To the editor:
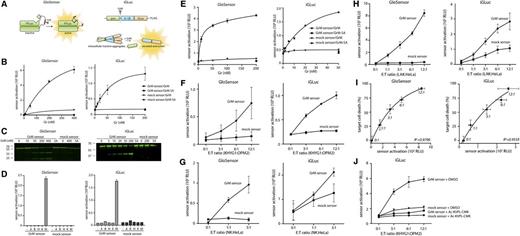
With great interest, we read the recent Blood article by Vrazo et al,1 who report on the use of luciferase-based biosensors to detect proteolytic activity of granzymes (Gr’s) A, B, and K. They show that these sensors can be used to profile Gr delivery by natural killer (NK) cells inside tumor cells in real time. However, humans express 5 Gr’s, and a specific sensor for GrM is lacking. GrM is expressed by lymphocytes of both the innate and adaptive immune system and plays a role in the control of cancer, viruses, and inflammation.2 Here, we report the development and comparison of 2 distinct variants of GrM gain-of-function biosensors: the GrM GloSensor, which is similar to the sensors described by Vrazo et al,1 and the GrM pro-interleukin (IL)-1β-gaussia luciferase (iGLuc) sensor3 (Figure 1A).
Intracellular profiling of GrM activity by bioluminescent gain-of-function biosensors. (A) Schematic overview of the GrM GloSensor (left) (based on Promega backbone) and the GrM iGLuc sensor (right). The GloSensor circularly permuted firefly luciferase (FLuc) molecule is locked in an inactive conformation by a short peptide linker containing a GrM-specific recognition sequence (AKMPL↓AAEEE). Upon cleavage of the linker by GrM, the FLuc subunits undergo a conformational change, resulting in the formation of active FLuc. The GrM iGLuc sensor is a fusion protein of pro–IL 1β and gaussia luciferase (GLuc). Upon cleavage of the AKMPL↓AAEEE recognition sequence by GrM, the prodomain is separated from the IL-1β and GLuc domains, leading to GLuc monomerization, resulting in sensor activation and secretion. (B) GrM GloSensors and iGLuc sensors were produced in vitro using a cell-free transcription/translation system (Protease-Glo Assay and TNT Sp6 High-Yield Wheat Germ Master Mix; Promega) and were subsequently incubated at 37°C for 30 minutes with increasing concentrations of purified GrM or GrM-SA (an inactive GrM mutant in which the catalytic Ser residue has been mutated to Ala) in 50 mM Tris, 150 mM NaCl, pH 7.4. Resulting relative luminescence units (RLUs) were detected with a Veritas Microplate Luminometer (Promega). (C) The GrM and mock GloSensors and iGLuc sensors were labeled with fluorescent green lysines (FluoroTect Green Lys; Promega) during in vitro transcription/translation and incubated at 37°C for 30 minutes with indicated concentrations of GrM or the maximal dose of GrM-SA. Samples were separated on SDS-PAGE and visualized on a Typhoon 9410 scanner (GE Healthcare). Full-length GloSensor (∼61 kDa) and its cleavage fragments (∼36 and 25 kDa) and full-length iGLuc sensor (∼60 kDa) and its largest cleavage fragment (∼40 kDa) are visible. (D) GrM GloSensors and iGLuc sensors were produced as in panel B and incubated at 37°C for 30 minutes with recombinant GrA, GrB, GrH, GrK, or GrM (50 nM). Gr’s were produced in Pichia pastoris, and Gr activity was verified as described previously.6 (E) GrM sensors were cloned into a lentiviral pLV plasmid for transduction of OPM-2 multiple myeloma cells. Sensor expression was verified with immunoblot and flow cytometry (using anti-FLuc and anti-FLAG antibodies). Freeze-thaw lysates (10 µg) of GloSensor- or iGLuc-transduced OPM-2 cells were treated with indicated concentrations of GrM or GrM-SA for 1 hour at 37°C, after which Bright-Glo or Stop&Glo reagent (Promega) was added and luminescence was measured. (F) GloSensor-transduced OPM-2 cells were cocultured with KHYG-1 cells for 2 hours in the presence of 300 µg/mL d-luciferin in increasing effector/target (E:T) ratios. iGLuc sensor-transduced OPM-2 cells were cocultured with KHYG-1 cells in increasing E:T ratios. After 2 hours, 4.4 µM coelenterazine was added, and luminescence was measured. (G) Peripheral blood mononuclear cells were isolated from whole blood using density gradient centrifugation. Sorting by fluorescence-activated cell sorter was then used to isolate CD3− CD56+ primary NK cells. NK cells were stimulated with IL-2 for 20 hours and subsequently cocultured with sensor-expressing HeLa target cells for 3 hours. Sensor activation was detected with LARII (Dual Luciferase kit; Promega) or coelenterazine for the GloSensors and iGLuc sensors, respectively. (H) Peripheral blood mononuclear cells were stimulated with IL-2 for 4 days to generate lymphokine-activated killer (LAK) cells. These were then cocultured with sensor-expressing HeLa cells as described in panel G. (I) Target cell death induced by coculture with LAK cells was measured after 16 hours using annexin V/propidium iodide flow cytometry. Annexin V/propidium iodide double-negative cells were considered viable (with viability set at 100% for untreated cells). (J) iGLuc-transduced OPM-2 cells and KHYG-1 cells were pretreated with 100 µM GrM-inhibitor Ac-KVPL-cmk or dimethyl sulfoxide (DMSO) (vehicle control) for 30 minutes, after which they were cocultured in the presence of Ac-KVPL-cmk or DMSO for 2 hours as in panel F. All data in this figure are depicted as mean ± standard deviation and represent at least 3 independent experiments.
Intracellular profiling of GrM activity by bioluminescent gain-of-function biosensors. (A) Schematic overview of the GrM GloSensor (left) (based on Promega backbone) and the GrM iGLuc sensor (right). The GloSensor circularly permuted firefly luciferase (FLuc) molecule is locked in an inactive conformation by a short peptide linker containing a GrM-specific recognition sequence (AKMPL↓AAEEE). Upon cleavage of the linker by GrM, the FLuc subunits undergo a conformational change, resulting in the formation of active FLuc. The GrM iGLuc sensor is a fusion protein of pro–IL 1β and gaussia luciferase (GLuc). Upon cleavage of the AKMPL↓AAEEE recognition sequence by GrM, the prodomain is separated from the IL-1β and GLuc domains, leading to GLuc monomerization, resulting in sensor activation and secretion. (B) GrM GloSensors and iGLuc sensors were produced in vitro using a cell-free transcription/translation system (Protease-Glo Assay and TNT Sp6 High-Yield Wheat Germ Master Mix; Promega) and were subsequently incubated at 37°C for 30 minutes with increasing concentrations of purified GrM or GrM-SA (an inactive GrM mutant in which the catalytic Ser residue has been mutated to Ala) in 50 mM Tris, 150 mM NaCl, pH 7.4. Resulting relative luminescence units (RLUs) were detected with a Veritas Microplate Luminometer (Promega). (C) The GrM and mock GloSensors and iGLuc sensors were labeled with fluorescent green lysines (FluoroTect Green Lys; Promega) during in vitro transcription/translation and incubated at 37°C for 30 minutes with indicated concentrations of GrM or the maximal dose of GrM-SA. Samples were separated on SDS-PAGE and visualized on a Typhoon 9410 scanner (GE Healthcare). Full-length GloSensor (∼61 kDa) and its cleavage fragments (∼36 and 25 kDa) and full-length iGLuc sensor (∼60 kDa) and its largest cleavage fragment (∼40 kDa) are visible. (D) GrM GloSensors and iGLuc sensors were produced as in panel B and incubated at 37°C for 30 minutes with recombinant GrA, GrB, GrH, GrK, or GrM (50 nM). Gr’s were produced in Pichia pastoris, and Gr activity was verified as described previously.6 (E) GrM sensors were cloned into a lentiviral pLV plasmid for transduction of OPM-2 multiple myeloma cells. Sensor expression was verified with immunoblot and flow cytometry (using anti-FLuc and anti-FLAG antibodies). Freeze-thaw lysates (10 µg) of GloSensor- or iGLuc-transduced OPM-2 cells were treated with indicated concentrations of GrM or GrM-SA for 1 hour at 37°C, after which Bright-Glo or Stop&Glo reagent (Promega) was added and luminescence was measured. (F) GloSensor-transduced OPM-2 cells were cocultured with KHYG-1 cells for 2 hours in the presence of 300 µg/mL d-luciferin in increasing effector/target (E:T) ratios. iGLuc sensor-transduced OPM-2 cells were cocultured with KHYG-1 cells in increasing E:T ratios. After 2 hours, 4.4 µM coelenterazine was added, and luminescence was measured. (G) Peripheral blood mononuclear cells were isolated from whole blood using density gradient centrifugation. Sorting by fluorescence-activated cell sorter was then used to isolate CD3− CD56+ primary NK cells. NK cells were stimulated with IL-2 for 20 hours and subsequently cocultured with sensor-expressing HeLa target cells for 3 hours. Sensor activation was detected with LARII (Dual Luciferase kit; Promega) or coelenterazine for the GloSensors and iGLuc sensors, respectively. (H) Peripheral blood mononuclear cells were stimulated with IL-2 for 4 days to generate lymphokine-activated killer (LAK) cells. These were then cocultured with sensor-expressing HeLa cells as described in panel G. (I) Target cell death induced by coculture with LAK cells was measured after 16 hours using annexin V/propidium iodide flow cytometry. Annexin V/propidium iodide double-negative cells were considered viable (with viability set at 100% for untreated cells). (J) iGLuc-transduced OPM-2 cells and KHYG-1 cells were pretreated with 100 µM GrM-inhibitor Ac-KVPL-cmk or dimethyl sulfoxide (DMSO) (vehicle control) for 30 minutes, after which they were cocultured in the presence of Ac-KVPL-cmk or DMSO for 2 hours as in panel F. All data in this figure are depicted as mean ± standard deviation and represent at least 3 independent experiments.
We previously characterized the extended substrate specificity of human GrM using complementary positional proteomics,4 identifying AKMPL↓AAEEE as an optimal cleavage recognition motif. Based on this sequence, we developed 2 GrM sensors and corresponding mock sensors (P1 Leu replaced by Ala). The GrM GloSensor is based on a circularly permuted FLuc,5 locked in an inactive state by a short peptide linker. Upon GrM-mediated cleavage of the linker, the FLuc becomes active. The GrM iGLuc sensor is a fusion of GLuc and murine pro-IL-1β.3 When GrM cleaves off the IL-1β prodomain, the GLuc monomerizes and becomes active.
We synthesized the GrM and mock sensors in vitro using cell-free transcription/translation and subsequently treated them with purified recombinant GrM or inactive GrM-SA mutant (Figure 1B). Both GrM sensors were activated by GrM in a concentration-dependent manner, but not by GrM-SA. The corresponding mock sensors were not activated, indicating that activation depends on GrM-mediated cleavage after the Leu residue in the peptide recognition sequences. Consistent with these data, visualization of the fluorescently labeled GrM sensors by SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) showed that the full-length Glo (∼61 kDa) and iGluc (∼60 kDa) sensors were cleaved by increasing concentrations of GrM, but not by GrM-SA, resulting in the formation of the expected subunits (∼36/∼25 and ∼40 kDa, respectively) (Figure 1C). To determine the specificity of the GrM sensors, they were incubated with all purified human Gr’s (Figure 1D).6 Of these, only GrM led to robust activation of the GrM sensors, and none of the Gr’s could activate the mock sensors.
We expressed the sensors in OPM-2 multiple myeloma cells and confirmed expression by immunoblot and flow cytometry (data not shown). Recombinant GrM activated the GrM sensors in freeze-thaw lysates of OPM-2 cells in a concentration-dependent manner, whereas GrM-SA did not (Figure 1E). Intracellular delivery of recombinant GrM via pore formation with streptolysin O in tumor cells also led to GrM sensor activation (data not shown).
To determine whether GrM activity can be profiled inside tumor cells that are attacked by cytotoxic lymphocytes, we cocultured GrM sensor-transduced OPM-2 cells with NK cells (KHYG-1), which express high levels of GrM.7 Increasing effector/target ratios resulted in increased activation of both GrM sensors (Figure 1F). Activation of the iGLuc sensor was much stronger, suggesting that the iGLuc sensor may outperform the GloSensor in a more physiological setting. The corresponding mock sensors were not activated. Activation of the sensors occurred within 2 hours, which is compatible with the proposed kinetics of Gr delivery.1,8 Similar results were obtained in HeLa cervix carcinoma cells (supplemental Figure 1, available on the Blood Web site). Primary NK and LAK cells also activated the GrM sensors (Figure 1G-H, respectively). In addition, sensor activation by LAK cells correlated with cell death induction in target cells (Figure 1I). To confirm that GrM sensor activation was attributable to GrM, iGLuc sensor–transduced OPM-2 cells and KHYG-1 NK cells were (pre-)incubated with the cell-permeable GrM-specific inhibitor Ac-KVPL-cmk.9 The tetrapeptide sequence KVPL is specific for GrM and is not recognized by the closely related neutrophil elastase and cathepsin G.10 Treatment with Ac-KVPL-cmk completely inhibited NK cell–mediated activation of the GrM iGLuc sensor (Figure 1J).
Our data nicely complement the recent data of Vrazo and coworkers,1 who developed GrA/K/B biosensors to profile tumor cell death kinetics induced by killer cells. We now report 2 new protease-cleavable biosensors that specifically track GrM proteolytic activity in the course of cytotoxic lymphocyte-induced cell death. This allows further studies to monitor entry and functional activity of all Gr’s in target cells during cancer development, inflammation, and virus infections.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Dr Veit Hornung (Institute of Molecular Medicine, University Hospital, University of Bonn, Bonn, Germany) for providing the basic iGLuc plasmid. This work was supported by grants from the Dutch Cancer Society (UU-2009-4302) (N.B.) and the Dutch Organization for Scientific Research (916.66.044) (N.B.).
Contribution: S.A.H.d.P. participated in the design of the study, performed experiments, analyzed and interpreted the data, and wrote the manuscript; E.A.v.E., J.M., R.B., M.C.O., and E.M.P.S. performed experiments; R.G. participated in the design of the study and interpreted the data; and N.B. participated in the design of the study, analyzed and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Niels Bovenschen, Department of Pathology, University Medical Center Utrecht, Heidelberglaan 100, Utrecht 3584 CX, The Netherlands; e-mail: n.bovenschen@umcutrecht.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal