Abstract
Diffuse large B-cell lymphoma (DLBCL) is a molecularly heterogeneous group of diseases consisting of gene translocations, amplifications, and mutations. G-quadruplex (G4) secondary DNA structures arising from at least 4 contiguous runs of guanines may facilitate this genomic instability. The rapidly progressing G4 field has demonstrated their involvement in nuclear processes, particularly replication and transcription. We previously demonstrated that DNA secondary structures within the MYC and BCL2 promoter regions serve as therapeutic targets for modulating oncogene expression. However, the prevalence and molecular associations of G4 formation within lymphoma tissues is unknown. In previous studies, cancer tissues, such as stomach and liver carcinomas, displayed higher G4 frequencies compared to their benign counterparts using a novel antibody. Here, we characterize the incidence of G4 structures within a highly annotated cohort of DLBCL.
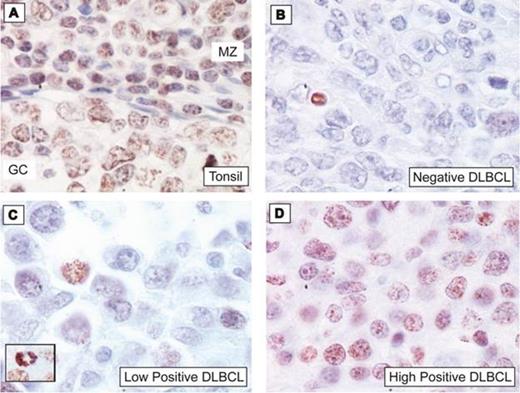
We successfully adapted a manual cell-based assay for G4 detection with a FLAG-tagged single-chain Fv antibody (BG4) to develop an automated colorimetric immunohistochemistry (IHC) protocol on a BenchMark® XT instrument (VMSI, Tucson AZ) for G4 visualization in DLBCL formalin-fixed paraffin-embedded tissues (FFPET). A non-malignant tonsil biopsy was used for IHC optimization and we observed BG4 positive staining of B-cells within normal germinal centers, mantle zones, and interfollicular regions (Figure 1A). We then applied our novel IHC technique to 86 DLBCL FFPET, evaluated the presence of G4 formation within a 150 cell count, and analyzed for associations with MYC and BCL2 genetic abnormalities (translocation, amplification, and chromosome polysomy), cell-of-origin (COO), and patient outcome collected in previous studies (Valentino et al, AJCP 2013; Kendrick et al, Human Path 2014; Lenz et al, NEJM 2008). The human tissues and clinical data for this study were kindly provided by the Lymphoma/Leukemia Molecular Profiling Project and use was approved from the University of Arizona Institutional Review Board in accordance with the Declaration of Helsinki.
Overall, G4 formation was detected in 85% (73/86) of the DLBCL FFPET (Figure 1B-D) with intense staining observed in mitotic nuclei (Figure 1C, inset). DLBCL with MYC amplification with or without chromosome 8 polysomy significantly associated with an increased presence of G4s (12/23 cases, P=0.02 and 19/23 cases, P=0.005, respectively). Notably, elevated G4 formation was also observed in DLBCL cases with dual BCL2/MYC amplification (6/18 cases, P=0.02), but not in BCL2 amplification positive only cases (26/54 cases, P=0.49). Amplification was defined as >44 gene copies according to previous publications. Only 2/24 cases were MYC translocation positive, both with low BG4 staining (0% and 4% positive cells), and 14/86 cases harbored a BCL2 translocation; however, BG4 staining did not differ from BCL2 translocation negative cases (46% v. 54% average positive cells, P=0.45). There was a trend (P=0.11) for a higher incidence of G4 formation in ABC-DLBCL (33/36 cases) in comparison to GCB-DLBCL (27/35 cases) with ABC cases also trending towards an increase in average positive cells (54% v. 39%, P=0.10). We then assessed whether BG4 staining correlated with overall survival by initially stratifying the patients into quartiles. We observed that the 3rd and 4th quartiles displayed steeper curves and thus we used a 75% cut-off threshold to dichotomize the patients. DLBCL cases with ≥75% BG4 positive cells tend to have worse survival (log rank test; P=0.08).
For the first time, we demonstrate G4 formation within DLBCL FFPET and that this phenomenon associates with concurrent gene amplification of two highly genetically altered and prognostically important oncogenes in DLBCL, BCL2 and MYC, as well as MYC amplification alone. G4 formation, if left unresolved by helicases, may lead to DNA replication errors resulting in increased genome instability and gene amplification. We are currently investigating this hypothesis by correlating PIF1, XBP, and XBD helicase expression with G4 prevalence. Our work greatly advances this field by providing a novel tool to directly measure G4 DNA within lymphoma tissue. With this new methodology, we can address the biological and clinical significance of these structures in contributing to the genomic instability and aggressiveness of DLBCL.
Rimsza:Nanostring: Patents & Royalties: U.S. Patent Application 61/900, 553; "Methods for Selecting and Treating Lymphoma Types"; Ventana Medical Systems, Inc.: Consultancy; Celgene: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal