor36 that efficiently knocked down miR-21 in macrophages of all 4 cell lines (Figure 4B) and measured the levels of the miR-21 substrate IL1β mRNA as well as secreted IL-1β. miR-21 knockdown overcame the pTyr-721 effect and led to a significant increase in IL-1β gene expression (Figure 4D) and IL-1β secretion (Figure 4E). In addition, the expression levels of mRNAs of proinflammatory molecules detected as miR-21 targets were shown to be significantly increased in LNA-miR-21 inhibitor–treated M−/−.WT and M−/−.3ABY721 cells (supplemental Table 8). Furthermore, the LNA-miR-21 inhibitor substantially increased IFN-γ– and LPS-induced elevation of iNOS, TNFα, and IL-6 in Y721-expressing cells (supplemental Figure 5). These results demonstrate that miR-21 directly mediates pTyr-721 suppression of M1 gene expression.
miR-21 mediates CSF-1R pTyr-721 enhancement of the repair (M2) genes
Our gene expression analysis did not reveal typical M2 markers as canonical miR-21 targets. However, to address whether the pTyr-721–regulated increase in miR-21 also regulates expression of M2 polarization markers, the abundance of 20 markers typically associated with macrophage polarization toward the M2 phenotype57 was measured by qRT-PCR. We found that although arginase 1, mannose receptor 1, IL-4Rα, and FIZZ mRNAs are not canonical miR-21 targets, their expression was significantly decreased upon LNA-mediated miR-21 knockdown (supplemental Figure 4), suggesting a positive role of miR-21 in the induction of M2 markers.
CSF-1R pTyr-721 regulates activation of ERK1/2 and NF-κB
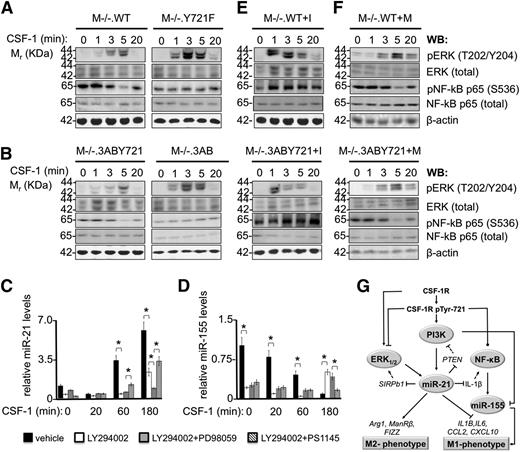
Our bioinformatic analysis of CSF-1R–modulated genes inferred a differential activation status of ERK1/2 and NF-κB downstream of pTyr-721 (supplemental Table 4). We sought to confirm this prediction by examining the kinetics of ERK1/2 and the NF-κB subunit p65 activation in CSF-1R Tyr-721 and CSF-1R Y721F cells. We observed a marked increase in the amplitude (∼3×) and duration (up to 20 minutes) of ERK1/2 phosphorylation in conjunction with a significantly lower, but constitutive activation of NF-κB p65 in both M−/−.Y721F (Figure 5A) and M−/−.3AB (Figure 5B) cells compared with their Tyr-721–expressing counterparts. Interestingly, consistent with the pTyr-721-mediated differential regulation of NF-κB targets (Figures 2 and 4; supplemental Table 4), we observed decreased NF-κB p65 activation at 5 minutes of stimulation in Tyr-721 cells (Figure 5A-B). These results indicate that pTyr-721 signaling attenuates ERK1/2 and NF-κB activation and is necessary for the transient suppression of NF-κB p65 phosphorylation.
CSF-1R pTyr-721/PI3K signaling regulates the amplitude and duration of ERK1/2 and NF-κB p65 activation and induces miR-21 expression. CSF-1–starved macrophages were stimulated with CSF-1 (120 ng/mL) for the indicated times and processed for western blotting (WB) or RNA extraction. (A) M−/−.WT and M−/−.Y721F macrophages and (B) M−/−.3ABY721 and M−/−.3AB macrophages were subjected to WB analysis with antibodies to the activated phosphorylated forms of ERK1/2 and NF-κB p65 and to the total ERK1/2 and NF-κB p65. (C) Relative qRT-PCR quantitation of miR-21 levels in M−/−.WT macrophages treated with PI3K inhibitor LY 2940002 alone (100 μM), in combination with either the ERK1/2 inhibitor PD98059 (50 μM) or the IKK inhibitor PS1145 (2 μM), or vehicle alone (1% dimethylsulfoxide). (D) miR-155 levels in M−/−.WT macrophages. Experiments performed as in panel A. Relative expression values in panels C-D indicate the fold-change of miRNA levels in CSF-1–treated cells relative to CSF-1–starved cells at the indicated times. Data are representative of 3 independent experiments (error bars, ±SD; *P ≤ .05). (E) M−/−.WT and M−/−.3ABY721 macrophages, treated for 48 hours with LNA-miR-21 inhibitor (I), or (F), an inhibitor mismatch control (M), were CSF-1–starved before CSF-1 stimulation for the indicated times and processed as described in panels A-B. Comparisons between matching cell lines (panels A-B; top 2 panels and bottom 2 panels of panels E-F) were made on the same blots (loading control, β-actin). (G) Schematic representation of pTyr-721–mediated signaling events leading to induction of miR-21 and suppression of inflammatory networks. Filled lines, relationships demonstrated in the present study; dashed lines, suggested interactions from the literature; arrows, activation; blunt arrows, inhibition; point arrows, kinetically regulated activation and inhibition. Ovals indicate molecule activity, not expression level. SIRPb1 and IL-1β are directly suppressed by miR-21 (supplemental Table 8), but are not necessarily the exclusive mediators of macrophage miR-21 effects on ERK1/2 and NF-κB. ERK1/2 activation is positively regulated through CSF-1R tyrosines 544, 559, and 807.28
CSF-1R pTyr-721/PI3K signaling regulates the amplitude and duration of ERK1/2 and NF-κB p65 activation and induces miR-21 expression. CSF-1–starved macrophages were stimulated with CSF-1 (120 ng/mL) for the indicated times and processed for western blotting (WB) or RNA extraction. (A) M−/−.WT and M−/−.Y721F macrophages and (B) M−/−.3ABY721 and M−/−.3AB macrophages were subjected to WB analysis with antibodies to the activated phosphorylated forms of ERK1/2 and NF-κB p65 and to the total ERK1/2 and NF-κB p65. (C) Relative qRT-PCR quantitation of miR-21 levels in M−/−.WT macrophages treated with PI3K inhibitor LY 2940002 alone (100 μM), in combination with either the ERK1/2 inhibitor PD98059 (50 μM) or the IKK inhibitor PS1145 (2 μM), or vehicle alone (1% dimethylsulfoxide). (D) miR-155 levels in M−/−.WT macrophages. Experiments performed as in panel A. Relative expression values in panels C-D indicate the fold-change of miRNA levels in CSF-1–treated cells relative to CSF-1–starved cells at the indicated times. Data are representative of 3 independent experiments (error bars, ±SD; *P ≤ .05). (E) M−/−.WT and M−/−.3ABY721 macrophages, treated for 48 hours with LNA-miR-21 inhibitor (I), or (F), an inhibitor mismatch control (M), were CSF-1–starved before CSF-1 stimulation for the indicated times and processed as described in panels A-B. Comparisons between matching cell lines (panels A-B; top 2 panels and bottom 2 panels of panels E-F) were made on the same blots (loading control, β-actin). (G) Schematic representation of pTyr-721–mediated signaling events leading to induction of miR-21 and suppression of inflammatory networks. Filled lines, relationships demonstrated in the present study; dashed lines, suggested interactions from the literature; arrows, activation; blunt arrows, inhibition; point arrows, kinetically regulated activation and inhibition. Ovals indicate molecule activity, not expression level. SIRPb1 and IL-1β are directly suppressed by miR-21 (supplemental Table 8), but are not necessarily the exclusive mediators of macrophage miR-21 effects on ERK1/2 and NF-κB. ERK1/2 activation is positively regulated through CSF-1R tyrosines 544, 559, and 807.28
CSF-1R pTyr-721–induced elevation of miR-21 is regulated via PI3K and ERK1/2
Previous studies have shown that phosphorylation of CSF-1R Tyr-721 mediates its association with and activation of PI3K to regulate macrophage motility.29 To determine the extent to which PI3K activation is critical for the CSF-1–induced expression of miR-21, CSF-1–starved M−/−.WT macrophages were treated with PI3K inhibitor LY294002, or vehicle, for 2 hours before stimulation with CSF-1 for different times and analysis of miR-21 levels by qRT-PCR (Figure 5C). As expected (Figure 4B), CSF-1 treatment resulted in a 3- and fivefold induction of miR-21 expression at 60 and 180 minutes of stimulation, respectively, relative to CSF-1–starved cells. In contrast, pharmacological inhibition of PI3K resulted in greater than 60% lower levels of CSF-1–induced miR-21 at all time points, indicating that the induction of miRNA-21 is in large part regulated by CSF-1R pTyr-721–activated PI3K.
Because pTyr-721 signaling also restricted the kinetics of ERK1/2 and NF-κB p65 activation and absence of pTyr-721 signaling enhanced the activation of ERK1/2 and NF-κB p65 (Figure 5A-B), we also examined the effect of combined inhibition of either PI3K and ERK1/2 (with LY294002 plus PD98059) or of PI3K and NF-κB p65 (with LY294002 plus PS114569 ) on CSF-1 induction of miR-21 (Figure 5C). Although combined inhibition of NF-κB p65 and PI3K resulted in miR-21 levels approximating those of PI3K inhibition alone, combined PI3K and ERK1/2 inhibition blocked miR-21 expression at levels comparable to those of CSF-1–starved cells, indicating that the activation of the PI3K/ERK1/2, but not NF-κB signaling, is critical for miR-21 expression and thus for suppression of the M1 phenotype. If the observed response is specific to miR-21, then miR-155, a typical M1 molecule, both regulated by and regulating the proinflammatory response,70-72 should not be induced by the CSF-1R pTyr-721/PI3K/ERK1/2 signaling pathway. Indeed, in vehicle-treated cells, we observed the CSF-1–induced suppression of miR-155 levels, with IEG kinetics (Figure 5D). Furthermore, although LY294002 treatment alone or followed by ERK1/2 inhibition suppressed the levels of miR-155 in unstimulated cells, they permitted its delayed induction, whereas combined inhibition of PI3K and NF-κB p65 completely suppressed induction of miR-155. These results indicate that the CSF-1/CSF-1R pTyr-721 axis coordinates the activation of PI3K, ERK1/2, and NF-κB p65 to respectively induce miR-21 and repress miR-155 to suppress the proinflammatory response. In addition, the pTyr-721–dependent transient decrease in NF-κB activation (Figure 5A-B), dependent on miR-21 induction (Figure 5E-F), could contribute to the suppression of the M1 response (Figure 5G).
Evidence for feedback regulation of CSF-1R pTyr-721/NF-κB/ERK1/2 signaling by miR-21
To further understand the circuitry connecting miR-21 with ERK1/2 and NF-κB p65 signaling downstream of pTyr-721, we addressed the existence of miR-21–mediated feedback loops. miR-21 is known to activate the PI3K/Akt axis through transcriptional downregulation of PTEN,68 suggesting its participation in a pTyr-721–initiated feed-forward loop activating PI3K/Akt (Figure 5G), whereas pTyr-721–activated PI3K29 may limit the observed ERK1/2 activation profile (Figure 5A-B). Moreover, although miR-21 suppresses the expression levels of a MEK/ERK1/2 pathway activator,73 SIRPb1 (supplemental Table 6), and of an NF-κB activator,51 IL-1β (Figure 4C; supplemental Table 6), miR-21 knock-down did not affect the levels of total ERK1/2 or NF-κB p65 protein (Figure 5E-F). However, pretreatment of pTyr-721–expressing macrophages with the LNA-miR-21 inhibitor (Figure 5E), but not mismatch control (Figure 5F), extended the duration and amplitude of ERK1/2 and NF-κB p65 activation in the inhibitor-treated cells that mimicked the behavior of CSF-1R Y721F cells (Figure 5A-B). Thus, these results are consistent with a negative feedback role for miR-21 on the activation of ERK1/2 and NF-κB p65. Although the comprehensive molecular network coordinating miR-21 induction and the observed kinetics of ERK1/2 or NF-κB p65 activation, downstream of pTyr-721, remains to be determined, possible mechanisms involved are shown schematically in Figure 5G.
Role of miR-21 in inflammatory processes in vivo
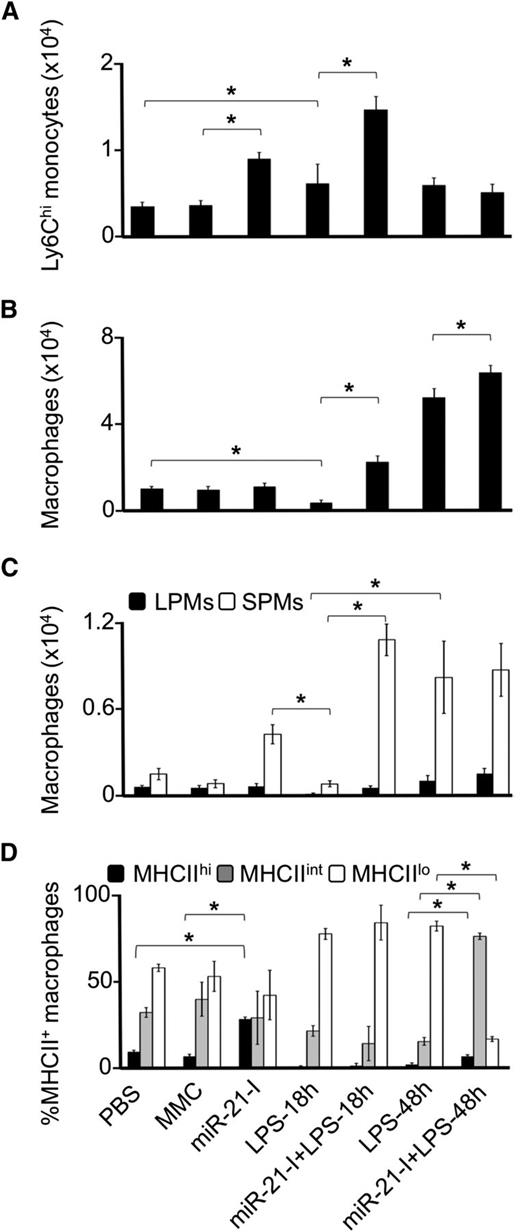
The response to intraperitoneal LPS involves an initial rapid recruitment of Ly6Chi (inflammatory) monocytes, which differentiate into M1 macrophages45 that accumulate within the cavity.45,74 Moreover, intraperitoneal injections of LPS cause a threefold elevation in circulating CSF-1 within 6 hours after injection.75 Because our in vitro experiments indicate that miR-21 suppresses the proinflammatory phenotype, we next examined the role of miR-21 during the peritoneal monocyte/macrophage response to LPS in vivo. Compared with control-treated mice, treatment with LNA-miR-21 inhibitor alone resulted in a twofold increase in Ly6Chi monocyte numbers, whereas there was a small increase in the number of peritoneal Ly6Chi monocytes at 18 hours post-LPS. Together, the inhibitor and LPS had an additive effect, whereas no significant effect of LPS, alone or with inhibitor, was observed at 48 hours post-LPS (Figure 6A). At 18 hours post-LPS treatment, total peritoneal macrophage numbers were decreased compared with mismatch or PBS control-injected mice (Figure 6B), as expected.45 Although treatment with miR-21 inhibitor alone had no significant effect, it synergistically enhanced the macrophage response to LPS at both 18 and 48 hours post-LPS (Figure 6B). These results indicate that miR-21 attenuates the recruitment of Ly6Chi monocytes to the peritoneal cavity, as well as the subsequent local accumulation of macrophages, during the local inflammatory response.
miR-21 suppresses the peritoneal monocyte/macrophage inflammatory response to LPS. Mice were intraperitoneally injected with endotoxin-free LNA-miR-21 inhibitor (LNA-miR-21-I) or mismatch inhibitor control (MMC) at days 1 and 3, followed by intraperitoneal injections of LPS or PBS at days 3 and 4. Peritoneal lavages were collected on day 5 at 18 or 48 hours post-LPS. Total viable CD11b+SSCAloLy6C+Ly6G− monocytes (A), CD11b+SSCAloLy6C−Ly6Glo/−F4-80+ macrophages (B), SPMs and large peritoneal macrophages (LPMs),45 (C) and the percent expression levels of the cell surface marker MHC II on the CD11b+SSCAloLy6C−Ly6Glo/-F4-80+ macrophages (D) were determined by flow cytometry. Data are representative of 2 independent experiments, 3 to 4 mice per group (error bars, ±SD; *P ≤ .05).
miR-21 suppresses the peritoneal monocyte/macrophage inflammatory response to LPS. Mice were intraperitoneally injected with endotoxin-free LNA-miR-21 inhibitor (LNA-miR-21-I) or mismatch inhibitor control (MMC) at days 1 and 3, followed by intraperitoneal injections of LPS or PBS at days 3 and 4. Peritoneal lavages were collected on day 5 at 18 or 48 hours post-LPS. Total viable CD11b+SSCAloLy6C+Ly6G− monocytes (A), CD11b+SSCAloLy6C−Ly6Glo/−F4-80+ macrophages (B), SPMs and large peritoneal macrophages (LPMs),45 (C) and the percent expression levels of the cell surface marker MHC II on the CD11b+SSCAloLy6C−Ly6Glo/-F4-80+ macrophages (D) were determined by flow cytometry. Data are representative of 2 independent experiments, 3 to 4 mice per group (error bars, ±SD; *P ≤ .05).
Previous work has demonstrated the accumulation of Ly6Chi-derived, MHC IIhi–expressing, M1-associated small peritoneal macrophages (SPMs) in the peritoneal fluid of mice treated with proinflammatory stimuli,45 from where they may migrate to lymph nodes and serve as antigen-presenting cells. However, regulators of SPM accumulation remain to be defined. Interestingly, we observed that treatment with LNA-miR-21 inhibitor alone increased recruitment of SPMs (Figure 6C) and the cell surface expression of M1 marker MHC II (Figure 6D)—and that LPS treatment enhanced these effects. Thus overall, our data indicate a miR-21 suppression of the proinflammatory response in vitro and in vivo.
Discussion
The role of the CSF-1R in promoting M2 polarization and mediating macrophage enhancement of tumor progression has been previously reported.26,76,77 Moreover, studies using CSF-1R Tyr/Phe mutants have underscored the role of distinct pTyr signaling pathways in the regulation of macrophage survival, proliferation, and motility.20,27-30 However, the regulatory networks downstream of the CSF-1R that control macrophage activation have not been reported. Here we focused on the macrophage CSF-1R pTyr-721 signaling pathway because in vitro coculture experiments have indicated its importance for enhancing tumor progression in vivo,29 suggesting a possible role in macrophage polarization. To understand the events downstream of this pathway, we took advantage of macrophage cell lines developed for CSF-1R structure/function studies27-29 and generated gene expression profiles (Figures 1 and 2; supplemental Figure 1) that were analyzed bioinformatically to predict processes, networks, and network directionality that were subsequently tested experimentally. We show that loss of a single CSF-1R tyrosine residue results in decreased expression of M2 markers and increased expression of M1 markers of macrophage polarization (Figures 1-3; supplemental Figures 1 and 2).
Loss of CSF-1R Tyr-721 was associated with loss of CSF-1 induction of miR-21 (Figure 4; supplemental Figure 4), which could be due to inefficient processing of miR-21 from its precursor forms,78-80 decreased stability of mature miR-21,81 or other processes.82 To investigate the regulation of M1/M2 functions by miR-21, the predicted mRNA targets were validated and shown to predominantly encode molecules promoting an M1 phenotype (Figure 4; supplemental Tables 6-8). Although miR-21 mimics could not be used to demonstrate suppression of M1 phenotype because of the activating effect of double-stranded nucleic acids on this response in macrophages83,84 (data not shown), an LNA-miR-21 inhibitor increased the M1 response (Figure 4). As far as the M2 response is concerned, our data (Figure 2, Figure 3D-F; supplemental Figure 4) do not allow us to further detail the pathways underlying the effects of CSF-1 alone or the combination of CSF-1 and IL-4 on the M2 response (Figure 3). However, as pTyr-721 signaling alone increased the expression of IL-4Rα (supplemental Tables 1-3) in the absence of differential secretion of IL-4, IL-10, or IL-13 (data not shown) and in addition, because our bioinformatics analysis predicted a pTyr-721–mediated repression of miR-223, an miRNA specific for IL-4/IL-4R signaling85 (supplementary Table 5), the CSF-1R pTyr-721/miR-21 pathway positively regulates expression of M2 markers in an IL-4/IL-10/IL-13–independent manner, while strongly priming the M2 response to exogenous IL-4, at least in part through induction of IL-4Rα. Possible mechanisms explaining the miR-21 activation of the M2 phenotype could involve miR-21 targeting of the expression of molecules that negatively regulate M2 transcript levels86 or miR-21 activation of mRNA stability factors87 that block miR-21 binding to noncanonical sites on M2 marker mRNAs.
TLR ligands induce miR-21 expression,63,88 and increased miR-21 levels have been reported in several inflammatory conditions such as osteoarthritis,89 ulcerative colitis,90 cardiac muscle injury,91 cardiac hypertrophy,92 and psoriasis93 as well as in H. pylori–induced gastric cancer95 and the inflamed lungs of LPS-treated mice.95 Many of these conditions were associated with alterations in the M1/M2 macrophage polarization state,93,96 thus leading to the hypothesis that miR-21 is an indicator of inflammation. However, inhibition of miR-21 downstream of pTyr-721 showed (Figure 4; supplemental Figures 4 and 5) that miR-21 decreases M1 and enhances M2 responses (summarized in Figure 5G). Our data suggest that the elevated miR-21 levels observed in these diseases reflect a feedback control mechanism that suppresses the M1 phenotype and promotes anti-inflammatory/repair programs in macrophages.
Our results show that there is a previously overlooked, CSF-1R–modulated molecular network that coordinately suppresses inflammatory responses and regulates the polarization state in macrophages (Figure 5). This network comprises CSF-1R pTyr-721, the downstream signaling kinases PI3K and ERK1/2, the p65 subunit of NF-κB and miR-21. In a rapid, positive feed-forward manner, the pTyr-721–mediated signaling activates the PI3K/Akt axis,29 restricts the duration and amplitude of ERK1/2 and NF-κB p65 activation (Figure 5A-B), and upregulates miR-21 (Figure 4B) in a PI3K- and ERK1/2- dependent, but IKK/NF-κB–independent manner (Figure 5C). Subsequently, upregulated miR-21 suppresses the expression of more than 30 proinflammatory genes (Figure 4C; supplemental Tables VI-VIII), including M1 markers (iNOS, TNFα, and IL-6; supplemental Figure 5), and induces transcription of 4 M2 genes (Arginase1, Mannose Receptor1, FIZZ, and IL-4Rα; supplemental Figure 4).
Additionally, we show that pTyr-721–induced miR-21 is necessary for a slower, positive feedback loop that restricts the early activation steps of ERK1/2 and NF-κB p65 (Figure 5E,F), possibly by transcriptional downregulation of miR-21 targets PTEN (a negative regulator of the PI3K/Akt axis68 ), SIRPb1 (an ERK1/2 activator73 ), and IL-1β (an NF-κB activator51 ). Our results indicate that miR-21 functions as a molecular hub that integrates signals from the CSF-1R pTyr-721/PI3K signaling axis to transcriptionally suppress the development of proinflammatory responses, to enhance M2 polarization, and, by feedback regulation, to increase its own expression (Figure 5G). Thus the miR-21 network provides a framework for targeting specific CSF-1R–mediated functions in macrophages.
Other studies have addressed the in vivo role of miR-21 in different contexts, focusing on its antiapoptotic or proinflammatory roles, alone or downstream of TLRs or IFN-γ, and observed no miR-21–mediated monocyte or macrophage recruitment.88,91-99 Our data show that intraperitoneal administration of the LNA-miR-21 inhibitor enhances Ly6Chi monocyte recruitment and M1 macrophage accumulation at an inflammatory site, suggesting miR-21–mediated suppression of M1 activities of these cells in vivo (Figure 6). Further investigation of the roles of miR-21 targets in these processes and in enhancement of the LPS response downstream of the CSF-1R are needed to understand the underlying mechanisms involved.
CSF-1 is expressed on many different tumor cell types (reviewed in Chitu et al100 ) and under the control of CSF-1, tumor-associated macrophages exert pro-tumoral (M2-like) responses in vivo.26,101,102 Furthermore, in vitro, the enhancement of tumor cell invasion by macrophages is CSF-1R pTyr-721–dependent,29 and a recent study demonstrates the importance of macrophage miRNA expression, including miR-21 and miR-29, during tumor cell proliferation and angiogenesis.102 M1-like macrophages mediate antitumoral responses by producing cytotoxic molecules or by modulating the adaptive immune response through presentation of MHC class II–bound peptides to cytotoxic T cells,103-107 which may then promote tumor cell destruction through the activation of tolerized T cells in tumor tissue. Consistent with these findings, we have shown that macrophage pTyr-721 signaling coordinates, via miR-21, the suppression of proinflammatory (antitumor) responses and that inhibition of miR-21 in vivo upregulates the cell surface expression of MHC II. Thus, the miR-21 network that we have identified represents a potential therapeutic target for the treatment of solid tumors.
Our data have identified miR-21 as a novel modulator of macrophage polarization and shown that miR-21 deficiency, by either genetic ablation of CSF-1R Tyr-721 or direct pharmacological inhibition of miR-21, skews macrophages toward an M1 phenotype. Because miR-21 is a known tumor-associated molecule108 and inhibition of the CSF-1R is relevant for many macrophage-mediated diseases, further studies on the role of miR-21 and its targets in the control of macrophage-mediated inflammation should provide important insights into the signaling mechanisms downstream of the CSF-1R that are relevant to both inflammation and cancer.
The microarray data reported in this article have been deposited into the Gene Expression Omnibus (accession number GSE62630).
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Gregoire Lauvau, Fiona J. Pixley, and Jeffrey E. Segall for critically reading the manuscript; Matthew Ritchie and Keith Satterley for advice in the initial stages of this work; Dr Arthur Skoultchi and Michael Wilcockson for mice; Shanisha Gordon for technical assistance; David M. Reynolds of the Einstein Genomics Facility for running the microarrays; and the Einstein Flow Cytometry Core Facility and Drs Jeffrey Pollard, Roy Noy, Laura Santambrogio, and Brian Scharf and Carolina Rodriguez for reagents.
This work was supported by grants from the National Institutes of Health, National Cancer Institute grants PO1 CA100324, CA32551, and CA26504 (E.R.S.), and 5P30-CA13330 (Cancer Center Grant to the Albert Einstein College of Medicine).
Authorship
Contribution: C.I.C. designed the research, performed research, analyzed and interpreted data, performed the Ingenuity Pathway Analysis and Cytoscape analyses, made figures, and wrote the manuscript; X.G. performed bioinformatics analyses and reviewed the manuscript; D.Z. performed bioinformatics analyses and reviewed the manuscript; L.T. performed research and analyzed data; T.D.B. contributed vital reagents and reviewed the manuscript; A.V. contributed vital reagents; and E.R.S. supervised and designed the research, carried out experiments, analyzed and interpreted data, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: E. Richard Stanley, Department of Developmental and Molecular Biology, Albert Einstein College of Medicine, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: richard.stanley@einstein.yu.edu.

![Figure 1. The CSF-1R pTyr721-regulated macrophage transcriptome. (A) Schematic depiction of the CSF-1Rs in the macrophage cell lines used to assess the necessity (loss of function, upper panels) and sufficiency (gain of function, lower panels) of pTyr-721 signaling in macrophages. The CSF-1R domains (extracellular [ECD]; transmembrane [TM], and tyrosine kinase [TK]) and the intracellular CSF-1R Tyr (Y, black) residues mutated to Phe (F, red) are indicated. (B) Pairwise comparisons of gene expression data for all 28 361 transcripts detected on microarrays, showing high Pearson correlation coefficients between biological replicates and the expected hierarchical clustering of samples, as a function of genotype and CSF-1 treatment. Results for M−/−.WT and M−/−.Y721F (upper panel) and M−/−.3ABY721 and M−/−.3AB (lower panel) are shown as heat maps illustrating the significant differences in gene expression between Tyr-721–expressing and Tyr-721–nonexpressing cells, each with an associated color key and histogram (−, without CSF-1; +, with 120 ng/mL of CSF-1; biological replicates are indicated by the numerals 1 and 2). (C-E) Venn diagrams, representing the size of the pTyr-721–regulated transcriptome, identified by each approach. To obtain the pTyr-721–regulated genes, the CSF-1–regulated transcripts expressed in either M−/−.WT or M−/−.3ABY721 cells were compared with those of the corresponding Y721F mutant cells. (C) The Y721F mutation (1904 CSF-1–regulated mRNAs) reduced the number of CSF-1–regulated mRNAs of M−/−.WT macrophages (2174) by 270 transcripts. However, of the CSF-1–regulated genes in both cell types, 1713 (∼9% of 19 000 total mouse genes) were differentially regulated in both data sets. (D) The addition of CSF1R Y721 to the 3AB background increased the number of CSF-1–regulated transcripts by 250 (from 870 to 1120). Of the CSF-1–regulated genes in both cell types, 244 (∼1.3% of 19 000 total mouse genes) were differentially regulated in both data sets. (E) Only 34 genes are common between the 2 datasets.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/8/10.1182_blood-2014-10-608000/5/m_e1f1.jpeg?Expires=1769100359&Signature=yFEgywB8iLYb~XLbyqa3pGhzxY1Qlw0C8fHV0mERwZzJdsVKaJTpxevZDuLQ5cJIIoeOQet-9qCbou5L1qi7x4iDd7sB2ca4dn1Y36BAcXy8EKXWhINn1BM~9bQ363Rzr0ohkOQ8XkO6dDQ1b~9rltZmQDySrR3CqAO2-4RIc-8YNce2sKsujGLy7vkiBx3KMYcfhAZSmTZk5zeAaPIrH53S7RqtnxN4lWQLrk1ioT5NOxBMMyk0dVP8pH7Llg~oRYm07vLsqXRMg9~-tFGTv4FwHmTSQFfxKjV8dijC39IxnjLDgGXlJHp4bsbSI-c20pWtgg4hIHCKfUk~xbMM2w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. CSF-1R pTyr-721 controls expression of a specific subset of IEGs and DEGs associated with suppression of inflammation and promotion of an M2 phenotype. (A) Bar charts representing the relative abundance of a subset of pTyr-721–induced IEGs (left panel) and DEGs (center and right panels), illustrating pTyr-721–mediated decreased abundance of transcripts encoding proinflammatory (M1, *) molecules in conjunction with increased abundance of anti-inflammatory (M2, **) molecules. Known IEGs or DEGs are shown in bold. CSF-1–regulated transcript abundance in M−/−.WT cells (ie, log2 fold change [FC] between the expression of the indicated RNA at the indicated time point and its measured expression at time 0) was normalized to CSF-1–regulated mRNA abundance in M−/−.Y721F macrophages calculated in the same manner. For a full set of pTyr-721–regulated genes, see supplemental Table 3. (B) Relative abundance of a subset of pTyr-721–induced genes in cells growing in CSF-1 (see supplemental Table 1) that encode secreted cytokines, illustrating the pTyr-721–mediated change in abundance of transcripts encoding M1 (*) and M2 (**) molecules (P < .05, FC > 1.5). (C-D) pTyr-721 suppresses secretion of proinflammatory molecules (*) and induces secretion of tumorigenic EGF, insulin-like growth factor-1 (IGF-1), and matrix metalloproteinase-3 (MMP-3). Conditioned medium from CSF-1–treated (120 ng/mL) and CSF-1–starved M−/−.WT and M−/−.Y721F macrophages was assayed for production of proinflammatory and proangiogenic factors using a mouse cytokine array permitting simultaneous measurements of 144 cytokines and soluble factors. For each cell line, signals detected for each cytokine produced by CSF-1–treated cells were integrated and normalized to the corresponding signal obtained from CSF-1–starved cells (duplicates ± range). (C) Molecules exhibiting pTyr-721–dependent increased secretion. (D) Molecules exhibiting increased secretion in the absence of pTyr-721 signaling. Error bars indicate the range of levels of biological duplicates.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/8/10.1182_blood-2014-10-608000/5/m_e1f2.jpeg?Expires=1769100359&Signature=DwdY9X4A7QAWfsOALqL5z9n0gSo2EbgspZbULWLxaHKTQ3UqZxQFU7j-i37V6T5Mhn51XdDSkHBmLlyHy-z7cXFQJsO9DLxaCKCzQgFGvUhz-LAUeJdRqkNYMEb-BBpq0yqfkk9znxOADH6G8uZYgy9PwC6xMJN1Z2qCMMbDzR0ATpMO2aA00XMLWSJSZIjXffJ8ptUbxaNSGIBUJK3wLBmWoUNhtVS1Va6VATX-OSQ0RWW34vYfPBqPZ4QfWxYstqsDRe9P04pf8PS3-7Fdo7NMvjYXIKPUJ9d9Wk6oiPkHd0JgvRV3Ptw2yn-U30IWRHDotTEQm-tlgY1TY9008w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)