Key Points
The HGF/c-MET pathway has a complex network to control KSHV+ PEL cell survival.
The c-MET inhibitor induces PEL apoptosis and suppresses tumor progression in vivo.
Abstract
Kaposi sarcoma–associated herpesvirus (KSHV) is a principal causative agent of primary effusion lymphoma (PEL) with a poor prognosis in immunocompromised patients. However, it still lacks effective treatment which urgently requires the identification of novel therapeutic targets for PEL. Here, we report that the hepatocyte growth factor (HGF)/c-MET pathway is highly activated by KSHV in vitro and in vivo. The selective c-MET inhibitor, PF-2341066, can induce PEL apoptosis through cell cycle arrest and DNA damage, and suppress tumor progression in a xenograft murine model. By using microarray analysis, we identify many novel genes that are potentially controlled by HGF/c-MET within PEL cells. One of the downstream candidates, ribonucleoside-diphosphate reductase subunit M2 (RRM2), also displays the promising therapeutic value for PEL treatment. Our findings provide the framework for development of HGF/c-MET–focused therapy and implementation of clinical trials for PEL patients.
Introduction
The oncogenic Kaposi sarcoma–associated herpesvirus (KSHV) is a principal causative agent of several human cancers including primary effusion lymphoma (PEL), which arises preponderantly in immunocompromised individuals, particularly AIDS patients.1 PEL usually comprises transformed B cells harboring KSHV episomes and presents as pleural, peritoneal, and pericardial neoplastic effusions, although some cases of extracavitary solid variants of PEL have been reported recently.2-4 PEL is a rare but aggressive malignancy, with a median survival time of ∼6 months even under conventional chemotherapy.5 Furthermore, the myelosuppressive effects of systemic cytotoxic chemotherapy synergize with those caused by antiretroviral therapy or immune suppression.5,6 Therefore, it is an urgent need to identify novel targets that can guide development of more effective therapeutic strategies against PEL.
The Met proto-oncogene encodes the receptor tyrosine kinase known as c-MET, and hepatocyte growth factor (HGF) is the only known ligand for c-MET.7,8 After its discovery in the mid-1980s, the HGF/c-MET pathway has gained considerable interest related to a variety of cancers due to the diversity of the cellular responses that follow HGF/c-MET pathway activation. HGF/c-MET interactions activate many downstream signaling intermediates, including the mitogen-activated protein kinase, phosphatidylinositol 3-kinase–AKT, v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (SRC), and signal transducer and activator of transcription.9,10 Moreover, an intricate network of cross-signaling involving the c-MET–epidermal growth factor receptor, c-MET–vascular endothelial growth factor receptor, and c-MET–Wnt pathways has also been reported in the past few years.11-13 Such cross-talk evokes a variety of pleiotropic biological responses leading to increased cell proliferation, survival, migration/invasion, angiogenesis, and metastasis in cancer cells.14 Deregulation and the consequent aberrant signaling of HGF/c-MET may occur by different mechanisms, including gene amplification, overexpression, activating mutations, increased autocrine or paracrine ligand-mediated stimulation, and interaction with other active cell surface receptors.15 Because of its pleotropic role in cellular processes important in oncogenesis and cancer progression, the HGF/c-MET pathway may be a viable target for anticancer therapies. Several molecules targeting HGF/c-MET have recently been evaluated in differential phase clinical trials, including small-molecule inhibitors, and monoclonal antibodies targeting either the ligand or the receptor.16-18
Despite this knowledge, roles of the HGF/c-MET pathway in virus-associated tumors remain largely unclear. For KSHV-related malignancies, only 1 study reports coexpression of HGF/c-MET in all of the KSHV+ PEL tumors tested (9 PEL cell lines and 4 primary specimens), whereas it was restricted to 1 of 34 high-grade B-cell non-Hodgkin lymphomas other than PEL (P < .001; χ2 test).19 Other studies report the coexpression of HGF/c-MET in some cases of diffuse large B-cell lymphoma and Hodgkin disease.20,21 However, the biological functions of the HGF/c-MET pathway in KSHV-infected host cells and/or tumor cells and their value as therapeutic “target” have not yet been addressed. Therefore, in the present study, we will determine the role of HGF/c-MET in PEL pathogenesis and underlying regulatory mechanisms by genomic analysis. We will also determine whether a selective c-MET inhibitor, PF-2341066, can suppress PEL progression in an established xenograft murine model.
Materials and methods
Cell culture and reagents
The PEL cell-line BCBL-1 (KSHV+/EBV−) was cultured as described previously.22 The other PEL cell lines BC-1 (KSHV+/EBV+) and BCP-1 (KSHV+/EBV−) were purchased from American Type Culture Collection (ATCC).22 Human umbilical vein endothelial cells (HUVECs) were grown in Dulbecco modified Eagle medium/F-12 50/50 medium (Cellgro) supplemented with 5% fetal bovine serum. All cells were cultured at 37°C in 5% CO2. All experiments were carried out using cells harvested at low (<20) passages. c-MET inhibitor PF-2341066 and RRM2 inhibitor 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP) were purchased from Selleck Chemicals and Sigma-Aldrich, respectively.
KSHV purification and infection
BCBL-1 cells were incubated with 0.6 mM valproic acid for 4 to 6 days, and KSHV was purified from the culture supernatants by ultracentrifugation at 20 000g for 3 hours, 4°C. The viral pellet was resuspended in 1/100 original volume in the appropriate culture media, and aliquots were frozen at −80°C. HUVECs were incubated with concentrated virus in the presence of 8 μg/mL Polybrene (Sigma-Aldrich) for 2 hours at 37°C. The concentration of infectious viral particles used in each experiment (multiplicity of infection [MOI]) was calculated as described previously.23,24
Cell proliferation assays
Cell proliferation was measured by using the WST-1 assays (Roche) according to the manufacturer’s instructions.
Patients and ethics statement
The study was approved by the Institutional Review Boards for Human Research (no. 8079) at Louisiana State University Health Science Center–New Orleans (LSUHSC-NO). All subjects were provided written informed consent in accordance with the Declaration of Helsinki. In the current study, a total of 27 HIV+ patients with antiretroviral treatment in our HIV Outpatient (HOP) Clinic are involved. There were 14 women and 13 men; the average age was 48.1 years (range, 21-63 years). The average CD4 T-cell counts were 530/mL (range, 35-1773/mL), and the average HIV viral loads were 5833 copies per mL (range, 25-66681 copies per mL).
Plasma preparation
Whole blood was collected in heparin-coated tubes, and plasma was isolated by centrifugation. The KSHV infection status was determined by using quantitative enzyme-linked immunosorbent assays (ELISAs) for identifying circulating immunoglobulin G (IgG) antibodies to KSHV proteins (LANA and K8.1) as previously described.25,26
Microarray
BC-1, BCP-1, and BCBL-1 cells were treated with vehicle or PF-2341066 (1.6 μM) for 24 hours, respectively. Total RNA was isolated using the Qiagen RNeasy kit (Qiagen), and 500 ng of total RNA was used to synthesize double-stranded complementary DNA. Biotin-labeled RNA was generated using the TargetAmp-Nano Labeling kit for Illumina Expression BeadChip (Epicentre), according to the manufacturer’s instructions, and hybridized to the HumanHT-12 v4 Expression BeadChip (Illumina).27,28 The microarray experiments were performed twice for each group and the average values were used for analysis. Common, similar, and unique sets of genes and enrichment analysis were performed using MetaCore software (Thompson Reuters) as previously reported.29
Flow cytometry
For detection of c-MET expression on the cell surface, PEL cells were resuspended in 3% bovine serum albumin in 1× phosphate-buffered saline (PBS), incubated on ice for 10 minutes, then incubated with primary antibodies diluted 1/50 recognizing c-MET (Santa Cruz Biotechnology), for an additional 30 minutes. Following 2 subsequent wash steps, cells were incubated for an additional 30 minutes with goat anti-rabbit IgG Alexa 647 using a dilution of 1/200 (Invitrogen). Control cells were incubated with secondary antibodies only. Cells were then resuspended in 1× PBS prior to analyses. Data were collected using a FACSCalibur 4-color flow cytometer (BD Biosciences) and FlowJo software (TreeStar). For apoptosis assays, the fluorescein isothiocyanate–Annexin V and propidium iodide (PI) Apoptosis Detection kit I (BD Pharmingen) was used.26 For cell cycle analysis, PEL cell pellets were fixed in 70% ethanol, and incubated at 4°C overnight. Cell pellets were resuspended in 0.5 mL of 0.05 mg/mL PI plus 0.2 mg/mL RNaseA and incubated at 37°C for 30 minutes. Cell cycle distribution was analyzed on a FACSCalibur 4-color flow cytometer (BD Biosciences).
ELISA
Concentrations of HGF in culture supernatants were determined using the human HGF ELISA kit (R&D Systems), according to the manufacturer’s instructions.
Immunoblotting
Total cell lysates (30 μg) were resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and immunoblotted using 100 to 200 µg/mL antibodies to cleaved caspase 3/9, phosphor (p)-ERK/total (t)-ERK, p-c-MET/t-c-MET, p-ALK/t-ALK, p-ROS1/t-ROS1, p-Cdc2, p-Chk1/2, p-Rb, Cyclin A2, Cyclin B1, Myt1, p-H2A.X/t-H2A.X, and p-p53/t-p53 (Cell Signaling). For loading controls, blots were incubated with antibodies detecting β-actin (Sigma-Aldrich). Immunoreactive bands were developed using an enhanced chemiluminescence reaction (PerkinElmer) and visualized by autoradiography.
Immunofluorescence assays
Cells were incubated in 1:1 methanol-acetone at −20°C for fixation and permeabilization. After blocking, cells were then incubated for 1 hour at 25°C with a 1/100 dilution of a mouse anti-p-p53 antibody or a rabbit anti-p-H2A.X antibody (Cell Signaling) followed by a 1/200 dilution of a goat anti-mouse or goat anti-rabbit secondary antibody conjugated with Texas Red (Invitrogen), respectively. For identification of nuclei, cells were subsequently counterstained with 0.5 µg/mL 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) in 180 mM Tris-HCl (pH 7.5).
CometAssay
The DNA damage was evaluated by using the reagent kit for single cell gel electrophoresis assay/CometAssay (Trevigen), according to the manufacturer’s instructions. The slides were viewed by using epifluorescence microscopy.
RNA interference
For RNA interference (RNAi) assays, ON-TARGET plus SMART pool small interfering RNA (siRNA) for RRM2 (Dharmacon), or negative control siRNA, were delivered using the DharmaFECT transfection reagent according to the manufacturer’s instructions.
Quantitative reverse transcription–polymerase chain reaction
Total RNA was isolated using the RNeasy Mini kit according to the manufacturer’s instructions (QIAGEN). Complementary DNA was synthesized from equivalent total RNA using the SuperScript III First-Strand Synthesis SuperMix kit (Invitrogen). Primers used for amplification of target genes are displayed in supplemental Table 1 (available on the Blood Web site). Amplification was carried out using an iCycler IQ Real-Time PCR Detection System, and calculated using automated iQ5 2.0 software (Bio-rad).
PEL xenograft model
Aliquots of 107 BCBL-1 cells were diluted in 200 μL of sterile PBS, and 6- to 8-week-old male nonobese diabetic/severe-combined immunodeficiency (NOD/SCID) mice (The Jackson Laboratory) received intraperitoneal (i.p.) injections with a single-cell aliquot. The PF-2341066 (20 mg/kg body weight), or vehicle alone, was administered using an insulin syringe for i.p. injection. Drug was administered either 24 hours or 28 days (allowed to establish tumor expansion) after BCBL-1 injection, once daily for 5 days per week. Two experiments, with 10 mice per group for each experiment, were performed. The PEL expansion in vivo was confirmed by testing the expression of cell surface markers including CD45, CD138, EMA, and viral protein LANA in nuclear within ascites tumor cells, using immunofluorescence assay and flow cytometry as described in our previous publications.22 Weights were recorded weekly as a surrogate measure of tumor progression, and ascites fluid volumes were tabulated for individual mice at the completion of each experiment. All protocols were approved by the Louisiana State University Health Science Center Animal Care and Use Committee in accordance with national guidelines (no. 3237).
Statistical analyses
Significance for differences between experimental and control groups was determined using the 2-tailed Student t test (Excel 8.0). The 50% inhibitory concentration (IC50) was calculated by using SPSS 20.0.
Results
KSHV activates the HGF/c-MET pathway in vitro and in vivo
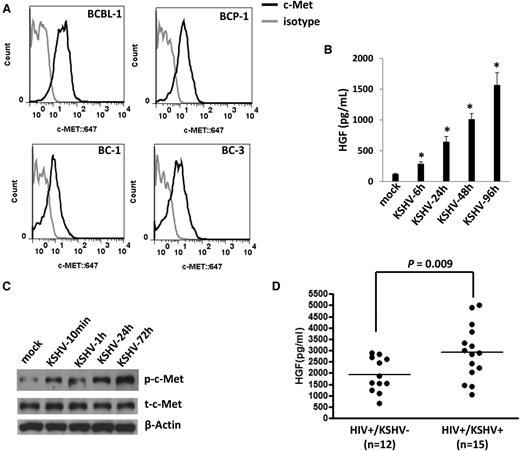
By using flow cytometry analysis, we first confirmed the expression of c-MET on the cell surface of 4 KSHV+ PEL cell lines (BCBL-1, BCP-1, BC-1, and BC-3) (Figure 1A), which is consistent with those reported previously.19 To further identify the ability of KSHV activating the HGF/c-MET pathway in vitro, we found that KSHV de novo infection greatly induced HGF production and the phosphorylation of c-MET from HUVECs in a time-course manner (Figure 1B-C). To explore the clinical relevance of HGF production within HIV-infected patients, we tested plasma HGF levels by ELISA in a small collection of our cohort HIV-infected patients with or without KSHV coinfection. KSHV infection status in these patients was determined as described previously.25,26 We found that the KSHV+ group (n = 15) had higher plasma HGF concentrations than those from the KSHV− group (n = 12) of these HIV-infected patients (Figure 1D). Taken together, our data strongly support the activation of the HGF/c-MET pathway by KSHV in vitro and in vivo.
KSHV activates the HGF/c-MET pathway in vitro and in vivo. (A) Four KSHV+ PEL cell lines were incubated with a monoclonal antibody recognizing the extracellular domain of c-MET (black line) or isotype antibody as a negative control (gray line), followed by a secondary antibody conjugated to Alexa 647. Surface expression of c-MET was quantified using flow cytometry. (B-C) HUVECs were infected by purified KSHV (MOI = 10), and the supernatant and cell lysates were collected at indicated time points. HGF concentrations in supernatant were determined by ELISA and protein expression was measured by immunoblots. Error bars represent the S.E.M. for 3 independent experiments. *P < .01. (D) The HGF concentrations in plasma from cohort HIV-infected patients were determined by ELISA, and KSHV infection status was determined by using ELISAs for identifying circulating IgG antibodies to KSHV proteins as described in “Methods.” S.E.M., standard error of the mean.
KSHV activates the HGF/c-MET pathway in vitro and in vivo. (A) Four KSHV+ PEL cell lines were incubated with a monoclonal antibody recognizing the extracellular domain of c-MET (black line) or isotype antibody as a negative control (gray line), followed by a secondary antibody conjugated to Alexa 647. Surface expression of c-MET was quantified using flow cytometry. (B-C) HUVECs were infected by purified KSHV (MOI = 10), and the supernatant and cell lysates were collected at indicated time points. HGF concentrations in supernatant were determined by ELISA and protein expression was measured by immunoblots. Error bars represent the S.E.M. for 3 independent experiments. *P < .01. (D) The HGF concentrations in plasma from cohort HIV-infected patients were determined by ELISA, and KSHV infection status was determined by using ELISAs for identifying circulating IgG antibodies to KSHV proteins as described in “Methods.” S.E.M., standard error of the mean.
c-MET inhibitor induces apoptosis for KSHV+ PEL cells potentially through causing cell cycle arrest and DNA damage
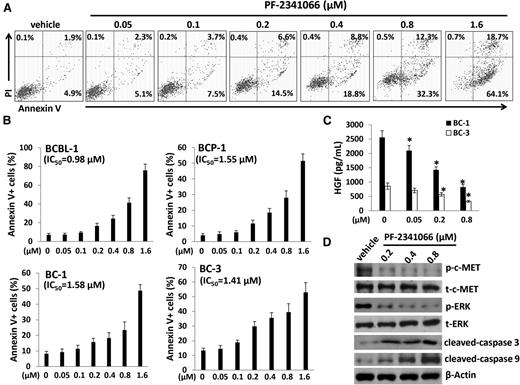
We next sought to understand the role of the HGF/c-MET pathway in PEL tumor cell survival/growth. Our data indicated that targeting HGF/c-MET by PF-2341066, a selective small-molecular c-MET inhibitor, induced significant apoptosis for all the 4 KSHV+ PEL cell lines we tested, in a dose-dependent manner (Figure 2A-B). By using the WST-1 assays, we found that PF-2341066 effectively reduced cell proliferation for these PEL cell lines (supplemental Figure 1). Moreover, PF-2341066 treatment greatly reduced HGF production from these lymphoma cells, although the underlying mechanisms remain unclear (Figure 2C). By using immunoblots, we found that PF-2341066 treatment increased the expression of cleaved-caspase 3 and 9, while reducing phosphor-c-MET and downstream phosphor-ERK (Figure 2D). To seek the potential mechanisms involved in c-MET inhibitor–induced PEL apoptosis, we found that PF-2341066 obviously caused cell cycle G2/M arrest when compared with vehicle control by flow cytometry analysis (Figure 3A). Further analysis indicated that PF-2341066 affected the expression of several checkpoint regulatory proteins (positive/negative), increasing Myt1, phosphor-Cdc2/Chk1/Chk2, while reducing Cyclin A2, Cyclin B1, and phosphor-Rb from BCBL-1 cells (Figure 3B). Interestingly, we found that PF-2341066 treatment also caused obvious DNA damage of PEL cells, as indicated by CometAssay and the upregulation of DNA damage markers phosphor-p53 and phosphor-H2A.X but not the total protein levels, respectively (Figure 3C-E). We have previously shown that several small-molecule compounds induced by PEL apoptosis are connected to reactivate KSHV lytic gene expression.30,31 However, here we only found a slight increase of viral latent/lytic gene expression (but with no statistical significance) within PF-2341066–treated BCBL-1 cells (supplemental Figure 2).
The c-MET–selective inhibitor PF-2341066 induces dose-dependent apoptosis for KSHV+ PEL cells. (A-B) A total of 4 PEL cell lines were incubated with the indicated concentrations of PF-2341066 for 24 hours, then cell apoptosis was quantified by Annexin V–PI/flow cytometry. BCBL-1 was shown as an example for the cell subpopulation diagram in panel A. (C) The HGF concentrations in supernatant were determined by ELISA. Error bars represent the S.E.M. for 3 independent experiments. *P < .01. (D) The protein expression in BCBL-1 was measured by immunoblots.
The c-MET–selective inhibitor PF-2341066 induces dose-dependent apoptosis for KSHV+ PEL cells. (A-B) A total of 4 PEL cell lines were incubated with the indicated concentrations of PF-2341066 for 24 hours, then cell apoptosis was quantified by Annexin V–PI/flow cytometry. BCBL-1 was shown as an example for the cell subpopulation diagram in panel A. (C) The HGF concentrations in supernatant were determined by ELISA. Error bars represent the S.E.M. for 3 independent experiments. *P < .01. (D) The protein expression in BCBL-1 was measured by immunoblots.
Targeting HGF/c-MET causes G2/M cell cycle arrest and DNA damage in KSHV+ PEL cells. (A) PEL cells were incubated with 0.4 µM PF-2341066 or vehicle control for 24 hours, then stained by PI and analyzed by flow cytometry. (B-D) The protein expression and DNA damage in BCBL-1 were measured by immunoblots and CometAssay, respectively. (E) The cellular expression of DNA damage markers phosphor-p53 and phosphor-H2A.X was detected by immunofluorescence, and the nuclear was shown by DAPI staining.
Targeting HGF/c-MET causes G2/M cell cycle arrest and DNA damage in KSHV+ PEL cells. (A) PEL cells were incubated with 0.4 µM PF-2341066 or vehicle control for 24 hours, then stained by PI and analyzed by flow cytometry. (B-D) The protein expression and DNA damage in BCBL-1 were measured by immunoblots and CometAssay, respectively. (E) The cellular expression of DNA damage markers phosphor-p53 and phosphor-H2A.X was detected by immunofluorescence, and the nuclear was shown by DAPI staining.
c-MET inhibitor suppresses PEL tumor progression in vivo
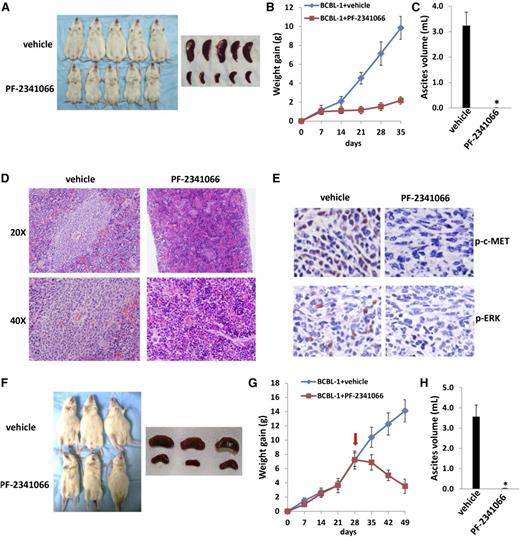
Next, we sought to determine whether PF-2341066 has the capacity to suppress PEL tumor growth in vivo using an established xenograft murine model.22 We administered PF-2341066 (or vehicle) i.p. within 24 hours of BCBL-1 cell injection and for a 5-week treatment. We found that PF-2341066 treatment dramatically suppressed PEL tumor progression including reducing ascites formation and spleen enlargement over this time frame (Figure 4A-C). By using hematoxylin-and-eosin staining, we observed huge tumor infiltration into the spleen of vehicle-treated mice, whereas only small tumor nodules were dispersed in the spleen of PF-2341066–treated mice (Figure 4D). By using immunohistochemistry staining, we further demonstrated the dramatically reduced expression of phosphor-c-MET and phosphor-ERK within spleen tissues from PF-2341066–treated mice, when compared with those from vehicle-treated mice (Figure 4E). Additional experiments were conducted wherein PF-2341066 therapy was initiated following establishment of PEL tumor expansion in mice (beginning 28 days after BCBL-1 cell injection). Using this approach, PF-2341066–treated mice still exhibited significant regression of PEL tumor burden relative to vehicle-treated mice (Figure 4F-G), and almost no ascites were found in these mice after 3-week treatment (Figure 4H).
The c-MET inhibitor PF-2341066 suppresses PEL progression in vivo. (A-C) NOD/SCID mice were injected i.p. with 107 BCBL-1 cells. Beginning 24 hours later, 20 mg/kg PF-2341066 or vehicle (n = 10 per group) were administered i.p., once daily, 5 days per week, for each of 2 independent experiments. Weights were recorded weekly. Images of representative animals and their spleens, as well as ascites fluid volumes, were collected at the conclusion of experiments on day 35. (D-E) Spleens from representative vehicle- or PF-2341066–treated mice were prepared for routine hematoxylin-and-eosin or immunohistochemistry staining. (F-H) NOD/SCID mice were injected i.p. with 107 BCBL-1 cells. Beginning 28 days later, 20 mg/kg PF-2341066 or vehicle (n = 10 per group) were administered i.p., once daily, 5 days per week. Images of representative animals and their spleens, as well as ascites fluid volumes, were collected at the conclusion of experiments on day 49. Error bars represent the S.E.M. for 1 of 2 independent experiments; *P < .01.
The c-MET inhibitor PF-2341066 suppresses PEL progression in vivo. (A-C) NOD/SCID mice were injected i.p. with 107 BCBL-1 cells. Beginning 24 hours later, 20 mg/kg PF-2341066 or vehicle (n = 10 per group) were administered i.p., once daily, 5 days per week, for each of 2 independent experiments. Weights were recorded weekly. Images of representative animals and their spleens, as well as ascites fluid volumes, were collected at the conclusion of experiments on day 35. (D-E) Spleens from representative vehicle- or PF-2341066–treated mice were prepared for routine hematoxylin-and-eosin or immunohistochemistry staining. (F-H) NOD/SCID mice were injected i.p. with 107 BCBL-1 cells. Beginning 28 days later, 20 mg/kg PF-2341066 or vehicle (n = 10 per group) were administered i.p., once daily, 5 days per week. Images of representative animals and their spleens, as well as ascites fluid volumes, were collected at the conclusion of experiments on day 49. Error bars represent the S.E.M. for 1 of 2 independent experiments; *P < .01.
Microarray analysis of the HGF/c-MET controlled network within KSHV+ PEL cells
There is a complex network for HGF/c-MET within cancer cells,9-13 although it still remains completely unknown to KSHV+ PEL cells. Therefore, we used the HumanHT-12 v4 Expression BeadChip (Illumina) which contains >47 000 probes derived from the NCBI RefSeq Release 38 and other sources to study the gene profile altered between vehicle- or PF-2341066–treated 3 KSHV+ PEL cell lines (BCBL-1, BC-1, and BCP-1). Intersection analysis indicated that there were 51 common genes significantly altered within all 3 PF-2341066–treated cell lines (up/down at least twofold and P < .05). There were 20 similar genes altered between BCBL-1 and BC-1, 188 similar between BCBL-1 and BCP-1, and 13 similar between BC-1 and BCP-1; 282 genes altered were unique to BCBL-1, 148 unique to BCP-1, and 52 unique to BC-1 (Figure 5A). Within the common gene set, the top 10 upregulated and/or downregulated candidate genes in PF-2341066–treated BCP-1, BC-1, and BCBL-1 cell lines are listed in Table 1, respectively, including gene description and the altered level of transcription in these 3 cell lines. Interestingly, we found that the functional role of most genes in PEL pathogenesis has never been reported, although some of them have been implicated in other types of malignancies. For example, Mucin-1 (MUC1) is a known tumor antigen and aberrantly overexpressed in various cancers with loss of its apical polarity.32-34 In addition, numerous cellular proteins implicated with MUC1 are involved in the malignancy of cancer cells and their resistance to chemotherapy.35 Notably, we found that some nuclear small RNA transcripts such as RN7SK, RNU1-4, RNVU1-18, and RNU1-1 are upregulated in PF-2341066–treated PEL cells (Table 1), although their contributions to PEL growth and pathogenesis remain unclear.
Microarray analysis of gene profile altered within PF-2341066–treated PEL cell lines. (A) The HumanHT-12 v4 Expression BeadChip (Illumina) was used to detect gene profile altered within PF-2341066–treated PEL cell lines (BCBL-1, BC-1, and BCP-1) when compared with vehicle-treated control. Intersection analysis of significantly altered genes (up/down at least twofold and P < .05) was performed using Illumina GenomeStudio software. Set I: Common genes altered in all the 3 cell lines. Set II: Similar genes altered in every 2 cell lines. Set III: Unique genes altered in each cell line. (B-C) The transcriptional levels of selected 4 candidate genes downregulated (B) or upregulated (C) as shown in microarray data were validated by using qRT-PCR, respectively. Error bars represent the S.E.M. for 3 independent experiments. (D-F) The enrichment analysis of gene profile (common, similar, and unique set as indicated) significantly altered by c-MET inhibitor was performed using the MetaCore software (Thompson Reuters) modules: Pathway Maps (D), Gene Ontology Processes (E), and Process Networks (F).
Microarray analysis of gene profile altered within PF-2341066–treated PEL cell lines. (A) The HumanHT-12 v4 Expression BeadChip (Illumina) was used to detect gene profile altered within PF-2341066–treated PEL cell lines (BCBL-1, BC-1, and BCP-1) when compared with vehicle-treated control. Intersection analysis of significantly altered genes (up/down at least twofold and P < .05) was performed using Illumina GenomeStudio software. Set I: Common genes altered in all the 3 cell lines. Set II: Similar genes altered in every 2 cell lines. Set III: Unique genes altered in each cell line. (B-C) The transcriptional levels of selected 4 candidate genes downregulated (B) or upregulated (C) as shown in microarray data were validated by using qRT-PCR, respectively. Error bars represent the S.E.M. for 3 independent experiments. (D-F) The enrichment analysis of gene profile (common, similar, and unique set as indicated) significantly altered by c-MET inhibitor was performed using the MetaCore software (Thompson Reuters) modules: Pathway Maps (D), Gene Ontology Processes (E), and Process Networks (F).
The top 10 “common” candidate genes downregulated or upregulated within PF-2341066–treated 3 KSHV+ PEL cell lines
| Gene symbol . | Description . | Fold change . | ||
|---|---|---|---|---|
| BCP-1 . | BC-1 . | BCBL-1 . | ||
| RRM2 | Ribonucleoside-diphosphate reductase subunit M2 | 0.38 | 0.41 | 0.21 |
| MUC1 | Mucin-1 | 0.53 | 0.26 | 0.32 |
| POLE2 | DNA polymerase ε subunit 2 | 0.39 | 0.47 | 0.26 |
| TMEM106C | Transmembrane protein 106C | 0.43 | 0.5 | 0.29 |
| FAM81A | Protein FAM81A | 0.46 | 0.5 | 0.3 |
| CHAC2 | Cation transport regulator-like protein 2 | 0.54 | 0.43 | 0.34 |
| TYMS | Thymidylate synthase | 0.42 | 0.52 | 0.39 |
| FBXO5 | F-box only protein 5 | 0.51 | 0.48 | 0.36 |
| ASPM | Abnormal spindle-like microcephaly-associated protein | 0.3 | 0.54 | 0.55 |
| DPAGT1 | UDP-N-acetylglucosamine–dolichyl-phosphate N-acetylglucosaminephosphotransferase | 0.52 | 0.54 | 0.33 |
| RN7SK | RNA, 7SK small nuclear transcript | 5.56 | 5.1 | 20.15 |
| HMGCS1 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 8.11 | 4.03 | 4.03 |
| RNU1-4 | RNA, U1 small nuclear 4 transcript | 2.47 | 4.49 | 7.84 |
| RNVU1-18 | RNA, variant U1 small nuclear 18 transcript | 2.2 | 4.36 | 7.43 |
| SNORD3A | Small nucleolar RNA, C/D box 3A transcript | 3.01 | 5.24 | 3.98 |
| RNU1-1 | RNA, U1 small nuclear 1 transcript | 2.15 | 3.11 | 6.4 |
| MSMO1 | Methylsterol monooxygenase 1 | 5.42 | 2.23 | 2.13 |
| PPP1R15A | Protein phosphatase 1 regulatory subunit 15A | 1.86 | 3.58 | 4.05 |
| HMGCR | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | 3.66 | 2.46 | 3.34 |
| PIK3IP1 | Phosphoinositide-3-kinase-interacting protein 1 | 3.31 | 3.07 | 3.03 |
| Gene symbol . | Description . | Fold change . | ||
|---|---|---|---|---|
| BCP-1 . | BC-1 . | BCBL-1 . | ||
| RRM2 | Ribonucleoside-diphosphate reductase subunit M2 | 0.38 | 0.41 | 0.21 |
| MUC1 | Mucin-1 | 0.53 | 0.26 | 0.32 |
| POLE2 | DNA polymerase ε subunit 2 | 0.39 | 0.47 | 0.26 |
| TMEM106C | Transmembrane protein 106C | 0.43 | 0.5 | 0.29 |
| FAM81A | Protein FAM81A | 0.46 | 0.5 | 0.3 |
| CHAC2 | Cation transport regulator-like protein 2 | 0.54 | 0.43 | 0.34 |
| TYMS | Thymidylate synthase | 0.42 | 0.52 | 0.39 |
| FBXO5 | F-box only protein 5 | 0.51 | 0.48 | 0.36 |
| ASPM | Abnormal spindle-like microcephaly-associated protein | 0.3 | 0.54 | 0.55 |
| DPAGT1 | UDP-N-acetylglucosamine–dolichyl-phosphate N-acetylglucosaminephosphotransferase | 0.52 | 0.54 | 0.33 |
| RN7SK | RNA, 7SK small nuclear transcript | 5.56 | 5.1 | 20.15 |
| HMGCS1 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | 8.11 | 4.03 | 4.03 |
| RNU1-4 | RNA, U1 small nuclear 4 transcript | 2.47 | 4.49 | 7.84 |
| RNVU1-18 | RNA, variant U1 small nuclear 18 transcript | 2.2 | 4.36 | 7.43 |
| SNORD3A | Small nucleolar RNA, C/D box 3A transcript | 3.01 | 5.24 | 3.98 |
| RNU1-1 | RNA, U1 small nuclear 1 transcript | 2.15 | 3.11 | 6.4 |
| MSMO1 | Methylsterol monooxygenase 1 | 5.42 | 2.23 | 2.13 |
| PPP1R15A | Protein phosphatase 1 regulatory subunit 15A | 1.86 | 3.58 | 4.05 |
| HMGCR | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | 3.66 | 2.46 | 3.34 |
| PIK3IP1 | Phosphoinositide-3-kinase-interacting protein 1 | 3.31 | 3.07 | 3.03 |
We next selected 4 genes from the top 10 upregulated or downregulated candidate list (Table 1) for validation of their transcriptional change by quantitative reverse transcription–polymerase chain reaction (qRT-PCR), respectively. Our results indicated that all 4 genes (RRM2, MUC1, TYMS, and FBXO5) were significantly downregulated in PF-2341066–treated PEL cells when compared with vehicle-treated control, whereas another 4 genes (HMGCS1, MSMO1, HMGCR, and PIK3IP1) were all significantly upregulated in PF-2341066–treated PEL cells (Figure 5B-C). Moreover, the altered transcriptional levels of these genes in all 3 KSHV+ PEL cell lines were comparable to those found in microarray data, demonstrating the credibility of our microarray analysis. We also performed enrichment analysis of these common, similar, and unique sets of genes using the Pathway Map, Gene Ontology (GO) Processes, and Process Network modules from Metacore software (Thompson Reuters).29 Our analysis showed that several major cellular functions were affected within PF-2341066–treated PEL cells, including the apoptosis/ER stress response pathway, epigenetic regulation of gene expression, cell cycle/checkpoint, and DNA damage–related proteins (Figure 5D-F). The top 2 scored pathway maps and protein networks based on the enrichment analysis of “common” gene set were also listed in supplemental Figures 3 and 4, respectively.
Targeting RRM2 induces PEL apoptosis through increasing DNA damage
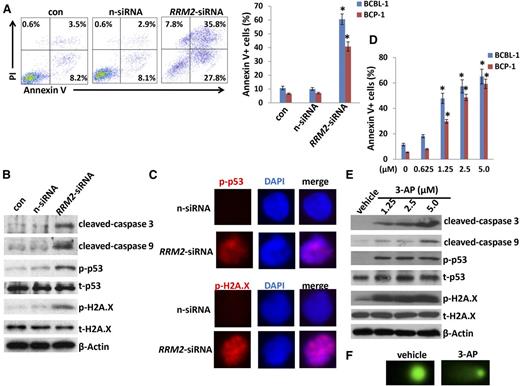
We next selected ribonucleoside-diphosphate reductase subunit M2 (RRM2), 1 of the significant downregulated genes in PF-2341066–treated PEL cells from microarray data, to determine its role in PEL pathogenesis. Actually, ribonucleoside-diphosphate reductase (RR) is an attractive target for anticancer agents given its central function in DNA synthesis, growth, metastasis, and drug resistance of cancer cells.36 Human RR is composed of α subunits (RRM1) that contain the catalytic site and 2 binding sites for enzyme regulators and β subunits (RRM2) with a binuclear iron cofactor that generates the stable tyrosyl radical necessary for catalysis.37 Here, we found that direct silence of RRM2 by RNAi induced caspase-dependent apoptosis within KSHV+ PEL cells (BCBL-1 and BCP-1), potentially through increasing DNA damage (Figure 6A-C; supplemental Figure 5). Furthermore, a selective RR inhibitor, 3-AP,38 also induced dose-dependent apoptosis and increased DNA damage by CometAssay and the expression of phosphor-H2A.X and phosphor-p53 but not the total proteins from KSHV+ PEL cells (Figure 6D-F). These data indicate that RRM2 may also represent a promising target for development of anti-PEL agents.
Directly targeting RRM2 induces PEL apoptosis through causing DNA damage. (A) BCP-1 and BCBL-1 were transfected with either negative control siRNA (n-siRNA) or RRM2-siRNA for 48 hours, then cell apoptosis was assessed using Annexin V-PI staining and flow cytometry analysis. (B-C) Protein expression was measured by immunoblots and immunofluorescence, respectively. (D-E) Cells were treated with indicated concentrations of RRM2 inhibitor, 3-AP, for 24 hours, then cell apoptosis and protein expression were measured as described in “Materials and methods.” Error bars represent the S.E.M. for 3 independent experiments; *P < .01. (F) Cells were treated with 5 μM 3-AP for 24 hours, then DNA damage was evaluated by using the CometAssay.
Directly targeting RRM2 induces PEL apoptosis through causing DNA damage. (A) BCP-1 and BCBL-1 were transfected with either negative control siRNA (n-siRNA) or RRM2-siRNA for 48 hours, then cell apoptosis was assessed using Annexin V-PI staining and flow cytometry analysis. (B-C) Protein expression was measured by immunoblots and immunofluorescence, respectively. (D-E) Cells were treated with indicated concentrations of RRM2 inhibitor, 3-AP, for 24 hours, then cell apoptosis and protein expression were measured as described in “Materials and methods.” Error bars represent the S.E.M. for 3 independent experiments; *P < .01. (F) Cells were treated with 5 μM 3-AP for 24 hours, then DNA damage was evaluated by using the CometAssay.
Discussion
In recent years, the HGF/c-MET pathway has become an attractive target for development of anticancer drugs in a variety of solid tumors. In contrast to this, little is known about the biological functions and therapeutic value of the HGF/c-MET pathway in virus-related malignancies such as PEL. To our knowledge, this is the first article to report that activation of the HGF/c-MET pathway is essential for KSHV+ PEL cell survival and that targeting this pathway successfully suppresses PEL progression in vivo. Interestingly, PF-2341066 has been shown as the inhibitor to several other receptor tyrosine kinases such as ALK and ROS1 in different types of cancer cells including anaplastic large-cell lymphoma and non-small-cell lung cancer.39,40 However, our immunoblots data indicate that the PEL cell lines we tested including BCBL-1 and BCP-1 do not express the p-ALK (Tyr1604), t-ALK, p-ROS1 (Tyr2274), or t-ROS1 reported in these previous studies,39,40 implying that these KHSV+ PEL cell lines may have unique expressional patterns of receptor tyrosine kinases (data not shown).
One of the remaining questions is the underlying mechanisms through which the HGF/c-MET pathway is activated in PEL cells. Previous studies have reported some activating mutations of c-MET in non-small-cell lung cancer, hereditary and spontaneous renal carcinomas, hepatocellular carcinomas, gliomas, gastric, squamous cell carcinoma of the head and neck, and breast cancers.41-46 Potentially oncogenic mutations involve mainly: (1) point mutations that generate an alternative splicing encoding a shorter protein that lacks exon 14, which encodes for the juxtamembrane domain of c-MET43,47 ; (2) point mutations in the kinase domain that render the enzyme constitutively active44 ; and (3) Y1003 mutations that inactivate the Cbl-binding site leading to constitutive c-MET expression.48-50 Therefore, we are now working on whether these similar mutations are also present in the c-MET sequence from KSHV+ PEL cell lines and/or primary specimens, resulting in constitutive activation of this pathway. In addition, we are interested in understanding which viral proteins (latent and/or lytic) are responsible for activating HGF/c-MET from PEL cells in future study.
Our data indicate that c-MET inhibitor–induced PEL apoptosis is potentially through increasing DNA damage and regulation of some DNA damage/repair–related proteins such as RRM2 (see discussion below). Interestingly, our recent data showed that silencing of 1 DNA damage–related gene, XRCC5 (x-ray repair cross-complementing protein 5, also known as Ku80) enhanced the induction of apoptosis and programmed cell death by low-dose concentrations of DNA-damage reagents such as doxorubicin.28 In fact, it has been reported recently that KSHV can activate the DNA damage response during de novo infection of primary endothelial cells and this plays a role in establishing latency.51 More recently, it has been demonstrated that lytic reactivation of KSHV leads to activation of the ataxia telangiectasia mutated (ATM) and DNA-dependent protein kinase DNA damage response kinases.52 Inhibition of ATM results in the reduction of overall levels of viral replication whereas inhibition of DNA-dependent protein kinase increases activation of ATM and leads to earlier viral release.52 However, our data indicate that targeting HGF/c-MET by PF-2341066 does not cause robust viral lytic gene expression, implying that PF-2341066–induced DNA damage is potentially through other mechanisms.
By using microarray analysis, we found that many downstream genes are altered in PF-2341066–treated PEL cells when compared with vehicle-treated control, although the roles of most of them in PEL pathogenesis remain unknown. One of candidate genes, RRM2, was found significantly downregulated in PF-2341066–treated PEL cells from the microarray data and subsequently validated by qRT-PCR. In fact, normal cells with a low proliferative status express very low levels of RR, whereas neoplastic cells overexpress RR to manufacture dNTP pools to support DNA synthesis and proliferation.36 Therefore, RR, especially the RRM2 subunit, is an important target for anticancer agents. Our data indicate that directly targeting RRM2 by either RNAi or the pharmacological inhibitor, 3-AP, greatly induces PEL apoptosis through increasing DNA damage. 3-AP is now used in clinical trials for a variety of advanced-stage solid tumors.53-55 Toxicities reported from the phase 1 trial were hypoxia, respiratory distress, and methemoglobulinemia, apparently due to iron chelation in the red blood cells of the patients.56 Recently, Zhou et al have developed a novel potent RR inhibitor, COH29, by using structure- and mechanism-based approaches, which displays a broad antitumor potential.36 Therefore, future work will focus on testing whether these RR inhibitors have antitumor effects by using our PEL xenograft model.
The microarray original data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE70594).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Center for Biomedical Research Excellence subaward P20-RR021970 (Z.Q.), SOM Research Enhancement Funding (Z.Q.), DOD Career Development Award CA140437 (Z.Q.), as well as awards from the National Natural Science Foundation of China (81272191, 81472547 [Z.Q.] and 81400164 [L. Dai]). HIV+ patient plasma samples were provided by the HIV Cancer Care Program Biorepository which is supported by from the National Institutes of Health grants UM1-CA181255 and R01-CA142362 (C.P.). This work was also funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E (D.W.). Funding sources had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: L. Dai and Z.Q. designed and performed experiments, analyzed results, and wrote the manuscript; J.T.-T., Y.C., K.B., L. Doyle, K.R., and J.Z. performed experiments; and L.D.V., D.W., and C.P. performed statistical analysis and provided critical input.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zhiqiang Qin, Louisiana Cancer Research Center, 1700 Tulane Ave, Suite 902, New Orleans, LA 70112; e-mail: zqin@lsuhsc.edu.