Key Points
NF-κB and AKT signaling prevent RAG-dependent DNA damage in cycling-transformed pre-B cells.
NF-κB activity negatively correlates with RAG expression in B-ALL patients.
Abstract
In developing lymphocytes, expression and activity of the recombination activation gene protein 1 (RAG1) and RAG2 endonuclease complex is tightly regulated to ensure ordered recombination of the immunoglobulin genes and to avoid genomic instability. Aberrant RAG activity has been implicated in the generation of secondary genetic events in human B-cell acute lymphoblastic leukemias (B-ALLs), illustrating the oncogenic potential of the RAG complex. Several layers of regulation prevent collateral genomic DNA damage by restricting RAG activity to the G1 phase of the cell cycle. In this study, we show a novel pathway that suppresses RAG expression in cycling-transformed mouse pre-B cells and human pre-B B-ALL cells that involves the negative regulation of FOXO1 by nuclear factor κB (NF-κB). Inhibition of NF-κB in cycling pre-B cells resulted in upregulation of RAG expression and recombination activity, which provoked RAG-dependent DNA damage. In agreement, we observe a negative correlation between NF-κB activity and the expression of RAG1, RAG2, and TdT in B-ALL patients. Our data suggest that targeting NF-κB in B-ALL increases the risk of RAG-dependent genomic instability.
Introduction
The adaptive immune system plays a crucial role in the defense against pathogens, functioning by virtue of highly specific antigen receptors expressed on B and T cells. Effective immunity requires a diverse repertoire of these antigen receptors, which is achieved by recombination of variable (V), diversity (D), and joining (J) gene segments of the immunoglobulin (Ig) and T-cell receptor (Tcr) loci.1 VDJ recombination requires the recombination activation gene proteins 1 and 2 (RAG1 and RAG2) to instigate DNA breaks in recombination signal sequences (RSSs) that flank the recombining gene segments.2,3 At the pro-B-cell stage, RAG1/2 initiates Ig heavy chain (Igh) recombination after which RAG is downregulated, followed by several rounds of cell division at the large pre-B-cell stage. Subsequently, pre-B cells exit the cell cycle, and RAG expression is upregulated resulting in Ig light chain (Igl) recombination. The functional expression of a tolerant (non-self) B-cell receptor (BCR) switches off RAG, whereas expression of an autoreactive BCR leads to prolonged RAG expression, thereby allowing secondary Igl recombinations in a process known as receptor editing.4,5
Signals emanating from the interleukin-7 receptor (IL7R) and the pre-B-cell receptor (pre-BCR) regulate the dynamic pattern of RAG expression, which involves phosphoinositide-3 kinase (PI3K) and protein kinase B (PKB, also known as AKT) impinging on forkhead box O (FOXO) transcription factors that are required for RAG expression.6,7 The interplay between these signals ensures a sharp demarcation between proliferation and Ig gene recombinations in order to conserve genomic stability in pre-B cells. Additionally, RAG2 protein is phosphorylated at threonine 490 (T490) by the cyclin A/cyclin-dependent kinase 2 (CDK2) complex, eliciting S phase kinase-associated protein 2 (SKP2) –mediated ubiquitination and protein degradation in S phase.8,9 A breach of this regulation results in genomic instability that activates a p53-dependent checkpoint, as was shown by the increased lymphomagenesis in p53-deficient RAG2-T490A mice.10
There is ample evidence for the involvement of RAG in chromosomal aberrations in lymphomas and leukemias, which underscores the importance of proper regulation of this potentially harmful recombination mechanism.11 Moreover, B-cell acute lymphoblastic leukemias (B-ALLs) show a developmental block at the pro- to pre-B cell stage and frequently display constitutive RAG, terminal deoxy-transferase (TdT) expression, and ongoing Ig gene recombinations.12,13 Recent genome-wide analyses of BCR-ABL-positive and ETV6-RUNX1-positive B-ALL have shown that breakpoints of secondary genetic events frequently map near RSS motifs, suggesting the involvement of RAG.14,15 Given its oncogenic potential, a deeper understanding of the regulation of RAG expression and activity is warranted. About 25% of adult B-ALL and 5% of childhood B-ALL patients carry the BCR-ABL1 fusion gene,16 a tyrosine kinase that mimics IL7R and pre-BCR signaling.17 Here, we made use of human BCR-ABL-positive B-ALL cell lines, Abelson-transformed (Abl) mouse pre-B cells, and IL7-dependent mouse pre-B cell cultures representing tractable models to study the regulation of RAG expression in (transformed) pre-B cells because inhibition and/or abrogation of BCR-ABL, Abl, or IL7 signaling induces differentiation that is accompanied by RAG expression and Igl recombination.18,19 In addition, we studied RAG expression in BCR-ABL-negative primary human B-ALL samples. We report the unexpected finding that nuclear factor κB (NF-κB) and AKT signaling suppresses RAG expression and activity in cycling-transformed mouse pre-B cells and in human B-ALL cells and show that inhibition of NF-κB and AKT signaling results in RAG-dependent DNA damage.
Materials and methods
Cell culture and small molecule inhibitors
Abl-transformed mouse pre-B cell lines generated from wild-type (WT) and RAG2−/− mice carrying an Eμ-Bcl2 transgene were kindly provided by Dr Craig Bassing (University of Pennsylvania School of Medicine, Philadelphia, PA). The human BCR-ABL-positive B-ALL cell lines BV173 and SUP-B15 were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Cells were treated with the following small molecule inhibitors at 106 cells per milliliter as indicated: STI571 (imatinib methanesulfonate, LC Laboratories, Woburn, MA), BMS-345541 (Sigma Aldrich), GSK-690693 (Selleckchem, Houston, TX), MLN120B (MCE MedChem Express, Princeton, NJ), CAL-101 (Idelalisib; Selleckchem), and PD-0332991 (Palbociclib; Selleckchem).
Immunoblotting
Protocols for immunoblotting experiments are available in the supplemental Data available at the Blood Web site.
Flow cytometry
PCR analysis and real-time reverse transcription PCR
Vκ6-23 to Jκ1 coding joins were determined in mouse Abl cells by semiquantitative polymerase chain reaction (PCR) by using previously published primers.22 PCR was performed on 100, 25, and 6.25 ng of DNA. For expression analysis by quantitative real-time reverse transcription PCR, RNA was isolated by using TRI Reagent (Sigma Aldrich), and equal amounts of RNA (0.5 μg) were first-strand transcribed into complementary DNA by using random primers (Promega Corp., Fitchburg, WI) and Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA). Gene expression levels were normalized by using RPLP0 (ribosomal protein, large, P0) and 18S ribosomal RNA (rRNA) housekeeping genes for human and mouse samples, respectively. Primer sequences are listed in supplemental Table 1.
Analysis of Jκ2-RSS signal-end DNA breaks in G1 and S phase cell cycle cells
NF-κB transcription factor activity assay
NF-κB transcription factor p65 and p50 DNA-binding activity was measured in 5 μg of nuclear extracts from stimulated BV173 and SUP-B15 cells by using a commercially available enzyme-linked immunosorbent assay kit (p50/p65 Transcription Factor Assay Kit; ab133128, Abcam, Cambridge, MA) according to the manufacturer’s instructions.
Expression constructs and retroviral transductions
The RAG-reporter construct was used as described previously.25 The retroviral RAG-reporter and IκBαSR (IκBα S32A/S36A) constructs were cotransfected with the pCL-ECO plasmid in Phoenix-A cells. For retroviral transduction, cells and retroviral supernatants were spun for 90 minutes at 2000g onto retronectin-coated 24-well plates (5 × 105 cells per well) according to the manufacturer’s instructions (Takara Bio Inc., Otsu, Shiga, Japan).
Primary mouse bone marrow pre-B-cell cultures
C57Bl/6 WT mice were housed in the animal research facility of the Academic Medical Center under specific pathogen-free conditions. Animal experiments were approved by the Animal Ethics Committee and were performed in agreement with national and institutional guidelines. Primary mouse pre-B-cell cultures were performed as described previously20 ; details are available in the supplemental Data.
Patient samples
Bone marrow aspirates and peripheral blood mononuclear cells from B-ALL patients containing >90% blasts were obtained and prepared according to ethical standards of our institutional medical ethical committee, as well as in agreement with the Helsinki Declaration of 1975, as revised in 1983. Cells were cultured for 48 hours in supplemented Iscove’s modified Dulbecco’s medium with 20% fetal calf serum, in the presence of 2.5 μM NF-κB kinase β (IKKβ) inhibitor (IKKβi) and 2.5 μM AKT inhibitor (AKTi), or vehicle (dimethylsulfoxide; untreated). Patient cytogenetic characteristics are listed in supplemental Table 2.
Gene expression analysis
Data from a previously published study in which the gene expression profiles of 207 untreated B-ALL patients were determined26,27 were reanalyzed using the R2 microarray analysis and visualization platform developed in our institute and publicly available (http://r2.amc.nl).
Statistics
The GraphPad Prism software package (GraphPad Software, La Jolla, CA) was used for statistical testing.
Results
AKT and NF-κB suppress RAG activity in Abl mouse pre-B cells
RAG activity in mouse Abl pre-B cells was assessed by using a green fluorescence protein (GFP)-based RAG activity reporter.25 Abl-transformed pre-B cells can be induced to undergo differentiation toward small pre-B cells with the Abl kinase inhibitor STI571.18,22 As expected, STI571 induced GFP expression in WT but not in RAG2−/− Abl cells (Figure 1A). The PI3K p110δ inhibitor CAL-101 (PI3Ki) and AKTi GSK-690693 increased RAG-reporter activity, showing that RAG activity is suppressed by PI3K and AKT (Figure 1B). Treatment with IKKβi BMS-345541 resulted in a modest but consistent increase in RAG activity. Strikingly, combined treatment with IKKβi and PI3Ki or AKTi synergistically increased RAG activity (Figure 1B-C). A structurally unrelated IKKβi (MLN120B) yielded similar results, demonstrating the specificity of this effect (supplemental Figure 1).
AKT and NF-κB regulate RAG activity in mouse Abl pre-B cells. (A) Representative fluorescence-activated cell sorter plots of the WT and RAG2−/− mouse Abl pre-B cells transduced with the retroviral RAG-reporter construct consisting of an antisense GFP complementary DNA flanked by a 12-bp spacer RSS and a 23-bp spacer RSS, followed by an IRES-RFP cassette. RAG activity mediates inversional recombination of the antisense GFP gene, which can be quantified by flow cytometry. WT and RAG2−/− mouse Abl pre-B cells were treated for 96 hours with 10 μM STI571. (B) WT mouse Abl pre-B cells transduced with the RAG-reporter construct stimulated with 5 μM PI3Ki, 2.5 μM AKTi, 2.5 μM IKKβi, or untreated (dimethylsulfoxide [DMSO] vehicle) for 96 hours. GFP histograms (RAG-reporter activity) of transduced cells are shown by gating on RFP+ cells. (C) Titration curve for IKKβi and for IKKβi plus 2.5 μM AKTi. RAG-reporter activity (y-axis) of WT mouse Abl pre-B cells is plotted against the concentration of IKKβi (x-axis). RAG-reporter activity of mouse Abl pre-B cells treated with 5 μM STI571 is shown. Cells were treated for 72 hours. A representative example of 3 independent experiments is shown, 4 replicate measurements were performed per experiment, and error bars show means ± standard deviation (SD).
AKT and NF-κB regulate RAG activity in mouse Abl pre-B cells. (A) Representative fluorescence-activated cell sorter plots of the WT and RAG2−/− mouse Abl pre-B cells transduced with the retroviral RAG-reporter construct consisting of an antisense GFP complementary DNA flanked by a 12-bp spacer RSS and a 23-bp spacer RSS, followed by an IRES-RFP cassette. RAG activity mediates inversional recombination of the antisense GFP gene, which can be quantified by flow cytometry. WT and RAG2−/− mouse Abl pre-B cells were treated for 96 hours with 10 μM STI571. (B) WT mouse Abl pre-B cells transduced with the RAG-reporter construct stimulated with 5 μM PI3Ki, 2.5 μM AKTi, 2.5 μM IKKβi, or untreated (dimethylsulfoxide [DMSO] vehicle) for 96 hours. GFP histograms (RAG-reporter activity) of transduced cells are shown by gating on RFP+ cells. (C) Titration curve for IKKβi and for IKKβi plus 2.5 μM AKTi. RAG-reporter activity (y-axis) of WT mouse Abl pre-B cells is plotted against the concentration of IKKβi (x-axis). RAG-reporter activity of mouse Abl pre-B cells treated with 5 μM STI571 is shown. Cells were treated for 72 hours. A representative example of 3 independent experiments is shown, 4 replicate measurements were performed per experiment, and error bars show means ± standard deviation (SD).
AKT and NF-κB regulate RAG transcription by inhibiting FOXO1
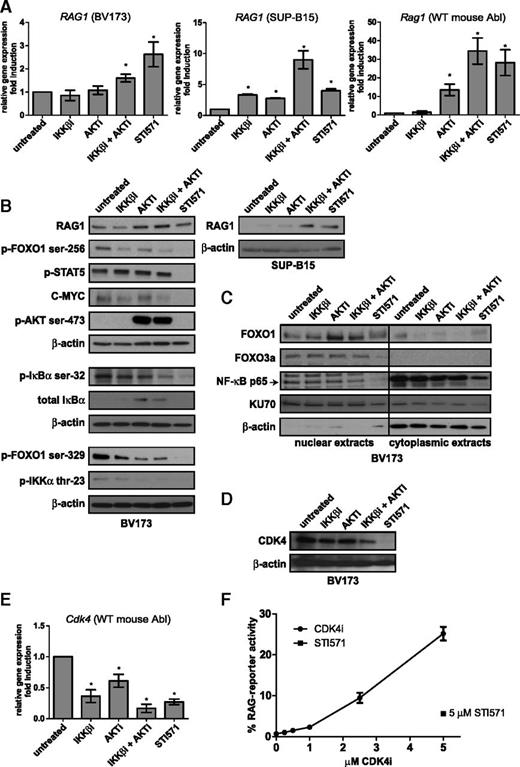
Combined treatment with IKKβi and AKTi induced RAG1 and RAG2 messenger RNA (mRNA) expression in Abl mouse pre-B cells and in two human BCR-ABL-positive B-ALL cell lines (BV173 and SUP-B15) at levels comparable to those observed after STI571 treatment (Figure 2A and supplemental Figure 2); RAG1 protein expression was correspondingly induced (Figure 2B). We next studied the expression of FOXO1 and FOXO3a, which are known regulators of RAG transcription.6,7 Nuclear FOXO1 levels were increased upon AKTi treatment and upon combined AKTi and IKKβi treatment, whereas FOXO3a levels remained unchanged (Figure 2C). Concomitantly, phosphorylation of FOXO1 on serine 256 and serine 329, which negatively regulate its stability, was decreased in cells treated with IKKβi and AKTi (Figure 2B). Recently, it was demonstrated that cyclin-dependent kinase 4 (CDK4) suppresses RAG expression in Myc-driven lymphomas by phosphorylating FOXO1 at serine 329.28 In line with this, we found that CDK4 protein and mRNA expression was decreased in IKKβi- and AKTi-treated cells (Figure 2D-E). Moreover, treatment with the CDK4-specific inhibitor PD-0332991 (CDK4i) increased RAG activity to a degree comparable to that with IKKβi and AKTi treatment (Figure 2F).
Transcriptional regulation of RAG1 by the AKT and NF-κB pathways. (A) Real-time reverse transcription PCR (RT-PCR) analysis of RAG1 mRNA in the human BCR-ABL-positive B-ALL cell lines BV173 (left) and SUP-B15 (middle) and Rag1 mRNA in the WT mouse Abl pre-B cell line (right). Cells were treated with 2.5 μM IKKβi, 2.5 μM AKTi, or 10 μM STI571 for 48 hours. (B) Immunoblot analysis of whole-cell extracts from BV173 and SUP-B15 cells treated as in (A). (C) Immunoblot analysis of nuclear and cytoplasmic extracts from BV173 cells treated as in (A). (D) Immunoblot analysis of CDK4 cells treated as in (A). β-actin was used as loading control in (B) and (D). KU70 is expressed in the nucleus and the cytoplasm and was used as loading control in (C). (E) Cdk4 real-time RT-PCR analysis of the WT mouse Abl pre-B cell line; cells were treated as in (A), and results were normalized to those in untreated cells (DMSO vehicle). Real-time RT-PCR results are presented relative to the expression of the housekeeping genes RPLPO (for human cells) and 18S ribosomal RNA (rRNA) (mouse cells); PCRs were performed at least in duplicate, and error bars show means ± SD of 3 independent experiments. (F) Titration curve of CDK4i. RAG-reporter activity of WT mouse Abl pre-B cells treated with 5 μM STI571 is plotted against the concentration of CDK4i. Cells were stimulated for 96 hours. A representative example of 3 independent experiments is shown, 4 replicate measurements were performed per experiment, and error bars represent means ± SD. *P < .05, as determined by the 1-sample Student t test.
Transcriptional regulation of RAG1 by the AKT and NF-κB pathways. (A) Real-time reverse transcription PCR (RT-PCR) analysis of RAG1 mRNA in the human BCR-ABL-positive B-ALL cell lines BV173 (left) and SUP-B15 (middle) and Rag1 mRNA in the WT mouse Abl pre-B cell line (right). Cells were treated with 2.5 μM IKKβi, 2.5 μM AKTi, or 10 μM STI571 for 48 hours. (B) Immunoblot analysis of whole-cell extracts from BV173 and SUP-B15 cells treated as in (A). (C) Immunoblot analysis of nuclear and cytoplasmic extracts from BV173 cells treated as in (A). (D) Immunoblot analysis of CDK4 cells treated as in (A). β-actin was used as loading control in (B) and (D). KU70 is expressed in the nucleus and the cytoplasm and was used as loading control in (C). (E) Cdk4 real-time RT-PCR analysis of the WT mouse Abl pre-B cell line; cells were treated as in (A), and results were normalized to those in untreated cells (DMSO vehicle). Real-time RT-PCR results are presented relative to the expression of the housekeeping genes RPLPO (for human cells) and 18S ribosomal RNA (rRNA) (mouse cells); PCRs were performed at least in duplicate, and error bars show means ± SD of 3 independent experiments. (F) Titration curve of CDK4i. RAG-reporter activity of WT mouse Abl pre-B cells treated with 5 μM STI571 is plotted against the concentration of CDK4i. Cells were stimulated for 96 hours. A representative example of 3 independent experiments is shown, 4 replicate measurements were performed per experiment, and error bars represent means ± SD. *P < .05, as determined by the 1-sample Student t test.
AKT and NF-κB inhibition does not interfere with Abl signaling
In Abl-transformed pre-B cells, the Abl kinase activity results in constitutive STAT5 phosphorylation, which is inhibited by STI571 (Figure 2B). However, phospho-STAT5 remained unchanged in cells treated with IKKβi and AKTi, showing that Abl kinase signaling was not abrogated. In addition, C-MYC was detectable in IKKβi- and AKTi-treated cells, whereas STI571 treatment led to the loss of C-MYC (Figure 2B) and the induction of a G1 cell cycle arrest (data not shown). The DNA binding activities of p65 and p50 NF-κB transcription factors were modestly but significantly decreased after IKKβi and AKTi treatment (∼50% reduction) (supplemental Figure 3), consistent with a decreased level of phosphorylated inhibitor of κBα (IκBα) and a modest decrease in nuclear p65 (Figure 2C). We also show that AKT is involved in NF-κB signaling because IκBα was increased whereas phosphorylation of IKKα on threonine 23 was decreased by AKTi treatment. Treatment with AKTi resulted in increased phosphorylation of AKT on serine 473 (Figure 2B) as a result of disturbed negative feedback regulation, which was previously described.29-31
AKT and NF-κB inhibit endogenous Iglκ recombination
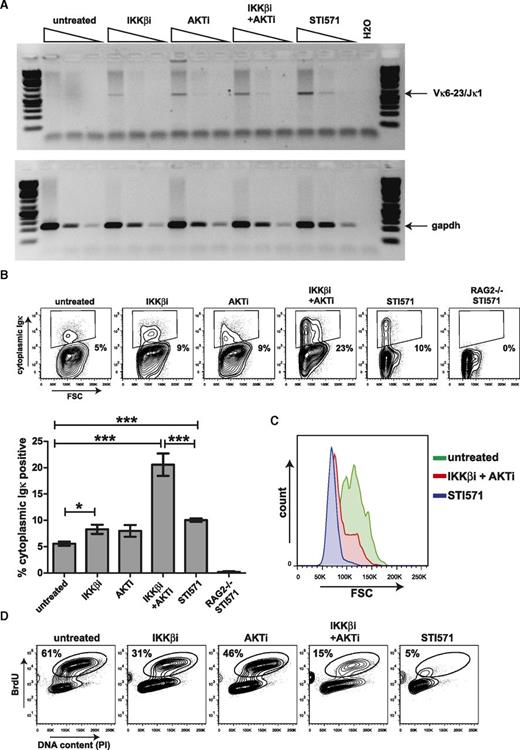
To determine whether endogenous Iglκ recombination was regulated by NF-κB, we assessed Vκ6-23 to Jκ1 coding joins by semiquantitative PCR. Coding joins were observed in Abl cells treated with STI571 and also in cells treated with IKKβi, AKTi, or IKKβi and AKTi combined (Figure 3A). In addition, cytoplasmic Igκ light chain expression was significantly induced in Abl cells treated with IKKβi and AKTi, exceeding induction by STI571 (Figure 3B). Cytoplasmic κ light chain expression upon treatment with STI571 was accompanied by the transition from large to small pre-B cells, as shown by forward scatter (FSC) analysis. However, a sizeable fraction of IKKβi- and AKTi-treated cells expressing cytoplasmic κ light chain remained large, suggesting that these cells did not exit the cell cycle (Figure 3C), which is in agreement with our finding that C-MYC is still detectable under these conditions (Figure 2B). In addition, a considerable portion of the cells (15%) incorporated BrdU. Although BrdU incorporation in the IKKβi- and AKTi-treated cultures was diminished compared with that in untreated cells (15% vs 61%), it was clearly higher than that in STI571-treated cells (15% vs 5%) (Figure 3D).
Inhibition of AKT and NF-κB signaling results in recombination of the endogenous Iglκ locus in mouse Abl pre-B cells. (A) Semiquantitative PCR analysis of Vκ6-23 to Jκ1 coding joins in mouse Abl pre-B cells treated with 2.5 μM IKKβi, 2.5 μM AKTi, or 10 μM STI571 for 72 hours. Semiquantitative PCR analysis for Gapdh was performed as a loading control. (B) Representative fluorescence-activated cell sorter (FACS) plots showing cytoplasmic Igκ expression vs forward scatter (FSC) in mouse Abl pre-B cells stimulated as in (A). Numbers below outlined gates indicate percentage of cells. Graph depicts percentages of cytoplasmic Igκ-positive cells in mouse Abl pre-B cell cultures stimulated for 72 hours. A representative example of 2 independent experiments is shown, 4 replicate measurements were performed per experiment, and error bars represent means ± SD. Statistical significances between groups were determined by 1-way analysis of variance (ANOVA) using a Bonferroni’s posttest. (C) Representative FACS plot showing overlay FSC histograms of cytoplasmic Igκ-positive mouse Abl pre-B cells treated as indicated. (D) Representative FACS plots showing BrdU incorporation vs DNA content as measured by propidium iodide (PI) staining in mouse Abl pre-B cells treated for 48 hours as indicated above FACS plots. Numbers above outlined gates indicate percentage of cells. *P < .05; ***P < .001.
Inhibition of AKT and NF-κB signaling results in recombination of the endogenous Iglκ locus in mouse Abl pre-B cells. (A) Semiquantitative PCR analysis of Vκ6-23 to Jκ1 coding joins in mouse Abl pre-B cells treated with 2.5 μM IKKβi, 2.5 μM AKTi, or 10 μM STI571 for 72 hours. Semiquantitative PCR analysis for Gapdh was performed as a loading control. (B) Representative fluorescence-activated cell sorter (FACS) plots showing cytoplasmic Igκ expression vs forward scatter (FSC) in mouse Abl pre-B cells stimulated as in (A). Numbers below outlined gates indicate percentage of cells. Graph depicts percentages of cytoplasmic Igκ-positive cells in mouse Abl pre-B cell cultures stimulated for 72 hours. A representative example of 2 independent experiments is shown, 4 replicate measurements were performed per experiment, and error bars represent means ± SD. Statistical significances between groups were determined by 1-way analysis of variance (ANOVA) using a Bonferroni’s posttest. (C) Representative FACS plot showing overlay FSC histograms of cytoplasmic Igκ-positive mouse Abl pre-B cells treated as indicated. (D) Representative FACS plots showing BrdU incorporation vs DNA content as measured by propidium iodide (PI) staining in mouse Abl pre-B cells treated for 48 hours as indicated above FACS plots. Numbers above outlined gates indicate percentage of cells. *P < .05; ***P < .001.
AKT and NF-κB signaling prevents RAG-dependent DNA damage in cycling-transformed pre-B cells
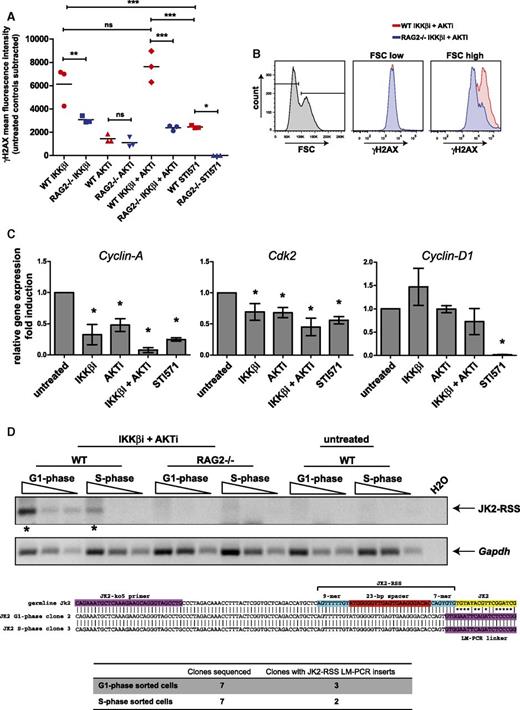
Flow cytometric analysis of phosphorylated H2AX (γH2AX) in WT and RAG2−/− Abl cells revealed that treatment with IKKβi and AKTi resulted in significantly higher levels of intracellular γH2AX in RAG-proficient cells, indicating that these cells experienced RAG-dependent DNA damage. RAG-dependent DNA damage was also increased in cells treated with IKKβi alone, but not with AKTi (Figure 4A). Strikingly, RAG-dependent γH2AX staining was exclusively observed in the large cells (FSC high), suggesting that the cycling cells sustained DNA damage (Figure 4B). In cycling cells, RAG2 protein expression is restricted by cyclin A/CDK2-mediated phosphorylation, whereas RAG1 is not regulated in a cell cycle–dependent manner.32,33 Interestingly, the levels of cyclin A and CDK2 mRNA expression were significantly decreased in IKKβi- and AKTi-treated Abl cells at a level comparable to that in STI571-treated cells, whereas the level of cyclin D1 was unaffected in contrast to STI571-treated cells (Figure 4C). The low levels of cyclin A/CDK2 could provide a window for RAG2 protein expression and RAG activity in cycling cells. To provide additional evidence that RAG is active in cycling cells under these conditions, we performed Jκ2-RSS signal-end LM-PCR on IKKβi- and AKTi-treated WT and RAG2−/− mouse Abl pre-B cells sorted according the G1 and S phase of the cell cycle. This approach detects transient RAG-dependent DNA breaks at the Jκ2-RSS upstream of the Jκ2 coding region. Upon visual inspection, Jκ2-RSS breaks appeared most prevalent in the G1 phase but were also detectable in S phase cells (Figure 4D). Cloning and sequencing of the PCR products obtained from G1 phase cells showed that 3 of 7 clones harbored the correct insert, whereas 2 of 7 clones obtained from S phase cells contained Jκ2-RSS linker fragments, indicating RAG activity in S phase. Negative clones harbored PCR products caused by promiscuity of the Jκ2-ko5 LM-PCR forward primer (data not shown). Jκ2-RSS LM-PCR products were not detected in RAG2−/− cells, confirming their RAG-dependence. Moreover, Jκ2-RSS breaks were not detected in untreated WT mouse Abl pre-B cells, consistent with the absence of endogenous Iglκ recombinations in untreated cells as shown in Figure 3A.
Inhibition of AKT and NF-κB signaling induces RAG-dependent DNA damage in mouse Abl pre-B cells. (A) Mean fluorescence intensity (MFI) of intracellular γH2AX staining in mouse Abl pre-B cells treated as indicated on x-axis. Red symbols represent γH2AX MFI in WT cells, and blue symbols represent γH2AX MFI in RAG2−/− cells. MFIs of untreated controls were subtracted to correct for background staining. Three independent experiments were performed, and horizontal lines represent means. Statistical significances between groups were determined by ANOVA using a Bonferroni’s posttest. (B) Overlay FACS histograms of intracellular γH2AX staining in IKKβi- and AKTi-treated cells. Small B cells (FSC low; left gate) vs large B cells (FSC high; right gate) are shown. (C) Real-time RT-PCR of Cyclin-A, Cdk2, and Cyclin-D1 mRNA expression in WT mouse Abl pre-B cells. Cells were treated with 2.5 μM IKKβi, 2.5 μM AKTi, or 10 μM STI571 for 48 hours. Real-time RT-PCR results are presented relative to the expression of the 18S rRNA housekeeping gene. Three independent experiments were performed, PCRs were performed at least in duplicate, and error bars represent means ± SD of 3 independent experiments. (D) Jκ2-RSS DNA breaks are detected by LM-PCR in cell-cycle sorted (G1 and S phase) WT mouse Abl pre-B cells that were treated with 2.5 μM IKKβi and 2.5 μM AKTi. Threefold dilutions of linker-ligated DNA were amplified with a Jκ2-specific forward primer and an LM-PCR linker-specific reverse primer. PCR products indicated with an asterisk were excised from the gel, cloned, and sequenced. The middle section of panel D shows the alignments of sequences obtained from G1 and S phase cells to the germline Jκ2 sequence. Jκ2-ko5 primer site, Jκ2-RSS, and the Jκ2 coding region are indicated by colored boxes. Sequencing results are listed at the bottom of panel D. *P < .05; **P < .01; ***P < .001. ns, not significant.
Inhibition of AKT and NF-κB signaling induces RAG-dependent DNA damage in mouse Abl pre-B cells. (A) Mean fluorescence intensity (MFI) of intracellular γH2AX staining in mouse Abl pre-B cells treated as indicated on x-axis. Red symbols represent γH2AX MFI in WT cells, and blue symbols represent γH2AX MFI in RAG2−/− cells. MFIs of untreated controls were subtracted to correct for background staining. Three independent experiments were performed, and horizontal lines represent means. Statistical significances between groups were determined by ANOVA using a Bonferroni’s posttest. (B) Overlay FACS histograms of intracellular γH2AX staining in IKKβi- and AKTi-treated cells. Small B cells (FSC low; left gate) vs large B cells (FSC high; right gate) are shown. (C) Real-time RT-PCR of Cyclin-A, Cdk2, and Cyclin-D1 mRNA expression in WT mouse Abl pre-B cells. Cells were treated with 2.5 μM IKKβi, 2.5 μM AKTi, or 10 μM STI571 for 48 hours. Real-time RT-PCR results are presented relative to the expression of the 18S rRNA housekeeping gene. Three independent experiments were performed, PCRs were performed at least in duplicate, and error bars represent means ± SD of 3 independent experiments. (D) Jκ2-RSS DNA breaks are detected by LM-PCR in cell-cycle sorted (G1 and S phase) WT mouse Abl pre-B cells that were treated with 2.5 μM IKKβi and 2.5 μM AKTi. Threefold dilutions of linker-ligated DNA were amplified with a Jκ2-specific forward primer and an LM-PCR linker-specific reverse primer. PCR products indicated with an asterisk were excised from the gel, cloned, and sequenced. The middle section of panel D shows the alignments of sequences obtained from G1 and S phase cells to the germline Jκ2 sequence. Jκ2-ko5 primer site, Jκ2-RSS, and the Jκ2 coding region are indicated by colored boxes. Sequencing results are listed at the bottom of panel D. *P < .05; **P < .01; ***P < .001. ns, not significant.
Expression of NF-κB superrepressor stimulates RAG activity
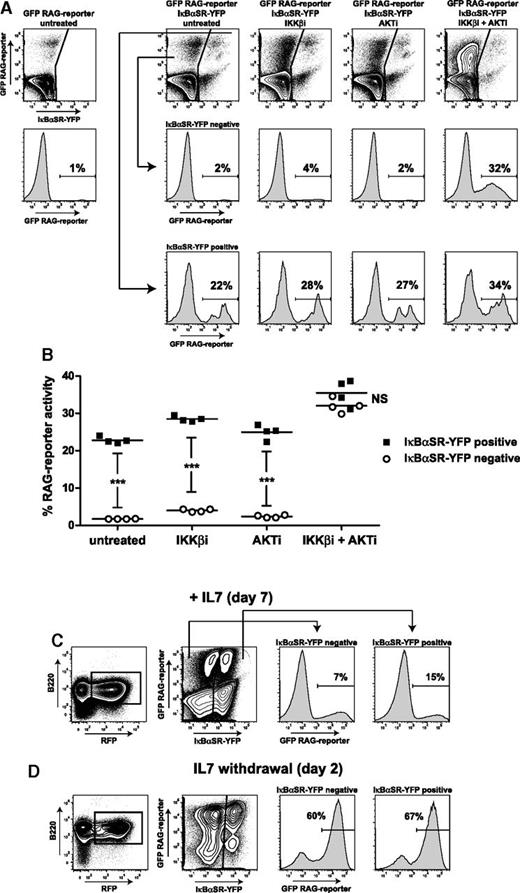
To provide further evidence that NF-κB signaling suppresses RAG activity, we introduced the NF-κB superrepressor IκBαSR-IRES-YFP cassette together with the RAG-reporter construct into the WT mouse Abl pre-B cell line. NF-κB signaling was effectively repressed as shown by diminished lipopolysaccharide-induced CD69 expression in IκBαSR-transduced cells (supplemental Figure 4). We found that IκBαSR-expressing cells (YFP+RFP+) had ∼10-fold higher RAG activity compared with the control cell population (YFP–RFP+) (mean of 24% vs 2% GFP+ cells) (Figure 5A). In agreement, Rag1 mRNA expression was ∼10-fold higher in fluorescence-activated cell sorter–purified IκBαSR-YFP+ cells compared with IκBαSR-YFP– cells (supplemental Figure 5). Treatment with IKKβi and AKTi increased RAG activity in the control cell population (increasing from a mean of 2% to 32% GFP+ cells), reaching a level similar to that in IκBαSR-transduced cells (Figure 5B), showing limited increase of RAG activity upon treatment (∼1.5-fold). We assessed whether NF-κB is also involved in suppressing RAG expression in untransformed primary mouse pro-B cells. In vitro expanded primary mouse pre-B cells were transduced with the IκBαSR and the RAG-reporter constructs and analyzed by flow cytometry 7 days after transduction. In the presence of IL7 and Flt3L, RAG activity was approximately twofold higher in the IκBαSR-transduced cells compared with the control cell population (15% vs 7% GFP+ cells) (Figure 5C). Withdrawal of IL7 increased RAG activity, upon which inhibition of NF-κB signaling had no substantial effect (60% vs 67% GFP+ cells) (Figure 5D).
NF-κB superrepressor augments RAG activity in mouse Abl pre-B cells and in IL7-dependent untransformed mouse pre-B cell cultures. (A) FACS plots of WT mouse Abl pre-B cells transduced with IκBαSR-YFP and RAG-reporter (GFP) IRES-RFP. Scatter plots show YFP (x-axis) vs GFP (y-axis) within RFP-positive gate. Histograms show RAG-reporter (GFP) in IκBαSR-YFP–negative cells (middle row) and in IκBαSR-YFP–positive cells (bottom row). Cells were treated for 96 hours as indicated above FACS plots. Numbers above outlined gates indicate percentage of cells. (B) Graph depicting percentage of RAG-reporter activity (GFP) in IκBαSR-YFP–positive cells and IκBαSR-YFP–negative cells. Horizontal lines represent means, and 4 independent experiments were performed. Statistical significances were determined by two-way ANOVA. (C) FACS scatter plots of untransformed mouse pre-B cells cultured for 7 days with IL7. Pre-B cells (B220+, IgM–CD43+) were retrovirally transduced with IκBαSR-YFP and RAG-reporter (GFP) IRES-RFP constructs. Gating strategy is shown (B220+RFP+). FACS histograms show RAG-reporter activity (GFP) in IκBαSR-YFP–negative and IκBαSR-YFP–positive cells. A representative example of 3 independent experiments is shown. (D) RAG-reporter activity (GFP) 2 days after IL7 withdrawal. Numbers above outlined gates indicate percentage of cells. A representative example of 2 independent experiments is shown. ***P < .001.
NF-κB superrepressor augments RAG activity in mouse Abl pre-B cells and in IL7-dependent untransformed mouse pre-B cell cultures. (A) FACS plots of WT mouse Abl pre-B cells transduced with IκBαSR-YFP and RAG-reporter (GFP) IRES-RFP. Scatter plots show YFP (x-axis) vs GFP (y-axis) within RFP-positive gate. Histograms show RAG-reporter (GFP) in IκBαSR-YFP–negative cells (middle row) and in IκBαSR-YFP–positive cells (bottom row). Cells were treated for 96 hours as indicated above FACS plots. Numbers above outlined gates indicate percentage of cells. (B) Graph depicting percentage of RAG-reporter activity (GFP) in IκBαSR-YFP–positive cells and IκBαSR-YFP–negative cells. Horizontal lines represent means, and 4 independent experiments were performed. Statistical significances were determined by two-way ANOVA. (C) FACS scatter plots of untransformed mouse pre-B cells cultured for 7 days with IL7. Pre-B cells (B220+, IgM–CD43+) were retrovirally transduced with IκBαSR-YFP and RAG-reporter (GFP) IRES-RFP constructs. Gating strategy is shown (B220+RFP+). FACS histograms show RAG-reporter activity (GFP) in IκBαSR-YFP–negative and IκBαSR-YFP–positive cells. A representative example of 3 independent experiments is shown. (D) RAG-reporter activity (GFP) 2 days after IL7 withdrawal. Numbers above outlined gates indicate percentage of cells. A representative example of 2 independent experiments is shown. ***P < .001.
NF-κB and AKT suppress RAG expression in primary human pre-B ALL cells
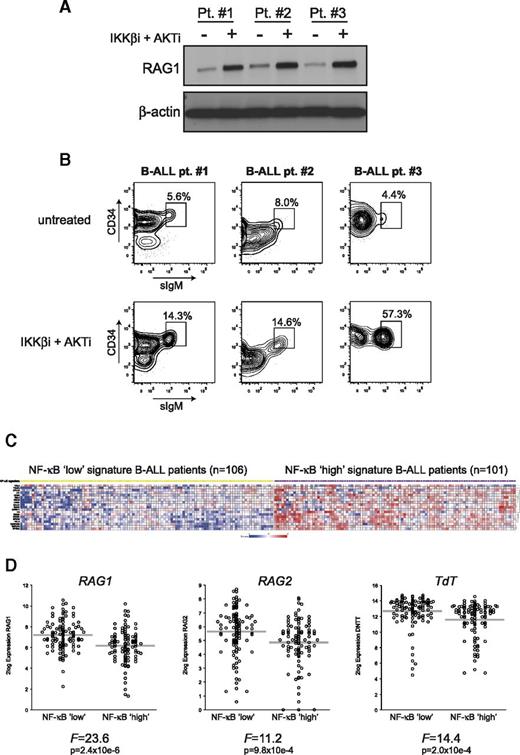
To confirm that the NF-κB and AKT pathways regulate RAG in primary human B-ALL cells, we stimulated B-ALL blasts in vitro from 3 (BCR-ABL–) patients with the combination of IKKβi and AKTi. RAG1 protein expression was increased in all 3 patients (Figure 6A). In addition, surface IgM expression on B-ALL blasts (CD19+CD10+CD34+/−) from these patients was increased (ranging from 1.8-fold to 13-fold) after treatment (Figure 6B), which may suggest increased RAG activity resulting in productive IgM expression in these cells.
Inhibition of AKT and NF-κB signaling induces RAG1 protein expression in primary human B-ALL cells, and NF-κB gene expression signature negatively correlates with RAG1, RAG2, and TdT mRNA expression in untreated B-ALL patients. (A) Immunoblot analysis of whole-cell extracts from primary (BCR-ABL-negative) B-ALL blasts (n = 3) cultured for 48 hours with 2.5 μM IKKβi and 2.5 μM AKTi or vehicle (DMSO; untreated). (B) FACS plots of primary human B-ALL cells from 3 patients cultured for 48 hours with 2.5 μM IKKβi and 2.5 μM AKTi or vehicle (DMSO; untreated). FACS plots show CD34 vs surface IgM (sIgM) expression within the CD19+ gate; gated events were CD10+ (data not shown). Numbers above outlined gates indicate percentage of cells. (C) Heatmap showing NF-κB low (yellow squares) and NF-κB high (purple squares) B-ALL patient subgroups based on a 19 NF-κB pathway/target classifier gene expression profile. Each column represents a B-ALL patient, and each row represents a unique gene classifying the 2 subgroups. (D) Relative RAG1, RAG2, and TdT mRNA expression in NF-κB low and NF-κB high signature B-ALL patients. Each circle represents an individual patient, 2log expression is depicted, and gray bars indicate means. The ANOVA F-statistic and P value are given below each graph.
Inhibition of AKT and NF-κB signaling induces RAG1 protein expression in primary human B-ALL cells, and NF-κB gene expression signature negatively correlates with RAG1, RAG2, and TdT mRNA expression in untreated B-ALL patients. (A) Immunoblot analysis of whole-cell extracts from primary (BCR-ABL-negative) B-ALL blasts (n = 3) cultured for 48 hours with 2.5 μM IKKβi and 2.5 μM AKTi or vehicle (DMSO; untreated). (B) FACS plots of primary human B-ALL cells from 3 patients cultured for 48 hours with 2.5 μM IKKβi and 2.5 μM AKTi or vehicle (DMSO; untreated). FACS plots show CD34 vs surface IgM (sIgM) expression within the CD19+ gate; gated events were CD10+ (data not shown). Numbers above outlined gates indicate percentage of cells. (C) Heatmap showing NF-κB low (yellow squares) and NF-κB high (purple squares) B-ALL patient subgroups based on a 19 NF-κB pathway/target classifier gene expression profile. Each column represents a B-ALL patient, and each row represents a unique gene classifying the 2 subgroups. (D) Relative RAG1, RAG2, and TdT mRNA expression in NF-κB low and NF-κB high signature B-ALL patients. Each circle represents an individual patient, 2log expression is depicted, and gray bars indicate means. The ANOVA F-statistic and P value are given below each graph.
To further explore whether the activity of the NF-κB pathway is related to RAG expression in primary B-ALL patients, we reanalyzed the gene expression profiles of a previously published cohort of BCR-ABL-negative childhood B-ALL patients.26,27 This heterogeneous patient group consists of 207 untreated high-risk B-ALL patients. We performed unsupervised clustering based on the expression of 19 NF-κB pathway/target genes by using a k-means clustering algorithm that identified an NF-κB low signature B-ALL patient group (n = 106) and an NF-κB high signature patient group (n = 101) (Figure 6C). Although there is considerable overlap between the patient groups that reflects their heterogeneous nature, the NF-κB low signature group displayed significantly increased RAG1, RAG2, and TdT mRNA expression compared with the NF-κB high signature group (Figure 6D), highlighting the negative correlation between NF-κB activity and RAG expression in primary B-ALL patients. Of interest, pathway analysis showed that the DNA replication pathway was significantly associated with the NF-κB signature groups (P < .0001), whereas the NF-κB low signature group showed higher expression of DNA replication genes and had significantly higher white blood cell counts (supplemental Figure 6A-B).
Discussion
In B-ALL, many secondary genetic events bear the hallmarks of being derived from aberrant RAG activity, suggesting that RAG is an important instigator of genomic instability.14,15 Aberrant RAG activity also contributes to subclonal diversification of B-ALL, which results in the development of therapy-resistant subclones.34 Despite these recent insights, the (mis)regulation of RAG expression in B-ALL remains poorly understood. During B-cell development, pre-B cell proliferation and RAG expression and/or activity are strictly separated, preventing genomic instability. Multiple layers of regulation safeguard this separation, which may be disrupted in proliferating leukemic cells that constitutively express RAG. In BCR-ABL-positive B-ALL, large cycling pre-B cells are transformed by the Abl kinase activity, preventing their differentiation toward the immature B-cell stage. Typically, proliferating BCR-ABL-positive B-ALL cells do not show RAG activity, indicating that the separation between proliferation and recombination is (still) enforced.19,35 Inhibition of the Abl kinase with STI571 leads to induction of RAG expression and activity accompanied by cell cycle exit, similar to that in normal pre-B or immature B cells.18,19 In proliferating pre-B cells, RAG expression is repressed by AKT signaling that negatively regulates FOXO1. Our results show that the NF-κB pathway contributes to the repression of RAG activity in cycling-transformed pre-B cells by regulating FOXO1. NF-κB was implicated in regulating Iglκ locus accessibility36 ; however, it is unlikely that increased RAG activity upon NF-κB inhibition can be ascribed to effects on locus accessibility because the RAG reporter used in this study measured RAG activity independently of Iglκ accessibility.
The role of AKT in the regulation of RAG expression in Abl cells is not entirely clear; previous results with an Abl cell line generated from a RAG1GFP knockin mouse showed that AKT inhibition by the AKTVIII inhibitor did not result in increased RAG1GFP expression,37 whereas the expression of constitutively active AKT suppressed RAG1GFP expression in untransformed B220+IgM– cells.7 We show that the specific AKTi GSK-69069329 increased RAG activity in Abl cells. Interestingly, the AKT-mediated RAG repression may operate by direct inhibition of FOXO1 and by feeding into NF-κB–mediated FOXO1 regulation, because simultaneous inhibition of the AKT and the NF-κB pathways resulted in increased levels of nuclear FOXO1, a concomitant decrease in FOXO1 phosphorylation, and a synergistic effect on RAG activity. In support, we show that AKT is involved in NF-κB activation by phosphorylating IKKα on serine 23, as was shown previously.38 In addition, we demonstrate that expression of IκBαSR had a stimulatory effect on RAG activity in Abl cells and in primary mouse pre-B cells, albeit to a lesser extent.
Several studies addressed the role of NF-κB in early B-cell development with discordant results. NF-κB was shown to be involved in Iglκ germline transcription but not RAG activity.36 Other studies suggest that NF-κB is dispensable for B-cell development.39,40 More recently, it was shown that NF-κB is active in pre-B cells and in immature B cells engaged in receptor editing during Iglλ recombination.41,42 Strikingly, in an earlier study, it was shown that NF-κB1/p50-deficient B cells had elevated RAG expression and activity.43 To add to the complexity, kinase-dead IKKα knockin mice showed defective B-cell development because of decreased PAX5 and IRF4 gene expression.44 It is likely that different modalities of NF-κB signaling exist in normal and transformed developing B cells that determine the outcome of this pleiotropic signaling pathway.
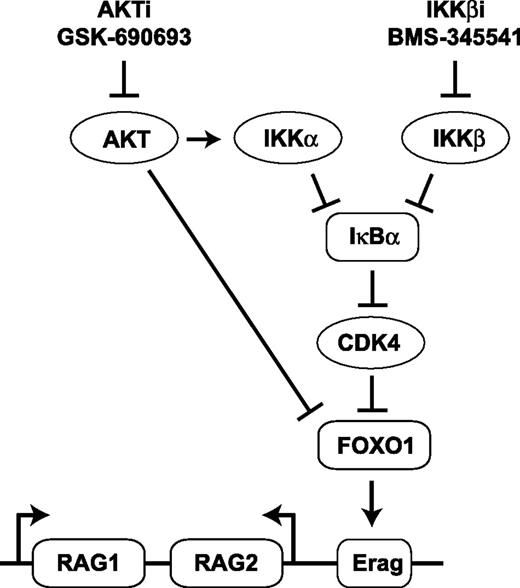
Recently, it was shown that CDK4 suppresses RAG expression in B-cell lymphomas in Eμ-Myc-transgenic mice. In that model, CDK4 deficiency accelerated lymphomagenesis in a RAG-dependent fashion by regulating FOXO1 levels. The authors showed that CDK4 phosphorylates FOXO1 on residue serine 329, which provokes nuclear exclusion.28 Of interest, CDK4 activity is regulated by NF-κB,45 and it was shown that IκBα binds to CDK4 thereby inhibiting its kinase function.46 We demonstrated that simultaneous inhibition of the NF-κB and the AKT pathways resulted in diminished Cdk4 mRNA and CDK4 protein levels and decreased FOXO1-serine 329 phosphorylation. In addition, specific inhibition of CDK4 increased RAG activity. On the basis of these observations, we speculate that IκBα stabilization could promote RAG expression by inhibition of CDK4, which acts as a negative regulator of FOXO1 (Figure 7). The NF-κB pathway is one of the major regulators of antiapoptotic gene expression during B-cell development, which makes it intrinsically difficult to assess other functions of this pathway because perturbation of NF-κB has detrimental effects on cell survival. It is possible that the antiapoptotic function of NF-κB is (partly) uncoupled from its other functions in Abl cells because Abl kinase activity can prevent apoptosis independently of NF-κB.47 Moreover, the mouse Abl cells used in this study harbor the Eμ-Bcl2 transgene and express the antiapoptotic BCL2 protein, which may have helped to uncover the role of NF-κB in the regulation of RAG expression. In support of this, treatment of the mouse and human BCR-ABL-positive cell lines with the IKKβi and AKTi resulted in only a minor increase in cell death (<10% specific cell death) over the course of the experiments described, whereas the primary human (BCR-ABL-negative) B-ALL samples displayed a much greater induction of cell death (ranging from 50% to 80% specific cell death) upon treatment (data not shown).
Model indicating the potential mechanism by which IKKβi and AKTi stimulate RAG expression in pre-B cells. Arrows represent positive and/or stimulatory effects. Horizontal bars represent inhibitory and/or repressive effects. Erag, conserved transcriptional enhancer in the RAG locus.
Model indicating the potential mechanism by which IKKβi and AKTi stimulate RAG expression in pre-B cells. Arrows represent positive and/or stimulatory effects. Horizontal bars represent inhibitory and/or repressive effects. Erag, conserved transcriptional enhancer in the RAG locus.
Our findings indicate that RAG expression induced after AKT and NF-κB inhibition does not coincide with cell cycle exit in Abl cells, which could have oncogenic potential because RAG activity during S phase is linked to lymphomagenesis.10 Consistent with this, our study shows that AKT and NF-κB inhibition leads to increased RAG-dependent DNA damage, most prominently in large cycling cells, which may provoke genomic instability. In agreement, our LM-PCR analysis confirms that RAG is active in S phase under these conditions.
Inhibition of NF-κB and AKT also increased RAG protein expression in primary BCR-ABL-negative human B-ALL cells, similar to that in mouse Abl cells and human BCR-ABL-positive B-ALL cell lines, suggesting that these pathways are important for suppressing RAG expression in primary human leukemic cells. Moreover, NF-κB and AKT inhibition resulted in increased surface IgM expression on leukemic blasts, which suggests that these cells may have experienced RAG activity. In support, by reanalysis of gene expression data, we found that NF-κB negatively correlated with RAG expression in primary BCR-ABL-negative B-ALL patients, although it must be noted that the differences between the NF-κB signature patient groups were small, reflecting the heterogeneity within these groups. In addition, NF-κB activity negatively correlated with the expression of DNA replication genes, and patients with an NF-κB low signature had higher white blood cell counts, suggesting that these patients may suffer from a more aggressive disease. These results indicate that NF-κB could have tumor-suppressive qualities in B-ALL, as was suggested previously for other cell types.48 Importantly, several novel therapies for leukemias and lymphomas are aimed at the NF-κB and PI3K-AKT pathways and at CDK4.49-52 In light of our data, it is conceivable that such treatments might have adverse effects in leukemia patients by unleashing the oncogenic potential of RAG, thus driving (sub)clonal diversification and genomic instability of leukemic pre-B cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Craig Bassing (University of Pennsylvania School of Medicine, Philadelphia, PA) for providing wild-type and RAG2−/− mouse Abl pre-B cell lines; Drs Inês Trancoso and Jocelyne Demengeot (Instituto Gulbenkian, Oeiras, Portugal) for the RAG-reporter (green fluorescence protein) IRES-RFP construct; Dr Hergen Spits (Academic Medical Center, Amsterdam, The Netherlands) for the IκBαSR-IRES-YFP construct; Prof Dr Ellen van der Schoot and Dr Christa Homburg (Sanquin Research Institute, Amsterdam, The Netherlands) and Dr Edwin Sonneveld (Children’s Oncology Foundation [within the Stichting Kinderoncologie Nederland], The Hague, The Netherlands) for sharing patient material and data; and Dr Carol Schrader (University of Massachusetts Medical School, Worcester, MA) for helpful discussions.
This work was supported by a fellowship from the Academic Medical Center and Innovational Research Incentives Scheme VIDI grant 016126355 from The Netherlands Organization for Scientific Research (both to J.E.J.G).
Authorship
Contribution: K.O.-M. and J.E.J.G. designed the research; K.O.-M., M.B., T.A.B., C.M., and J.E.J.G. performed the research; A.M.d.B. provided mouse materials and reagents; K.O.-M., M.B., T.A.B., R.J.B., C.J.M.v.N., and J.E.J.G. analyzed the data; K.O.-M., M.B., and J.E.J.G. wrote the manuscript; and all authors edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeroen E. J. Guikema, Department of Pathology, Academic Medical Center, Meibergdreef 9, 1105 AZ, Amsterdam, The Netherlands; e-mail: j.e.guikema@amc.uva.nl.
References
Author notes
K.O.-M. and M.B. contributed equally to this work.

![Figure 1. AKT and NF-κB regulate RAG activity in mouse Abl pre-B cells. (A) Representative fluorescence-activated cell sorter plots of the WT and RAG2−/− mouse Abl pre-B cells transduced with the retroviral RAG-reporter construct consisting of an antisense GFP complementary DNA flanked by a 12-bp spacer RSS and a 23-bp spacer RSS, followed by an IRES-RFP cassette. RAG activity mediates inversional recombination of the antisense GFP gene, which can be quantified by flow cytometry. WT and RAG2−/− mouse Abl pre-B cells were treated for 96 hours with 10 μM STI571. (B) WT mouse Abl pre-B cells transduced with the RAG-reporter construct stimulated with 5 μM PI3Ki, 2.5 μM AKTi, 2.5 μM IKKβi, or untreated (dimethylsulfoxide [DMSO] vehicle) for 96 hours. GFP histograms (RAG-reporter activity) of transduced cells are shown by gating on RFP+ cells. (C) Titration curve for IKKβi and for IKKβi plus 2.5 μM AKTi. RAG-reporter activity (y-axis) of WT mouse Abl pre-B cells is plotted against the concentration of IKKβi (x-axis). RAG-reporter activity of mouse Abl pre-B cells treated with 5 μM STI571 is shown. Cells were treated for 72 hours. A representative example of 3 independent experiments is shown, 4 replicate measurements were performed per experiment, and error bars show means ± standard deviation (SD).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/11/10.1182_blood-2015-01-621623/4/m_1324f1.jpeg?Expires=1770857402&Signature=EyCEw-IGNzQ7f9W2DT1zmsVWzf~~0pE5DV3rBdMfD-BDMZpg~g3Gno~voaRm0DJy5T66wduQcl4Zc8EMtshDV5yj3~K8Mdqrz~9eLC41VRd3uMMKHpNCWSLMJPp9R~zI5~Dth1M47PI8IrSRYW3wFQxYnc194W1bZMyTXH1STwD8QYOIXwuJ3vjskr1Pi1AKM3c5miQ7I5DrqpOdDQzl-v~UiK8eBFYkEomCoMMI5d0ut8oVYW-BpQ2Z8tyefLgS~xItPRnY3G9hz-vi-yf90nY2MSYH2bLdRoQqse1MBdhrH7TmnbOGLNCvfCXVP52oC1tpI4L6hGYWrhYh7U71kw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal