Abstract
Introduction: The myelodysplastic syndromes (MDS) are a group of clonal diseases derived from hematopoietic stem cells (HSC). Colony-forming unit cell (CFU-C) assay is an effective method to study the number and the function of HSC in vitro. In this study, we focus on the characteristics and the prognostic value of CFU-C in patients with MDS.
Patients and Method: CFU-C assays were performed according to the protocol of MethoCultTM H4435 Enriched (STEMCELL Technologies). A colony was defined as an aggregate of >40 cells. Clusters consisted of 4 to 40 cells. 560 consecutive newly-diagnosed, untreated subjects with MDS diagnosed from March, 2001 to April, 2013 were studied. All subjects were reclassified according to the 2008 WHO criteria. 535 subjects with evaluable cytogenetics were classified using the International Prognostic Scoring System (IPSS) and the revised International Prognostic Scoring System (IPSS-R) criteria. Follow-up data were available for 470£¨84%£©subjects. Median follow-up of survivors was 26 months (range, 1-170) months. Subjects receiving an allotransplants were censored in survival analyses. Erythroid and myeloid colonies were isolated from each subject with one cytogenetic abnormality such as del(5/5q-) or +8. Cytogenetic abnormalities of each colony were analyzed by fluorescence in situ hybridization (FISH). SPSS 17.0 software was used to make statistical analysis.
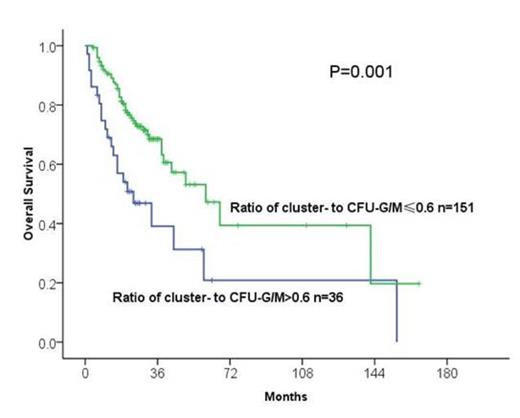
Results: Frequencies of burst-forming units-erythroid (BFU-E), colony forming unit-erythroid (CFU-E) and colony forming unit-granulocytes/macrophages (CFU-G/M) were significantly lower than normals (P<0.05) (Table 1). Subjects classified as lower risk in IPSS and IPSS-R had significantly higher numbers of BFU-E and CFU-E (P<0.05) but similar numbers CFU-G/M and clusters-G/M compared with higher risk subjects (Table 2). In 11 subjects with del(-5/5q-) or +8 identified by G- and/or R-banding, both normal and abnormal CFU-Cs were identified in 8 subjects studied by FISH. A high ratio of cluster- to CFU-G/M (>0.6) was associated with poor-risk cytogenetics (Table 2) and with worse overall survival in univariable (Figure 1, P=0.001) and multivariable analyses (HR 1.748, [1.01-3.0]; P=0.046) after adjusting for IPSS.
Conclusions: These data suggest abnormalities of proliferation and differentiation of erythroid and myeloid precursor cells in vitro parallel the ineffective hematopoiesis typical of MDS and may be useful in predicting outcomes of patients with MDS.
CFU-C in MDS subtypes
| . | N . | BFU-E . | CFU-E . | CFU-G/M . | N . | Ratio of cluster- to CFU-G/M . |
|---|---|---|---|---|---|---|
| RA | 21 | 8 (0-44) | 40 (0-134) | 14 (0-127)1 | 6 | 0.25 (0.40-1.00) |
| RT | 4 | 18 (4-55) | 75 (60-90)1 | 30 (18-70)1 | 2 | 2 |
| RARS | 27 | 12 (0-33) | 35 (1-140) | 12 (0-70)1 | 10 | 0.45 (0.17-0.80) |
| RCMD | 275 | 10 (0-80) | 33 (0-178) | 14 (0-100) | 126 | 0.35 (0-0.83) |
| RAEB1 | 112 | 10 (0-258) | 32 (0-312) | 14 (0-89) | 53 | 0.47 (0-1.00) |
| RAEB2 | 103 | 9 (0-46) | 25 (0-120) | 13 (0-72) | 42 | 0.37 (0-1.00) |
| MDS-U | 15 | 4 (0-58) | 25 (0-161) | 10 (0-43) | 3 | 2 |
| Del(5q) | 3 | 2 (2-4) | 15 (0-20) | 5 (5-41)1 | 1 | 2 |
| . | N . | BFU-E . | CFU-E . | CFU-G/M . | N . | Ratio of cluster- to CFU-G/M . |
|---|---|---|---|---|---|---|
| RA | 21 | 8 (0-44) | 40 (0-134) | 14 (0-127)1 | 6 | 0.25 (0.40-1.00) |
| RT | 4 | 18 (4-55) | 75 (60-90)1 | 30 (18-70)1 | 2 | 2 |
| RARS | 27 | 12 (0-33) | 35 (1-140) | 12 (0-70)1 | 10 | 0.45 (0.17-0.80) |
| RCMD | 275 | 10 (0-80) | 33 (0-178) | 14 (0-100) | 126 | 0.35 (0-0.83) |
| RAEB1 | 112 | 10 (0-258) | 32 (0-312) | 14 (0-89) | 53 | 0.47 (0-1.00) |
| RAEB2 | 103 | 9 (0-46) | 25 (0-120) | 13 (0-72) | 42 | 0.37 (0-1.00) |
| MDS-U | 15 | 4 (0-58) | 25 (0-161) | 10 (0-43) | 3 | 2 |
| Del(5q) | 3 | 2 (2-4) | 15 (0-20) | 5 (5-41)1 | 1 | 2 |
1No significant difference compared with normals.
2Too few cases to analyze.
Associations between CFU-C and clinical and laboratory variables
| . | N . | BFU-E . | P . | CFU-E . | P . | CFU-G/M . | P . | Number . | Ratio of cluster- to CFU-GM . | P . |
|---|---|---|---|---|---|---|---|---|---|---|
| IPSS | 0.064 | 0.006 | 0.361 | 0.089 | ||||||
| Low | 30 | 13 (0-44) | 60 (0-169) | 19 (0-45) | 10 | 0.44 (0.24-0.70) | ||||
| Int-1 | 361 | 10 (0-258) | 33 (0-312) | 14 (0-127) | 150 | 0.33 (0-1.00) | ||||
| Int-2 | 115 | 9 (0-61) | 30 (0-137) | 14 (0-72) | 52 | 0.45 (0-1.00) | ||||
| High | 29 | 7 (0-34) | 21 (0-93) | 12 (0-67) | 12 | 0.44 (0-1.00) | ||||
| IPSS-R | 0.003 | 0.003 | 0.125 | 0.209 | ||||||
| Very low | 7 | 16 (9-25) | 30 (15-120) | 18 (5-33) | 2 | 0.29 (0.10-0.49) | ||||
| Low | 130 | 14 (0-80) | 42 (0-178) | 17 (0-70) | 48 | 0.31 (0-0.77) | ||||
| Intermediate | 173 | 10 (0-66) | 34 (0-161) | 13 (0-127) | 81 | 0.37 (0-1.00) | ||||
| High | 139 | 9 (0-259) | 29 (0-312) | 11 (0-89) | 51 | 0.33 (0-1.00) | ||||
| Very high | 86 | 8 (0-61) | 25 (0-137) | 14 (0-91) | 42 | 0.47 (0-1.00) | ||||
| Cytogenetics (IPSS) | 0.867 | 0.055 | 0.290 | 0.007 | ||||||
| Good | 327 | 10 (0-258) | 36 (0-312) | 15 (0-89) | 133 | 0.33 (0-1.00) | ||||
| Intermediate | 133 | 10 (0-69) | 30 (0-162) | 12 (0-127) | 63 | 0.45 (0-1.00) | ||||
| Poor | 75 | 10 (0-61) | 25 (0-137) | 14 (0-91) | 28 | 0.42 (0-1.00) | ||||
| Cytogenetics (IPSS-R) | 0.990 | 0.090 | 0.676 | 0.022 | ||||||
| Very good | 7 | 11 (4-20) | 48 (1-110) | 14 (8-28) | 2 | 0.49 (0.43-0.56) | ||||
| Good | 324 | 10 (0-258) | 35 (0-312) | 15 (0-89) | 132 | 0.33 (0-1.00) | ||||
| Intermediate | 129 | 10 (0-69) | 30 (0-162) | 12 (0-127) | 62 | 0.45 (0-1.00) | ||||
| Poor | 27 | 10 (0-61) | 35 (0-137) | 16 (0-48) | 8 | 0.36 (0.15-1.00) | ||||
| Very poor | 48 | 11 (0-42) | 22 (0-120) | 14 (0-91) | 20 | 0.53 (0-1.00) |
| . | N . | BFU-E . | P . | CFU-E . | P . | CFU-G/M . | P . | Number . | Ratio of cluster- to CFU-GM . | P . |
|---|---|---|---|---|---|---|---|---|---|---|
| IPSS | 0.064 | 0.006 | 0.361 | 0.089 | ||||||
| Low | 30 | 13 (0-44) | 60 (0-169) | 19 (0-45) | 10 | 0.44 (0.24-0.70) | ||||
| Int-1 | 361 | 10 (0-258) | 33 (0-312) | 14 (0-127) | 150 | 0.33 (0-1.00) | ||||
| Int-2 | 115 | 9 (0-61) | 30 (0-137) | 14 (0-72) | 52 | 0.45 (0-1.00) | ||||
| High | 29 | 7 (0-34) | 21 (0-93) | 12 (0-67) | 12 | 0.44 (0-1.00) | ||||
| IPSS-R | 0.003 | 0.003 | 0.125 | 0.209 | ||||||
| Very low | 7 | 16 (9-25) | 30 (15-120) | 18 (5-33) | 2 | 0.29 (0.10-0.49) | ||||
| Low | 130 | 14 (0-80) | 42 (0-178) | 17 (0-70) | 48 | 0.31 (0-0.77) | ||||
| Intermediate | 173 | 10 (0-66) | 34 (0-161) | 13 (0-127) | 81 | 0.37 (0-1.00) | ||||
| High | 139 | 9 (0-259) | 29 (0-312) | 11 (0-89) | 51 | 0.33 (0-1.00) | ||||
| Very high | 86 | 8 (0-61) | 25 (0-137) | 14 (0-91) | 42 | 0.47 (0-1.00) | ||||
| Cytogenetics (IPSS) | 0.867 | 0.055 | 0.290 | 0.007 | ||||||
| Good | 327 | 10 (0-258) | 36 (0-312) | 15 (0-89) | 133 | 0.33 (0-1.00) | ||||
| Intermediate | 133 | 10 (0-69) | 30 (0-162) | 12 (0-127) | 63 | 0.45 (0-1.00) | ||||
| Poor | 75 | 10 (0-61) | 25 (0-137) | 14 (0-91) | 28 | 0.42 (0-1.00) | ||||
| Cytogenetics (IPSS-R) | 0.990 | 0.090 | 0.676 | 0.022 | ||||||
| Very good | 7 | 11 (4-20) | 48 (1-110) | 14 (8-28) | 2 | 0.49 (0.43-0.56) | ||||
| Good | 324 | 10 (0-258) | 35 (0-312) | 15 (0-89) | 132 | 0.33 (0-1.00) | ||||
| Intermediate | 129 | 10 (0-69) | 30 (0-162) | 12 (0-127) | 62 | 0.45 (0-1.00) | ||||
| Poor | 27 | 10 (0-61) | 35 (0-137) | 16 (0-48) | 8 | 0.36 (0.15-1.00) | ||||
| Very poor | 48 | 11 (0-42) | 22 (0-120) | 14 (0-91) | 20 | 0.53 (0-1.00) |
Overall survival in subjects with cluster- to CFU-G/M ratios ¡Ü or > 60%.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal