Key Points
HSCs emerge, engraft, and differentiate in the absence of cdh5.
HSCs emerging from Cdh5−/−GFP+/+ endothelium of mouse chimeric embryos are functional.
Abstract
Rare endothelial cells in the aorta-gonad-mesonephros (AGM) transition into hematopoietic stem cells (HSCs) during embryonic development. Lineage tracing experiments indicate that HSCs emerge from cadherin 5 (Cdh5; vascular endothelial–cadherin)+ endothelial precursors, and isolated populations of Cdh5+ cells from mouse embryos and embryonic stem cells can be differentiated into hematopoietic cells. Cdh5 has also been widely implicated as a marker of AGM-derived hemogenic endothelial cells. Because Cdh5−/− mice embryos die before the first HSCs emerge, it is unknown whether Cdh5 has a direct role in HSC emergence. Our previous genetic screen yielded malbec (mlbbw306), a zebrafish mutant for cdh5, with normal embryonic and definitive blood. Using time-lapse confocal imaging, parabiotic surgical pairing of zebrafish embryos, and blastula transplantation assays, we show that HSCs emerge, migrate, engraft, and differentiate in the absence of cdh5 expression. By tracing Cdh5−/−green fluorescent protein (GFP)+/+ cells in chimeric mice, we demonstrated that Cdh5−/−GFP+/+ HSCs emerging from embryonic day 10.5 and 11.5 (E10.5 and E11.5) AGM or derived from E13.5 fetal liver not only differentiate into hematopoietic colonies but also engraft and reconstitute multilineage adult blood. We also developed a conditional mouse Cdh5 knockout (Cdh5flox/flox:Scl-Cre-ERT) and demonstrated that multipotent hematopoietic colonies form despite the absence of Cdh5. These data establish that Cdh5, a marker of hemogenic endothelium in the AGM, is dispensable for the transition of hemogenic endothelium to HSCs.
Introduction
During fetal development, rare aortic endothelial cells of the aorta-gonad mesonephros (AGM) region, termed hemogenic endothelial cells, transition to hematopoietic stem cells (HSCs). In zebrafish, the first definitive HSCs emerge from AGM between 30 and 36 hours postfertilization (hpf) and peak at 36 to 72 hpf. These HSCs migrate to caudal hematopoietic tissue (CHT) and later to kidney marrow and thymus.1 Similarly, adult-type definitive HSCs first emerge in the aortic endothelium of the mouse AGM at embryonic day 10.5 (E10.5) and enter into circulation to seed the fetal liver where the production of hematopoietic progenitor cells occurs. Then, hematopoiesis shifts to the bone marrow and spleen, and the liver becomes the hub of metabolism.1 Lineage tracing experiments have revealed that HSCs emerge from cadherin 5+ (Cdh5+) endothelial precursors.2,3 Isolated populations of Cdh5+ cells from the E11.5 AGM of both murine embryos and mouse embryonic stem (mES) cells differentiate into hematopoietic cells.4-7 Thus, Cdh5 is a marker of AGM-derived hemogenic endothelial cells transitioning to HSCs.
Sca1, Runx1, and Cdh5 are all markers of HSCs that emerge from hemogenic endothelial cells. Sca1 is itself dispensable for HSC emergence,8 whereas Runx1 is essential for the formation of erythroid/myeloid progenitors from the yolk sac and HSCs from the hemogenic endothelium.9 Cdh5, a cell-adhesion molecule, regulates endothelial permeability10 as well as vascular integrity.11 Although Cdh5 is present in virtually all active HSCs derived from E11.5 AGM, no difference was observed in multilineage differentiation capacity between vascular endothelial (VE)-cadherin+CD45+ and VE-cadherin−CD45+ HSCs isolated from the E13.5 liver.7 In fact, there are conflicting reports suggesting that Cdh5 expression declines either in E13.5 fetal liver6,7 or in E16.5 fetal liver HSCs.12 However, there is a consensus that Cdh5 is absent from adult bone marrow HSCs.6,7,12 Prior studies have established that loss or truncation of Cdh5 does not significantly impair mouse primitive hematopoiesis in the E8.5 yolk sacs.13 However, Cdh5 mutant embryos die at and beyond E9.13,14 Because HSCs emerge from hemogenic endothelium in the AGM around E10.5,1,15 the direct role of Cdh5 in endothelial emergence and the development of HSCs as well as in adult hematopoiesis has not yet been firmly established. In this manuscript, we have addressed this question using zebrafish and murine systems.
Methods
Animal models
All procedures were approved by the animal care and use committee of Boston Children’s Hospital. The malbec (mlbbw306) mutant was recovered from our unbiased chemical mutagenesis screen. C57BL6/J, CD1, and CD45.1 (SJL) mice were purchased from The Jackson Laboratory. Cdh5flox/flox mice were provided by Dietmar Vestweber (Max Planck Institute, Münster, Germany) and Scl-Cre-ERT mice by Stuart H. Orkin (Boston Children’s Hospital, Boston, MA).
Mutational analysis and candidate verification
Bioinformatics tools were used to identify and validate cdh5 as the candidate gene disrupted in the mlb locus on the zebrafish chromosome.16,17 The complementary DNA prepared from wild-type (WT) and mlb embryos were sequenced and the mutation in the exon 3 sequence was verified using an allele-specific oligohybridization technique.18 We performed quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) using TaqMan gene expression assays (zf-cdh5: Dr03089732_g1 spanning exon 3-4, Dr03089733_m1 spanning exon 4-5, Dr03089737_m1 spanning exon 8-9, Dr03089729_m1 spanning exon 11-12, and zf-hprt: Dr03138604_m1; Applied Biosystems) to measure transcript levels of cdh5 along the length of the gene. Morpholinos (Gene Tools) against splice sites (TACAAGACCGTCTACCTTTCCAATC) and translational (CCTCCTGGCACACTGTTTCATCATC) sites of cdh5 were designed and injected into WT embryos to analyze a loss-of-function phenotype. Thirty embryos were pooled in each experiment (n = 5) to isolate RNA for qRT-PCR analysis
Confocal analysis
We crossed cd41:eGFP19 with flk1:mCherry20,21 zebrafish and runx1+23:NLS-mCherry22 with flk1:eGFP23 zebrafish and injected cdh5-morpholino in their transgenic embryos. We mounted cdh5-silenced transgenic embryos in low-melting-point agarose and used a spinning-disk confocal microscope to perform time-lapse confocal imaging of cd41:eGFP+ HSCs and runx1+23:NLS-mCherry+ hematopoietic stem and progenitor cells (HSPCs) emerging from flk1+ endothelium from 30 to 42 hpf.22,24 We analyzed 15 cdh5-silenced cd41:eGFP::flk1:mCherry embryos (vs 13 control) and 23 cdh5-silenced runx1:mCherry::flk1:eGFP embryos (vs 11 control).
Parabiotic surgery of zebrafish embryos
We injected cdh5-morpholino premixed with Dextran blue dye in 1- to 2-cell-stage-old cd41:eGFP and runx1+23:NLS-mCherry transgenic embryos, and allowed them to grow until their high to 30% epiboly stage. We used custom-made pulled glass needles to fuse the blastula of cdh5-silenced transgenic embryos with WT casper embryos. We allowed these fused embryos to grow in high-calcium Ringer's solution containing antibiotics, and then tested for the efficacy of fusions 24 hours past surgery.25 Fused embryos viable after 24 hours of surgery were allowed to grow and were mounted in low-melting-point agarose to test the efficacy of HSC emergence in the morphant embryos. Their migration and engraftment into the WT CHT region between 32 and 48 hpf, and the subsequent differentiation into platelets between days 4 and 5 were captured using confocal microscopes. All 10 of 10 WT parabiots survived whereas 30 of 35 cdh5-silenced parabiots survived.
Blastula transplantation assay
We injected cdh5-morpholino, premixed with rhodamine-dextran to label all of the donor cells, in lcr:eGFP transgenic embryos at the 1-cell stage and allowed them to grow until they were between high or dome stage. We harvested the blastula from cdh5-silenced transgenic embryos (donor) and injected ∼20 cells into age-matched WT embryos (recipient).26,27 We let these embryos grow, and then tested for lcr:GFP+ erythroid cells in both morphant and transplanted (recipient) embryos at 36, 48, and 72 hpf (n = 14 recipients).
Electroporation of CRISPR/CAS9 in mouse GFP ES cells
We designed clustered regularly interspaced short palindromic repeat (CRISPR) single guide RNA (sgRNA) sequences flanking exon 1 and exon 12 of the Cdh5 gene using CRISPR Design software (http://crispr.mit.edu/; supplemental Table 1, see supplemental Data available at the Blood Web site). We cloned these sequences in a vector containing Cas9.28,29 We electroporated the plasmids targeting both the forward and reverse sgRNAs along with the H2B-mKO vector in mouse green fluorescent protein (GFP) embryonic stem (ES) cells (GFP-mESC LB10; GlobalStem) using the GenePulser II (Bio-Rad).30 We coelectroporated CRISPR constructs with the mH2B-KO vector to allow us to isolate mES cells carrying CAS9, which provides a much greater screening efficiency. We sorted the GFP and orange-positive cells, plated them on mouse embryonic fibroblasts, and genotyped 1869 clones in order to detect a clone containing a homozygous deletion of the Cdh5 gene using the primers listed in supplemental Table 2.
Karyotyping
We performed karyotyping of candidate Cdh5-deleted clones to eliminate a possibility of aneuploidy in their chromosomes during genetic manipulations.31
Development of chimeric mice
After engineering mES cells with ubiquitous expression of GFP, and identifying the Cdh5−/−GFP+ mES clone, we karyotyped them to exclude aneuploidy. We injected these Cdh5−/−GFP+ mES cells (∼35-50) into the C57/BL6-derived WT blastocysts to generate chimeric embryos, which were transferred to the oviduct of CD1 foster mothers.32 Of the 318 injected embryos of 46 foster mothers, 24 chimeric embryos survived up to E11.5 and 11 chimeric embryos survived up to E13.5. In addition, we used 6 E10.5 chimeric embryos to perform immunohistochemistry of Runx1 expression in the AGM region. Extensive detail on numbers of chimeric mice embryos profiled at the E11.5 and E13.5 developmental stage is provided in supplemental Table 3.
Immunohistochemistry
We harvested E10.5 chimeric mouse embryos, embedded them in a paraffin block, performed transverse sections, and immunostained with Runx1, GFP, and 4,6 diamidino-2-phenylindole (DAPI) antibodies in order to detect their expression in the E10.5 AGM region.33-35
FACS and CFU analyses
We harvested E11.5 AGM and E13.5 fetal liver tissues from chimeric mouse embryos and used their single-cell suspension for fluorescence-activated cell sorter (FACS), colony-forming unit capacity (CFU), and transplant analyses.4,33,35 Specifically, we detected expression of GFP, CD41 (phycoerythrin-Cy7), c-Kit (allophycocyanin), CD45.2 (Pacific Blue) in the E11.5 AGM as well as Sca1 (phycoerythrin), c-Kit (allophycocyanin) expression in E13.5 fetal liver. Background staining and negative control for conjugated antibodies are provided in supplemental Figure 8. We also plated 2 embryo equivalent whole E11.5 AGM as well as sorted Cdh5-deleted GFP+ or sorted control GFP− E13.5 fetal liver cells on M3434 media (StemCell Technologies) and analyzed their capacity to make granulocyte, erythroid, macrophage, megakaryocyte (GEMM), granulocyte macrophage, macrophage, and erythroid colonies.4,35
Transplant analysis
We transplanted 2 embryo equivalent (e.e.) chimeric E11.5 whole AGM cells into irradiated (9 cGy, 2 split dose at an interval of 2-3 hours) CD45.1 (SJL) mice. In contrast, we sorted GFP+ (Cdh5−/−) and GFP− (WT) cells from E13.5 fetal liver cells and transplanted them (∼250 000 cells) separately into irradiated CD45.1 mice. We used CD45.1 splenic helper cells for AGM and fetal liver transplants. All GFP+ transplants were positive for GFP expression. We then tested the efficiency of these transplanted cells to engraft and reconstitute B cells (CD19), T cells (CD3), and macrophages (Gr1 or Mac1) beginning after 4 weeks of transplantation and up to 16 weeks using FACS analysis.4,33,35 Extensive detail on numbers of the E11.5 AGM and E13.5 fetal liver transplanted in irradiated SJL mice is provided in supplemental Table 3.
Development of a conditional Cdh5 knockout
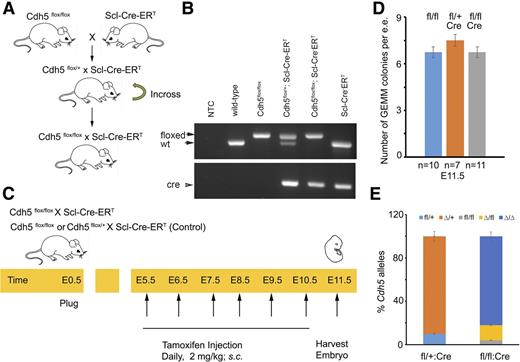
We developed Cdh5flox/flox:Scl-Cre-ERT (as described in Figure 6A) and verified its genotype (as described in Figure 6B). We injected Tamoxifen (2 mg/kg, subcutaneously [s.c.]) in pregnant Cdh5flox/flox:Scl-Cre-ERT every day from E5.5 to E10.5 (see Figure 6C). At E11.5, we harvested the AGM, seeded its single-cell suspension in M3434 media for CFU analyses, followed by the genotyping of colonies to analyze the Cdh5 deletion. Sequences for the Cdh5 and Cre primers are listed in supplemental Table 4.
Statistical analyses were performed by Student t tests. Significance was set at P < .05.
Results
cdh5 is disrupted in the malbec locus
To investigate the role of cadherin 5 in blood development, we studied mlb, an embryonic lethal zebrafish mutant that emerged from an unbiased ethylnitrosourea screen for anemic embryos.16 We performed chromosomal walking to demonstrate cadherin 5 (cdh5, VE-CAD) as the most probable candidate gene for the mlb locus. To identify the genetic mutation in the mlb locus, we sequenced the cdh5 gene and found an A267T stop codon in exon 3 (supplemental Figure 1A). We then used qRT-PCR (supplemental Figure 1B) and whole-mount in situ hybridization (WISH; supplemental Figure 1C) to validate loss of the cdh5 messenger RNA (mRNA) levels in mlb embryos, suggesting that the A267T stop codon resulted in nonsense-mediated decay of cdh5 mRNA. As an antibody cross-reacting with zebrafish cdh5 protein was not available, we were unable to directly assay the cdh5 protein levels in mlb embryos.
To further verify the loss-of-function phenotype for cdh5, we injected 2 independently derived cdh5 antisense splice-blocking and translational-blocking morpholinos (MOs)17 to knockdown cdh5 expression in zebrafish embryos. The cdh5-silenced embryos (morphants) phenocopied mlb’s anemia (supplemental Figure 1D-E). We further found a loss of cdh5 mRNA levels in the morphant embryos, suggesting that the splice-blocking MOs accurately targeted cdh5 (supplemental Figure 1F). Injection of cdh5 complementary RNA is toxic to malbec embryos, so it is not technically feasible to study the gain-of-function phenotype. Our phylogenetic analysis and peptide alignments further demonstrate human (CDH5, chromosome 16) and mouse (Cdh5, chromosome 8) orthologs for cdh5 (supplemental Figure 1G). These findings, along with positional cloning, mutational analysis, and qRT-PCR, as well as WISH data provide convincing evidences that cdh5 is mutated in the mlb locus.
malbec has normal primitive and definitive blood formation despite circulatory defects
The cdh5 knockdown zebrafish embryos have impaired cardiac function and circulatory arrest due to poor formation of endocardial junctions, which results in leakage across the endothelial layer and a reduction in the density of the cardiac jelly.36 We followed the circulation of lcr:eGFP+ erythroid cells using time-lapse confocal imaging of the lcr:eGFP::flk1:mCherry zebrafish embryos between 19 and 32 hpf, and observed no active circulation in mlb embryos despite the initiation of a heartbeat. We ruled out necrosis of hematopoietic cells using acridine orange staining of mlb embryos (supplemental Figure 2A). We thus concluded that the observed anemic phenotype of mlb results from pooling of erythroid cells in blood vessels (supplemental Figure 2B) in the absence of adequate blood circulation.
To study the role of cdh5 in zebrafish hematopoiesis, we first analyzed the expression of markers for primitive and definitive hematopoiesis. Unlike the mouse Cdh5 mutant, mlb zebrafish embryos survive, despite circulatory defects, past the stage when definitive hematopoiesis emerges. We found normal expression of markers for primitive erythroid progenitors (gata1) and myeloid (mpo, mpx) cells in the mlb or cdh5-MO embryos (Figure 1A-C). Analyzing these markers of definitive hematopoiesis, we detected normal HSCs (runx1 and c-Myb mRNA expression, mlb::c-myb:eGFP+ cells and mlb::cd41:eGFPlow levels) as well as definitive erythroid (slc4a1), lymphoid (rag1), and thrombocytic (cd41) cells (Figure 1D-I; supplemental Figure 2B), indicating that mlb has intact primitive and definitive hematopoiesis. Normal expression of runx1 and c-myb (supplemental Figure 2C-E) in cdh5-silenced embryos further demonstrates that the cdh5 morphant phenocopies definitive HSC formation as seen in mlb.
malbec (mlbbw306) has normal embryonic and definitive hematopoiesis despite disruption of the cdh5 gene. (A) The expression of erythro-myeloid progenitors (gata1) is normal in wt and mlb embryos at 20 somite stage (ss). wt: n = 80/80; mlb: n = 78/80. (B) WISH analysis of the surrogate marker for myeloid cell expression shows normal mpo levels in mlb embryos at 20 ss. wt: n = 80/80; mlb: n = 75/80. (C) FACS analysis of mpx:eGFP+ neutrophils in control and cdh5-silenced zebrafish embryos. Twenty embryos were used in each experiment; n = 5. (D) qRT-PCR analysis of runx1 and c-Myb mRNA expression in control and malbec embryos. Forty embryos were used in each experiment; n = 5. (E) Normal runx1 mRNA expression in the AGM region of 36-hpf-old wt and mlb embryos. wt: n = 220/220; mlb: n = 220/220. (F) A progeny of mlb crossed with c-myb:eGFP has comparable levels of mlb::c-myb:eGFP+ HSCs cells in malbec embryos; n = 60. (G) A progeny of mlb crossed with cd41:eGFP has comparable levels of mlb::cd41:eGFPlow HSCs (FACS analysis), indicating that mlb produces sufficient amounts of HSCs. Forty embryos pooled in each sample; n = 5. (H) Normal rag-1 mRNA expression in the thymus of a mlb embryo (examples indicated with arrows). wt: n = 80/80; mlb: n = 80/80. (I) Progeny of mlb crossed with cd41:eGFP shows mlb::cd41:eGFP+ platelets (examples indicated with arrows) at day 4 postfertilization. wt: n = 160/160; mlb: n = 156/160. *P < .05 (t test, error bars indicate standard error of the mean [s.e.m.]).
malbec (mlbbw306) has normal embryonic and definitive hematopoiesis despite disruption of the cdh5 gene. (A) The expression of erythro-myeloid progenitors (gata1) is normal in wt and mlb embryos at 20 somite stage (ss). wt: n = 80/80; mlb: n = 78/80. (B) WISH analysis of the surrogate marker for myeloid cell expression shows normal mpo levels in mlb embryos at 20 ss. wt: n = 80/80; mlb: n = 75/80. (C) FACS analysis of mpx:eGFP+ neutrophils in control and cdh5-silenced zebrafish embryos. Twenty embryos were used in each experiment; n = 5. (D) qRT-PCR analysis of runx1 and c-Myb mRNA expression in control and malbec embryos. Forty embryos were used in each experiment; n = 5. (E) Normal runx1 mRNA expression in the AGM region of 36-hpf-old wt and mlb embryos. wt: n = 220/220; mlb: n = 220/220. (F) A progeny of mlb crossed with c-myb:eGFP has comparable levels of mlb::c-myb:eGFP+ HSCs cells in malbec embryos; n = 60. (G) A progeny of mlb crossed with cd41:eGFP has comparable levels of mlb::cd41:eGFPlow HSCs (FACS analysis), indicating that mlb produces sufficient amounts of HSCs. Forty embryos pooled in each sample; n = 5. (H) Normal rag-1 mRNA expression in the thymus of a mlb embryo (examples indicated with arrows). wt: n = 80/80; mlb: n = 80/80. (I) Progeny of mlb crossed with cd41:eGFP shows mlb::cd41:eGFP+ platelets (examples indicated with arrows) at day 4 postfertilization. wt: n = 160/160; mlb: n = 156/160. *P < .05 (t test, error bars indicate standard error of the mean [s.e.m.]).
HSCs emerge from cdh5-silenced hemogenic endothelium
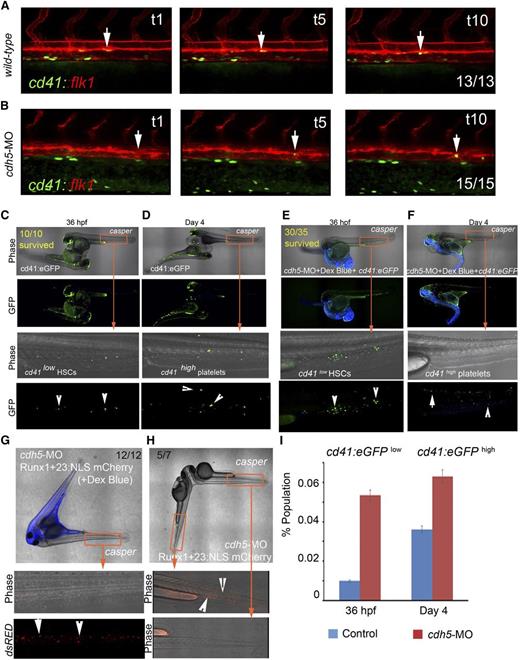
To demonstrate whether HSCs would emerge from cdh5-silenced hemogenic endothelium, we first performed time-lapse confocal imaging of cd41:eGFP+ HSC formation from flk1:mCherry+ endothelium in zebrafish blood vessels. We found that cd41:eGFP+ HSCs19,37 are continuously being formed in the vascular endothelium of the dorsal aorta in cdh5-silenced cd41:eGFP::flk1:mCherry embryos (Figure 2A-B; supplemental Videos 1-2). We also found that arterial endothelium is intact in cdh5-silenced embryos (supplemental Figure 2F). To analyze whether cd41:eGFP+ HSCs emerging from cdh5-silenced cd41:eGFP+ embryos are null for cdh5 expression, we sorted cd41:eGFP+ cells, measured cdh5 mRNA levels, and found that cd41:eGFP+ HSCs indeed lack cdh5 expression (supplemental Figure 2G). These data indicate that arterial endothelium continues to transition to HSCs in the absence of cdh5. We independently verified these observations by tracking continuous endothelial emergence of runx1+23:NLS-mCherry+ HSPCs22 from cdh5-silenced flk1:eGFP+ vascular endothelium (supplemental Figure 2H-J).
cdh5 is dispensable for endothelial emergence, migration, engraftment, and differentiation of HSCs. (A) Time-lapse confocal imaging of cd41:eGFP+ HSCs emerging from flk1:mCherry+ endothelial cells in control embryos between 30 and 42 hpf (examples are demonstrated with arrows); n = 13/13. (B) cdh5 silencing in cd41:eGFP::flk1:mCherry embryos does not affect endothelial emergence of cd41:eGFP+ HSCs (examples are demonstrated with arrows); n = 15/15. (C) cd41:eGFP transgenic embryo fused to transparent casper embryo (wt control) using parabiotic surgery (imaged at 36 hpf), indicates that cd41:eGFPlow HSCs emerging from the control transgenic embryo migrates to casper CHT to engraft and divide (examples are demonstrated with arrows). All 10 of 10 WT parabiots survived. (D) Parabiotic surgery of cd41:eGFP transgenic embryo to transparent casper embryo shows cd41:eGFPhigh platelets in circulation in transgenic and casper embryos (imaged at day 4; examples are demonstrated with arrows). (E) cdh5-MO and dextran blue–injected cd41:eGFP transgenic embryo fused to transparent casper embryo shows the emergence of cd41:eGFPlow HSCs in cdh5-silenced embryo (imaged at 36 hpf), which migrate to casper CHT via passive circulation exchange to engraft and further differentiate (examples are demonstrated with arrows). Thirty of 35 cdh5-silenced parabiots survived. (F) Four-day-old cdh5-silenced cd41:eGFP embryo fused to casper embryo, indicating that morphant cd41:eGFPlow HSCs in casper CHT differentiated into circulating cd41:eGFPhigh platelets (imaged at day 4; examples with arrows). (G) Surgical fusion of cdh5-silenced and dextran blue–injected runx1+23:NLS-mCherry with casper embryo (imaged at 36 hpf) shows runx1+23:NLS-mCherry+ HSPCs emerging from cdh5-silenced transgenic embryo and engrafting to casper’s CHT (examples are demonstrated with arrows). Twelve of 12 parabiots survived. (H) Lateral body-to-head fusion of cdh5-silenced ruxn1+23:NLS-mCherry embryo with casper embryo, showing a lack of circulation exchange between the 2 embryos impedes migration of cdh5-silenced runx1+23:NLS-mCherry+ HSPCs to casper (examples are demonstrated with arrows). Five of 7 parabiots survived. (I) Flow analyses of cd41:eGFPlow HSCs at 36 hpf and cd41:eGFPhigh platelets at day 4 in the control and cdh5-silenced cd41:eGFP embryos show that differentiation into platelets continues despite the lack of cdh5. Twenty embryos were used in each experiment; n = 5. *P < .05 (t test; error bars indicate s.e.m.).
cdh5 is dispensable for endothelial emergence, migration, engraftment, and differentiation of HSCs. (A) Time-lapse confocal imaging of cd41:eGFP+ HSCs emerging from flk1:mCherry+ endothelial cells in control embryos between 30 and 42 hpf (examples are demonstrated with arrows); n = 13/13. (B) cdh5 silencing in cd41:eGFP::flk1:mCherry embryos does not affect endothelial emergence of cd41:eGFP+ HSCs (examples are demonstrated with arrows); n = 15/15. (C) cd41:eGFP transgenic embryo fused to transparent casper embryo (wt control) using parabiotic surgery (imaged at 36 hpf), indicates that cd41:eGFPlow HSCs emerging from the control transgenic embryo migrates to casper CHT to engraft and divide (examples are demonstrated with arrows). All 10 of 10 WT parabiots survived. (D) Parabiotic surgery of cd41:eGFP transgenic embryo to transparent casper embryo shows cd41:eGFPhigh platelets in circulation in transgenic and casper embryos (imaged at day 4; examples are demonstrated with arrows). (E) cdh5-MO and dextran blue–injected cd41:eGFP transgenic embryo fused to transparent casper embryo shows the emergence of cd41:eGFPlow HSCs in cdh5-silenced embryo (imaged at 36 hpf), which migrate to casper CHT via passive circulation exchange to engraft and further differentiate (examples are demonstrated with arrows). Thirty of 35 cdh5-silenced parabiots survived. (F) Four-day-old cdh5-silenced cd41:eGFP embryo fused to casper embryo, indicating that morphant cd41:eGFPlow HSCs in casper CHT differentiated into circulating cd41:eGFPhigh platelets (imaged at day 4; examples with arrows). (G) Surgical fusion of cdh5-silenced and dextran blue–injected runx1+23:NLS-mCherry with casper embryo (imaged at 36 hpf) shows runx1+23:NLS-mCherry+ HSPCs emerging from cdh5-silenced transgenic embryo and engrafting to casper’s CHT (examples are demonstrated with arrows). Twelve of 12 parabiots survived. (H) Lateral body-to-head fusion of cdh5-silenced ruxn1+23:NLS-mCherry embryo with casper embryo, showing a lack of circulation exchange between the 2 embryos impedes migration of cdh5-silenced runx1+23:NLS-mCherry+ HSPCs to casper (examples are demonstrated with arrows). Five of 7 parabiots survived. (I) Flow analyses of cd41:eGFPlow HSCs at 36 hpf and cd41:eGFPhigh platelets at day 4 in the control and cdh5-silenced cd41:eGFP embryos show that differentiation into platelets continues despite the lack of cdh5. Twenty embryos were used in each experiment; n = 5. *P < .05 (t test; error bars indicate s.e.m.).
HSCs migrate, engraft, and differentiate in the absence of cdh5 expression
We performed parabiotic surgery25 to fuse cdh5-silenced cd41:eGFP morphant embryos with transparent casper embryos, which were otherwise WT. We found that cd41:eGFP+ HSCs emerged from cdh5-silenced aortic endothelium and then migrated to the CHT (a region analogous to mammalian fetal liver) of the casper recipients. The cdh5-silenced cd41:eGFP+ HSCs engrafted, divided, and subsequently differentiated into circulating cd41:eGFP+ platelets at day 4 postfertilization (Figure 2C-F; supplemental Figure 2G; supplemental Videos 3-4). Similarly, we fused cdh5-silenced runx1+23:NLS-mCherry+ morphant embryos with transparent casper embryos, and found that runx1+23:NLS-mCherry+ HSPCs formed in the cdh5 morphant embryos migrated to the casper WT CHT for engraftment and differentiation (Figure 2G). These data demonstrate that cdh5 is dispensable for HSC formation in the zebrafish, and that cdh5-silenced HSCs can migrate, engraft, and differentiate into definitive hematopoietic lineages in a WT hematopoietic niche.
As both cdh5 morphant and mlb mutant embryos have circulation defects, we further analyzed whether the migration of runx1:mCherry+ HSPCs from cdh5 morphants to the WT niche was due to a partial recovery of cardiac function and circulation, or due to the exchange of the blastula cells early in development. Because surgically paired embryos appeared to have 1 fused heart, we hypothesized that partial or complete recovery of cardiac function from the WT zebrafish was responsible for the migration of runx1:mCherry+ HSPCs from cdh5 morphants to the casper niche (Figure 2G). To test this, we fused 2 zebrafish embryos along the lateral body and head, such that the cdh5 morphant zebrafish could be directly observed to lack active circulation. Indeed, we found that the runx1: mCherry+ HSPCs could not migrate from cdh5 morphant to casper CHT in these pairs (Figure 2H). These data reinforce the conclusion that runx1+23:NLS- mCherry+ hematopoietic cells are formed in the AGM of cdh5 morphant zebrafish embryos and subsequently migrate via passive circulation to the casper CHT to engraft and differentiate (Figure 2G-H). We independently verified our time-lapse confocal imaging data by documenting levels of cd41:eGFPlow HSCs at 36 hpf and their differentiation into cd41:eGFPhigh platelets at day 4 in cdh5-morphant embryos (Figure 2I). However, we could not confirm whether the increase in cd41 expression translates into an increase in functional HSCs. Thus, HSCs are being formed in the absence of cdh5, and manifest their stem cell properties in a casper CHT.
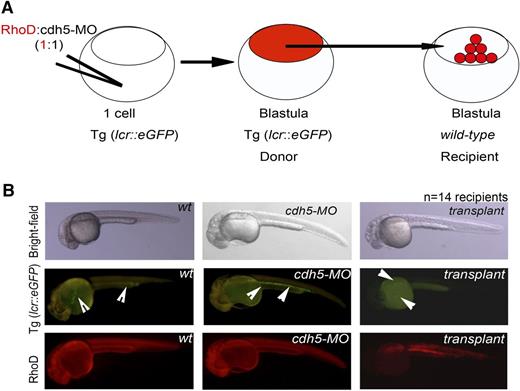
To further investigate the cell-intrinsic roles of cdh5 in HSC differentiation, we silenced cdh5 in erythroid transgenic lcr:eGFP embryos. We then allowed these morphants to develop up to high or dome stage, and then transplanted the blastula cells from cdh5-silenced transgenic embryo into WT recipients. We not only found that cdh5-morphant embryos produced lcr:eGFP+ erythroid cells, but also that cdh5-silenced blastula transplant (donor) cells contributed to lcr:eGFP+ erythroid cells in WT recipients at the definitive hematopoiesis stage of embryonic development (Figure 3). These data demonstrate that blastula cells lacking cdh5 expression differentiate into erythroid cells.
cdh5-silenced blastula transplant successfully differentiates into red cells. (A) Schematic representation of the blastula transplantation strategy. Injections of equal amounts of rhodamine and cdh5-MO into erythroid transgenic embryo, lcr:eGFP, followed by transplantation of cdh5-silenced blastula (donor) cells of transgenic embryo between high and dome stage into age-matched casper-recipient embryo (wild-type) to analyze whether donor cells can contribute to transgenic erythroid cells in a recipient embryo. (B) Transplantation of cdh5-silenced transgenic blastula reconstituted into lcr:eGFP+ erythroid cells into casper recipient embryos, indicating that the cell-intrinsic role of cdh5 is dispensable for blastula differentiation into erythroid cells (examples are demonstrated with arrows); n = 14 recipients.
cdh5-silenced blastula transplant successfully differentiates into red cells. (A) Schematic representation of the blastula transplantation strategy. Injections of equal amounts of rhodamine and cdh5-MO into erythroid transgenic embryo, lcr:eGFP, followed by transplantation of cdh5-silenced blastula (donor) cells of transgenic embryo between high and dome stage into age-matched casper-recipient embryo (wild-type) to analyze whether donor cells can contribute to transgenic erythroid cells in a recipient embryo. (B) Transplantation of cdh5-silenced transgenic blastula reconstituted into lcr:eGFP+ erythroid cells into casper recipient embryos, indicating that the cell-intrinsic role of cdh5 is dispensable for blastula differentiation into erythroid cells (examples are demonstrated with arrows); n = 14 recipients.
HSC development in Cdh5−/−GFP+/+:Cdh5+/+GFP−/− chimeric mice embryos
As Cdh5 mutant mouse embryos die before first HSCs emerge in AGM, we set out to analyze chimeric mice (in which Cdh5−/−GFP+ ES cells injected into Cdh5+/+GFP− blastocysts) to test whether functional HSCs emerge from Cdh5-deleted hemogenic endothelium (Figure 4A).
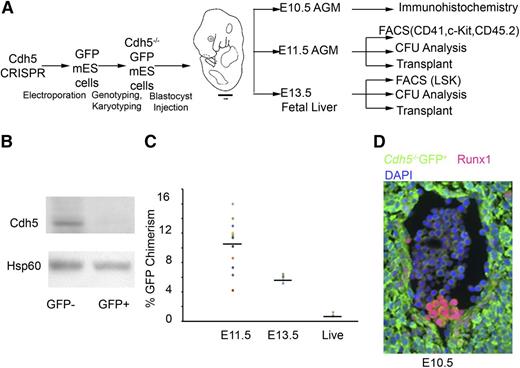
Development and analysis of Cdh5−/−GFP+/+: Cdh5+/+GFP−/− chimeric mice embryos. (A) Schematic representation of the Cdh5 chimeric mouse development and analysis. Identification of Cdh5−/−GFP+/+ mES cells after electroporation of Cdh5 CRISPRs targeting exons 1 to 12 of the Cdh5 gene followed by karyotyping to exclude aneuploidy. Injection of Cdh5−/−GFP+ mES cells into WT mouse embryo of foster mothers and harvesting chimeric AGM at E10.5 for immunohistochemistry (GFP, Runx1, DAPI) and at E11.5 for cd41/c-Kit/cd45.2 expression, CFU analysis, transplant into sublethally irradiated mice as well as at E13.5 fetal liver for Lineage−Sca1+c-Kit+ expression, CFU analysis, and reconstitution capacity of fetal liver transplants. (B) Western analysis for Cdh5 and Hsp60 protein expression in E13.5 Cdh5−/−GFP+:Cdh5+/+GFP−-fetal liver–derived GFP− and GFP+ cells. To demonstrate the absence of Cdh5 at protein level, we sorted GFP+ and GFP− cells from fetal liver of E13.5 Cdh5−/−GFP+/+:Cdh5+/+GFP−/− chimeric mice embryos. Our western blot data confirmed the absence of Cdh5 protein in GFP+ cells. (C) The percentage of GFP chimerism in 11 E11.5 AGM, 6 E13.5 fetal liver, and 3 live pups. (D) Immunostaining of GFP (green), Runx1 (red), and DAPI (violet) in transverse sections of E10.5 AGM region, indicating that Cdh5−/− GFP+ cells produced Runx1+ GFP+ hematopoietic clusters in the ventral wall of the E10.5 dorsal aorta. Magnification, 40×.
Development and analysis of Cdh5−/−GFP+/+: Cdh5+/+GFP−/− chimeric mice embryos. (A) Schematic representation of the Cdh5 chimeric mouse development and analysis. Identification of Cdh5−/−GFP+/+ mES cells after electroporation of Cdh5 CRISPRs targeting exons 1 to 12 of the Cdh5 gene followed by karyotyping to exclude aneuploidy. Injection of Cdh5−/−GFP+ mES cells into WT mouse embryo of foster mothers and harvesting chimeric AGM at E10.5 for immunohistochemistry (GFP, Runx1, DAPI) and at E11.5 for cd41/c-Kit/cd45.2 expression, CFU analysis, transplant into sublethally irradiated mice as well as at E13.5 fetal liver for Lineage−Sca1+c-Kit+ expression, CFU analysis, and reconstitution capacity of fetal liver transplants. (B) Western analysis for Cdh5 and Hsp60 protein expression in E13.5 Cdh5−/−GFP+:Cdh5+/+GFP−-fetal liver–derived GFP− and GFP+ cells. To demonstrate the absence of Cdh5 at protein level, we sorted GFP+ and GFP− cells from fetal liver of E13.5 Cdh5−/−GFP+/+:Cdh5+/+GFP−/− chimeric mice embryos. Our western blot data confirmed the absence of Cdh5 protein in GFP+ cells. (C) The percentage of GFP chimerism in 11 E11.5 AGM, 6 E13.5 fetal liver, and 3 live pups. (D) Immunostaining of GFP (green), Runx1 (red), and DAPI (violet) in transverse sections of E10.5 AGM region, indicating that Cdh5−/− GFP+ cells produced Runx1+ GFP+ hematopoietic clusters in the ventral wall of the E10.5 dorsal aorta. Magnification, 40×.
We first used CAS9 nuclease genome editing to target sequences flanking exon 1 and exon 12 of the Cdh5 gene in mice (supplemental Figure 3A; supplemental Table 1). We engineered mouse ES cells with ubiquitous expression of GFP and identified a Cdh5−/−GFP+ mES clone (supplemental Table 2; supplemental Figure 3B). After verifying loss of Cdh5 gene expression (supplemental Figure 3B) and confirming their normal karyotype (supplemental Figure 3C), we generated blastocyst chimeras by injecting the Cdh5−/−GFP+ mES cells into WT embryos, and then transplanted these chimeric embryos into CD1 foster mothers (Figure 4A). By western blot, we confirmed the absence of Cdh5 expression in GFP+ cells isolated by flow cytometric sorting from the E13.5 fetal livers of chimeric embryos (Figure 4B). The percentage of GFP chimerism in 11 E11.5 AGM, 6 E13.5 fetal liver, and peripheral blood of 3 live pups is provided in Figure 4C.
We recovered E10.5 chimeric embryos, embedded in paraffin, and performed sections. By immunohistochemistry,33,34 we found that Runx1+GFP+ cells emerge from the aortic endothelium (Figure 4D; supplemental Figure 3D-E) in a pattern similar to Runx1+ expression in E10.5 WT mice embryos, thus demonstrating the emergence of hematopoietic clusters from Cdh5-deleted mammalian hemogenic endothelial cells.
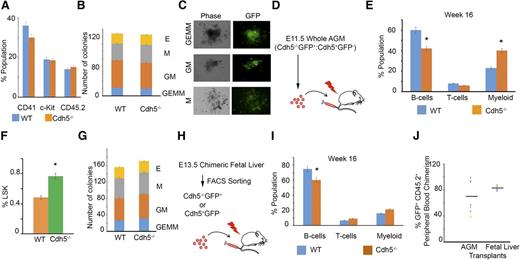
To prove the emergence of functional HSCs, we isolated the AGM from E11.5-old chimeric embryos and analyzed the hematopoietic marker-expression profile, colony formation, and reconstitution capacity of Cdh5−/−GFP+ or Cdh5+/+GFP− AGM-derived cells. These cells expressed CD45.2, c-Kit, and CD41 (Figure 5A; supplemental Figures 4A-D and 5A-D), and differentiated into multipotential GEMM progenitors, granulocyte-macrophage progenitors (CFU-GM, CFU-M, CFU-G), and erythroid colonies (CFU-E) (Figure 5B-C). These data demonstrate that the Cdh5 is dispensable for CD41, c-Kit, and CD45.2 constitutive expression as well as the hematopoietic colony-forming capacity of Cdh5−/−GFP+ HSCs. We, however, could not distinguish whether hematopoietic progenitors in E11.5 AGM are from yolk sac or AGM. In addition, transplantation of 2 e.e. of chimeric E11.5 AGM resulted in the long-term (16 week) multilineage engraftment of Cdh5−/−GFP+ B cells, macrophages, and T cells in irradiated B6.SJL-PtprcaPep3b/BoyJ (CD45.1) recipient mice4,35 (Figure 5D-E; supplemental Figures 4E-G and 5E-H; Figure 5J).
Cdh5 is dispensable for HSC development in E11.5 AGM and E13.5 fetal liver and reconstitution of multilineage adult hematopoiesis. (A) Quantitative flow analysis of CD41, c-Kit, and CD45.2-positive cells in E11.5 Cdh5−/−GFP+ and WT (Cdh5+/+GFP−) cells, indicating that loss of Cdh5 is dispensable for CD41, c-Kit, and CD45.2 expression. Eleven AGM were used for FACS analyses. (B) CFU analysis of sorted E11.5 AGM-derived WT and Cdh5−/− GFP+ cells, indicating that loss of Cdh5 has no influence on hematopoietic colony differentiation. Twelve AGM were used for CFU analyses; 2 e.e. per 3.5 mL of M3434 media. (C) Photographs of green fluorescent GFP+ hematopoietic colonies formed from the E11.5 AGM-derived Cdh5−/−GFP+ HSCs. Magnification, ×4. (D) Schema showing the transplant of 2 e.e. E11.5 whole chimeric AGM in irradiated SJL mice. (E) Quantitative analysis of peripheral blood from mice transplanted with 2 e.e. E11.5 chimeric whole AGM, indicating that Cdh5−/−GFP+ HSCs reconstitute to multilineage adult blood up to 16 weeks of transplant. Twelve AGM were transplanted in 6 SJL recipients. (F) Quantitative analysis of the percentage of Lineage−Sca1+c-Kit+ (LSK) cells in WT (Cdh5+/+GFP−) and Cdh5−/−GFP+ compartments of E13.5 fetal liver, indicating that loss of Cdh5 has no intrinsic influence on development of LSK HSCs. Six fetal livers were used for FACS analyses. (G) CFU analysis of sorted E13.5 fetal liver–derived sorted WT (Cdh5+/+GFP−) and Cdh5−/−GFP+ cells, indicating that loss of Cdh5 has no influence on hematopoietic colony differentiation. Sorted cells from 10 fetal liver were used for CFU analyses. (H) Schema showing the transplant of E13.5 fetal liver–derived GFP+ or GFP− sorted cells in irradiated SJL mice. (I) Quantitative analysis of peripheral blood from mice transplanted with E13.5 fetal liver–derived WT and Cdh5−/−GFP+ sorted cells, indicating that Cdh5−/−GFP+ HSCs reconstitute multilineage adult blood up to 16 weeks of transplant; n = 6 recipients. (J) Percentage GFP+CD45.2+ peripheral blood chimerism from the Cdh5 knockout cells in E11.5 AGM and E13.5 fetal liver–transplanted recipients. Each dot represents an individual recipient; n = 6; *P < .05 (t test, error bars indicate s.e.m.).
Cdh5 is dispensable for HSC development in E11.5 AGM and E13.5 fetal liver and reconstitution of multilineage adult hematopoiesis. (A) Quantitative flow analysis of CD41, c-Kit, and CD45.2-positive cells in E11.5 Cdh5−/−GFP+ and WT (Cdh5+/+GFP−) cells, indicating that loss of Cdh5 is dispensable for CD41, c-Kit, and CD45.2 expression. Eleven AGM were used for FACS analyses. (B) CFU analysis of sorted E11.5 AGM-derived WT and Cdh5−/− GFP+ cells, indicating that loss of Cdh5 has no influence on hematopoietic colony differentiation. Twelve AGM were used for CFU analyses; 2 e.e. per 3.5 mL of M3434 media. (C) Photographs of green fluorescent GFP+ hematopoietic colonies formed from the E11.5 AGM-derived Cdh5−/−GFP+ HSCs. Magnification, ×4. (D) Schema showing the transplant of 2 e.e. E11.5 whole chimeric AGM in irradiated SJL mice. (E) Quantitative analysis of peripheral blood from mice transplanted with 2 e.e. E11.5 chimeric whole AGM, indicating that Cdh5−/−GFP+ HSCs reconstitute to multilineage adult blood up to 16 weeks of transplant. Twelve AGM were transplanted in 6 SJL recipients. (F) Quantitative analysis of the percentage of Lineage−Sca1+c-Kit+ (LSK) cells in WT (Cdh5+/+GFP−) and Cdh5−/−GFP+ compartments of E13.5 fetal liver, indicating that loss of Cdh5 has no intrinsic influence on development of LSK HSCs. Six fetal livers were used for FACS analyses. (G) CFU analysis of sorted E13.5 fetal liver–derived sorted WT (Cdh5+/+GFP−) and Cdh5−/−GFP+ cells, indicating that loss of Cdh5 has no influence on hematopoietic colony differentiation. Sorted cells from 10 fetal liver were used for CFU analyses. (H) Schema showing the transplant of E13.5 fetal liver–derived GFP+ or GFP− sorted cells in irradiated SJL mice. (I) Quantitative analysis of peripheral blood from mice transplanted with E13.5 fetal liver–derived WT and Cdh5−/−GFP+ sorted cells, indicating that Cdh5−/−GFP+ HSCs reconstitute multilineage adult blood up to 16 weeks of transplant; n = 6 recipients. (J) Percentage GFP+CD45.2+ peripheral blood chimerism from the Cdh5 knockout cells in E11.5 AGM and E13.5 fetal liver–transplanted recipients. Each dot represents an individual recipient; n = 6; *P < .05 (t test, error bars indicate s.e.m.).
Similarly, the fetal liver of E13.5 chimeric embryos had Lin−Sca+c-Kit+GFP+ hematopoietic cells (Figure 5F; supplemental Figures 6A-C and 7A-C), which differentiated into CFU-GEMM, CFU-GM, CFU-M, and CFU-E hematopoietic colonies (Figure 5G). The transplantation of E13.5 Cdh5−/−GFP+ or Cdh5+/+GFP− fetal liver cells into irradiated CD45.1 mice also produced engraftment with B cells, macrophages, and T cells, indicating production of functional HSCs from Cdh5-deficient cells (Figure 5H-I; supplemental Figures 6D-F and 7D-E). To analyze the percentage of donor Cdh5−/−GFP+ cells contributing to adult lineages, we analyzed the percentage of GFP+CD45.2+ peripheral blood chimerism in E11.5 AGM or E13.5 fetal liver transplants (Figure 5J). We also measured the percentage of GFP−CD45.2+ peripheral blood chimerism in E11.5 AGM or E13.5 fetal liver transplants to measure WT donor cells contributing to adult lineages (supplemental Figure 7F). To exclude reversion of Cdh5 expression, we also sorted GFP+CD45.2+ cells from E13.5 fetal liver transplanted recipients and analyzed Cdh5 gene expression (supplemental Figure 7G). We found deletion of Cdh5 in sorted cells, suggesting that GFP+ blood lineages in transplants are derived from Cdh5-null GFP+ mES cells.
Cdh5 deletion in Scl+ cells did not alter hematopoietic colony formation
After generating a conditional mouse Cdh5 knockout (Cdh5flox/flox:Scl-Cre-ERT; Figure 6A-B), we deleted Cdh5 floxed alleles beginning at E5.5 using tamoxifen-regulated Cre driven from Scl regulatory sequences. Tamoxifen was injected into pregnant dams from E5.5 to E10.5 to execute the Cdh5 deletion (Figure 6C). We harvested E11.5 embryos, dissected the AGM, and enumerated CFU colonies in methylcellulose colony-forming assays. We compared the number of GEMM colonies in the Cdh5fl/fl:Scl-Cre-ERT embryos with the Cdh5fl/+:Scl-Cre-ERT embryos for potential Cre toxicity (Figure 6D). We assessed the deletion of the Cdh5 alleles in individual CFU colonies from Cdh5fl/fl: Scl-Cre-ERT and Cdh5fl/+:Scl-Cre-ERT by PCR (Figure 6E). We found that the deletion of Cdh5 did not alter numbers of GEMM colonies (Figure 6D). Therefore, Cdh5 is dispensable for hematopoietic colony formation from Scl+ cells, further strengthening the conclusion that Cdh5 is dispensable within blood lineages.
Development of a conditional mouse Cdh5 knockout. (A) Breeding strategy to develop Cdh5flox/flox: Scl-Cre-ERT mouse. (B) PCR-based strategy to genotype WT, Cdh5flox/flox, Cdh5flox/+:Scl-Cre-ERT, Cdh5flox/flox: Scl-Cre-ERT, and Scl-Cre-ERT using primers listed in supplemental Table 4. (C) Schema to delete Cdh5 in Scl+ cells in Cdh5flox/flox:Scl-Cre-ERT. Tamoxifen was injected in pregnant dam from E5.5 to E10.5 and AGM were harvested on E11.5 for CFU analyses. (D) Average number of GEMM colonies per 1 e.e. AGM derived from Cdh5 flox/flox (n = 10), Cdh5 flox/+:Scl-Cre-ERT (n = 7), and Cdh5 flox/flox:Scl-Cre-ERT (n = 11), respectively. (E) Percentage Cdh5 alleles in colonies derived from Cdh5 flox/+:Scl-Cre-ERT (n = 55 were either fl/+ or ∆/+), and Cdh5 flox/flox:Scl-Cre-ERT (n = 72 were fl/fl, ∆/+, or ∆/∆). t test; error bars indicate s.e.m.
Development of a conditional mouse Cdh5 knockout. (A) Breeding strategy to develop Cdh5flox/flox: Scl-Cre-ERT mouse. (B) PCR-based strategy to genotype WT, Cdh5flox/flox, Cdh5flox/+:Scl-Cre-ERT, Cdh5flox/flox: Scl-Cre-ERT, and Scl-Cre-ERT using primers listed in supplemental Table 4. (C) Schema to delete Cdh5 in Scl+ cells in Cdh5flox/flox:Scl-Cre-ERT. Tamoxifen was injected in pregnant dam from E5.5 to E10.5 and AGM were harvested on E11.5 for CFU analyses. (D) Average number of GEMM colonies per 1 e.e. AGM derived from Cdh5 flox/flox (n = 10), Cdh5 flox/+:Scl-Cre-ERT (n = 7), and Cdh5 flox/flox:Scl-Cre-ERT (n = 11), respectively. (E) Percentage Cdh5 alleles in colonies derived from Cdh5 flox/+:Scl-Cre-ERT (n = 55 were either fl/+ or ∆/+), and Cdh5 flox/flox:Scl-Cre-ERT (n = 72 were fl/fl, ∆/+, or ∆/∆). t test; error bars indicate s.e.m.
Discussion
Lineage tracing,2,3 in vitro differentiation,4,5 and various mouse studies have demonstrated that HSCs emerge from AGM-derived Cdh5+ endothelial precursors,38 implicating Cdh5 as a marker for the hemogenic endothelial cells that transition to HSCs.39-41 Because Cdh5 mouse mutant embryos die13,14 before AGM-derived HSCs emerge at E10.5,1,15 whether Cdh5 is essential for differentiation of HSCs into definitive blood lineages has remained unclear.12 In this study, we demonstrate that functional HSCs emerge from Cdh5-deficient AGM-hemogenic endothelium during zebrafish and murine fetal development, and show that Cdh5-deleted HSCs engraft and reconstitute multilineage adult blood. Thus, we have established that Cdh5 is dispensable for HSC formation and differentiation (supplemental Figure 9).
Creation of genetic mosaics (chimeras) between WT and genetically modified cells is a well-established and classical strategy for demonstrating cell-intrinsic effects of a gene on cell functions.42,43 Creation of zebrafish chimeras has been used to study the cell-intrinsic functions of cloche and bloodless genes in HSPC proliferation, survival, and differentiation.26,27,44 Here, we demonstrated that transplanted cdh5-silenced blastula cells differentiate into red cells in WT zebrafish embryos. To test the cell-intrinsic role of Cdh5 in mammalian blood development and corroborate our findings from zebrafish, we bypassed the lethality of Cdh5 mutant mouse embryos by creating chimeras of WT mouse embryos with GFP+Cdh5-mutant ES cells. A similar approach has been used to study cell-intrinsic roles of Ascl2 and Eed.45,46 Although we did not perform comprehensive functional analyses of Cdh5-deficient blood cells by competitive transplantation, we observed that Cdh5−/−GFP+ cells isolated from E11.5 AGM and E13.5 fetal liver express a normal range of hematopoietic markers, differentiate normally into hematopoietic colonies, and reconstitute normal levels of adult B cells, macrophages, and T cells following transplantation into irradiated SJL mice, indicating the production of functional HSCs from Cdh5-deficient cells. We also developed a conditional mouse knockout of Cdh5 (Cdh5flox/flox:Scl-Cre-ERT) and independently validated that multipotent hematopoietic colonies form from Scl+ hemogenic endothelial cells despite the absence of Cdh5. Therefore, our zebrafish, mouse chimera, and conditional mouse knockout data demonstrate that although Cdh5 is a marker of definitive hemogenic endothelium, it need not be expressed within the blood lineage to enable HSC emergence and function. However, our data do not exclude the possibility that an unknown molecule might compensate for the function of Cdh5 critical to hematopoietic specification. Our analyses provide a refined view of the molecular features of hemogenic endothelium and illustrate the power of combining model organisms in order to investigate the factors regulating HSC development and differentiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Kathryn Crosier, Philip Crosier, Jeff Cooney, Iman Schultz, Matthew King, Yi Zhou, Christian Lawrence, Eric L. Pierce, Benjanim S. Brigham, Jessica Collantonio, Spencer K. Heggers, and Michelle Cassim for their technical help and/or for providing reagents.
D.I.S. is supported by grants from the American Society of Hematology, the Cooley’s Anemia Foundation, and the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants (K01DK085217 and R03DK100672). H.A. is supported by a fellowship from the Sigrid Juselius Foundation. E.J.H. is supported by a fellowship from the Helen Hay Whitney Foundation. P.N.G.R. is supported by a fellowship from the American Society of Hematology. O.J.T. is supported by a fellowship from the American Society of Hematology. G.A.M. is supported by a fellowship from the Alex’s Lemonade Stand Foundation. D.E.B. is supported by a grant from the NIH, NIDDK (K08DK093705). B.H.P. is supported by a grant from the NIH, National Heart, Lung, and Blood Institute (P01HL032262). M.F. and D.V. are supported by grants from the Deutsche Forschungsgemeinschaft and the Max Planck Society. L.I.Z. is supported by grants from the NIH National Cancer Institute (R01CA103846) and National Heart, Lung, and Blood Institute (P01HL032262 and R01HL04880). S.H.O. is supported by grants from the NIH NIDDK (P30DK049216) and National Heart, Lung, and Blood Institute (R01HL032259 and P01HL032262). G.Q.D. is supported by grants from the NIH National Heart, Lung and Blood Institute (Progenitor Cell Biology Consortium UO1-HL100001) and NIDDK (R24DK092760), the Alex’s Lemonade Foundation, and the Boston Children’s Hospital Stem Cell Program.
L.I.Z., S.H.O., and G.Q.D. are investigators of the Howard Hughes Medical Institute.
Authorship
Contribution: D.I.S. originally conceived the project, designed, supervised, and performed the experiments, analyzed data, and wrote the manuscript; H.A. identified the Cdh5 mutation, and performed in situ and blastula transplantation; T.C.P. performed confocal imaging and parabiotic experiments and helped with chimeric mouse experiments; H.A., T.C.P., E.J.H., K.A.S., O.J.T., K.H., N.C.H., J.P.K., and G.V. helped with zebrafish husbandry, imaging, and analyses; T.C.P., P.N.G.R., P.G.K., M.J.C., G.A.M., M.N., and Y.F. helped with mice colony management, injection of mES cells, transplantation of hematopoietic tissues, and FACS analyses; D.E.B. helped to design CRISPR constructs; M.F. and D.V. provided Cdh5flox/flox mice; and D.V., B.H.P., L.I.Z., S.H.O., and G.Q.D. analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliations are: H.A., University of Helsinki, Helsinki, Finland; K.A.S., University of Buffalo School of Medicine and Biomedical Sciences, Buffalo, NY; M.F., Uppsala University, Uppsala, Sweden; K.H., Max Planck Institute for Molecular Biomedicine, Münster, Germany; J.P.K., The Medical Foundation, Boston, MA; and G.V., Institut de Cancerologie de Lorraine, Vandoeuvre-lès-Nancy, France.
Correspondence: Dhvanit I. Shah, Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Harvard Stem Cell Institute, Boston, MA 02115; e-mail: dshah@research.bwh.harvard.edu.
References
Author notes
H.A. and T.C.P. contributed equally to this work, and are co-first authors.

![Figure 1. malbec (mlbbw306) has normal embryonic and definitive hematopoiesis despite disruption of the cdh5 gene. (A) The expression of erythro-myeloid progenitors (gata1) is normal in wt and mlb embryos at 20 somite stage (ss). wt: n = 80/80; mlb: n = 78/80. (B) WISH analysis of the surrogate marker for myeloid cell expression shows normal mpo levels in mlb embryos at 20 ss. wt: n = 80/80; mlb: n = 75/80. (C) FACS analysis of mpx:eGFP+ neutrophils in control and cdh5-silenced zebrafish embryos. Twenty embryos were used in each experiment; n = 5. (D) qRT-PCR analysis of runx1 and c-Myb mRNA expression in control and malbec embryos. Forty embryos were used in each experiment; n = 5. (E) Normal runx1 mRNA expression in the AGM region of 36-hpf-old wt and mlb embryos. wt: n = 220/220; mlb: n = 220/220. (F) A progeny of mlb crossed with c-myb:eGFP has comparable levels of mlb::c-myb:eGFP+ HSCs cells in malbec embryos; n = 60. (G) A progeny of mlb crossed with cd41:eGFP has comparable levels of mlb::cd41:eGFPlow HSCs (FACS analysis), indicating that mlb produces sufficient amounts of HSCs. Forty embryos pooled in each sample; n = 5. (H) Normal rag-1 mRNA expression in the thymus of a mlb embryo (examples indicated with arrows). wt: n = 80/80; mlb: n = 80/80. (I) Progeny of mlb crossed with cd41:eGFP shows mlb::cd41:eGFP+ platelets (examples indicated with arrows) at day 4 postfertilization. wt: n = 160/160; mlb: n = 156/160. *P < .05 (t test, error bars indicate standard error of the mean [s.e.m.]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/26/10.1182_blood-2015-07-659276/4/m_2811f1.jpeg?Expires=1769089565&Signature=ntYefGGQkQl~isv0YEdFFSalM-MAoMAfULHYOClKA63sWkfJwfpG1aEDQhLjOWT1Yx2o8wwtxtqjsgJ-u~1T40Y9Hj7YojRXRjiOlHGYYIu7BEFvvcXadrqYBe3ZKlk3NitJUb~V7gKRQpp5m9yXXejuwYv3r8NLxs3-wo58DRcXkuTJG9UCqaSv42b5C4Df0DvFR3ZqqO7OlOHlKyUJQuruEyxWbd9gQs9evhofihdbNSWQrNsV4OwLhWDd4om4WH0NfVWt~CBcviflhbFD6n9zmDAKge2lRJzXEz9~txOqlqtYXtogdWY5u1l0320pfB98gEdOx8LTBSxtr1q6yQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal