Key Points
NF-κB family members RelB and cRel are coordinately activated by BAFF and provide distinct survival signals.
In vivo and in vitro B-cell developmental defects are observed when both RelB and cRel are deleted.
Abstract
Targeted deletion of BAFF causes severe deficiency of splenic B cells. BAFF-R is commonly thought to signal to nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB)–inducing kinase dependent noncanonical NF-κB RelB. However, RelB-deficient mice have normal B-cell numbers. Recent studies showed that BAFF also signals to the canonical NF-κB pathway, and we found that both RelB and cRel are persistently activated, suggesting BAFF signaling coordinates both pathways to ensure robust B-cell development. Indeed, we report now that combined loss of these 2 NF-κB family members leads to impaired BAFF-mediated survival and development in vitro. Although single deletion of RelB and cRel was dispensable for normal B-cell development, double knockout mice displayed an early B-cell developmental blockade and decreased mature B cells. Despite disorganized splenic architecture in Relb−/−cRel−/− mice, generation of mixed-mouse chimeras established the developmental phenotype to be B-cell intrinsic. Together, our results indicate that BAFF signals coordinate both RelB and cRel activities to ensure survival during peripheral B-cell maturation.
Introduction
B-cell development originates in the bone marrow, where hematopoietic stem cell precursors commit to the B-cell lineage and immunoglobulin heavy-chain gene rearrangements occur.1,2 If rearrangement is successful, differentiation into the transitional B-cell compartment occurs. Cells that generate functional B-cell antigen receptors eventually leave the bone marrow and migrate to the spleen to complete their maturation process.3,4 The first B cells to arrive are referred to as transitional 1 (T1) B cells.5,6 T1 B cells are still subject to negative selection, where strong antigenic signals lead to apoptosis. In later transitional stages, some of the transitional B cells (transitional 2 [T2]) are allowed to develop into either mature follicular (FO) B cells, which can recirculate in the periphery, or marginal zone (MZ) B cells, which remain largely sessile.7,8
The B-cell activation factor receptor belonging to the tumor necrosis factor (TNF) superfamily (BAFF-R, BR3) provides critical survival signals to all splenic B-cell subsets. Targeted deletion of BAFF ligand or BAFF-R results in a partial block at the T1 to T2 transition, resulting in severe deficiency of mature B cells.9,10 BAFF initiates the noncanonical nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) pathway via TRAF3, resulting in the stabilization of NF-κB–inducing kinase (NIK) and activation of a NF-κB essential modulator (NEMO)-independent IKK1 kinase complex. This mediates p100 processing, and nuclear translocation of RelB:p52 dimers.11 Recent human studies have shown that patients with germ-line mutations in NFKB2 have immunodeficiency. In some of the patients, there is a loss of B cells.12-14 It is likely that some of these B-cell developmental defects in the patients result from impaired BAFF-R signaling because of their nonprocessable p100. BAFF has also been reported to activate the canonical NF-κB pathway.15,16 Gene-targeted deletion of NFkB1 (p50), the primary binding partner of RelA and cRel, results in defective survival of B cells in response to BAFF.17 Although neither cRel−/− nor Rela−/− mice show a phenotype in B-cell numbers, doubly deficient B-cell precursors fail to develop the full mature subsets.18 This raises the question of whether the noncanonical NF-κB pathway and RelB play any role at all in safeguarding B-cell development. However, we note that RelA/cRel-deficiency is also known to diminish RelB expression and noncanonical signaling.19-22 The same considerations apply to interpreting other severe knockouts of the canonical pathway such as B-cell-specific NEMO or IKK2 knockouts.23,24 The fact that the Nfkb1−/−Nfkb2−/− mouse shows a phenotype similar to BAFF/BAFF-R–deficient mice (unlike either single mutant) suggests that both pathways may be redundant.11 However, studies of a compound knockout of the 2 transcriptional activators that mediate canonical and noncanonical pathways, respectively, have not been reported.
Here, we show that only cRel and RelB show persistent activation in response to BAFF, and we therefore examine the physiological consequence of their deletion singly or in combination. We find that both provide survival signals, albeit via distinct gene expression programs, and that these complement each other, such that only the doubly deficient mouse shows severe B-cell developmental deficiencies. Deficiencies in mature B-cell subsets are based not solely on survival defects but also a block in differentiation block at the transitional T1 stage that is cell autonomous and can be observed in an ex vivo differentiation assay.
Materials and methods
Cell isolation and culture
Spleens were harvested from C57BL/6 wild-type, cRel−/−, Relb−/−, Relb−/−cRel−/−, or IκBα−/−IκBε−/−cRel−/−TNF−/− mice. B-cell isolation performed by anti-CD43 (Ly-48) microbeads and separated on LS column (Miltenyi Biotec) as in previous studies.25-28 Purity was confirmed to be between 92% and 95% (data not shown). Marginal Zone and Follicular B Cell Isolation Kit (Miltenyi Biotec) was used for separation of follicular B cells from whole splenocytes, with a purity of 83% to 87% CD23+ follicular B cells (supplemental Figure 1A, available on the Blood Web site). Complete media consisted of RPMI-1640, 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 1 mM sodium pyruvate, 1 mM nonessential amino acids, 0.055 mM β-mercaptoethanol, 100 U penicillin/streptomycin, and 0.3 mg/mL glutamine.
Fluorescence-activated cell sorter (FACS) analysis of survival and B-cell development
Purified B cells were cultured in complete media with or without 50 ng/mL recombinant mouse BAFF/BLyS/TNFSF13B (R&D systems 2106BF). At indicated time points, B cells were collected and stained with 7-aminoactinomycin D (7AAD) (Invitrogen A1310). Cells were analyzed for survival using a C6 Accuri flow cytometer (BD Biosciences). B-cell development was obtained from single-cell suspensions of spleens incubated with fluorescently labeled antibodies for 30 minutes at 4°C in staining buffer (phosphate-buffered saline [PBS] with 0.5% bovine serum albumin or 2.5% fetal calf serum). Data were collected on a FACSCalibur or LSR II flow cytometer (BD Biosciences). FACS analysis was performed using FlowJo software (Tree Star Inc.). Antibodies used in this study from eBioscience (unless specified) included reagents specific for the following: CD21 (7G6), B220 (RA3-6B2), CD23 (B3B4), Ly5.1 (A20), Ly5.2 (RA3-6B2), CD93 (AA4.1), immunoglobulin (Ig) D (11-26), and IgM (115-096-020; Jackson Immunoresearch).
Immunohistochemistry
Spleens from wild-type or Relb−/−cRel−/− mice were frozen in optimum cutting temperature compound above liquid nitrogen and kept at −80°C. Five-micron sections were cut from frozen spleens, fixed in cold 4% paraformaldehyde, and allowed to dry. Sections were then hematoxylin and eosin stained or subjected to immunofluorescent labeling. Sections were blocked with 5% fetal bovine serum in PBS and incubated with anti-Moma1, anti-CD3 fluorescein isothiocyanate, and anti-B220 allophycocyanin (eBioscience) for 3 hours or overnight. Washed sections were mounted with Fluoromount-G (eBioscience), and images were captured on a CRI Nuance Multispectral Imaging (Caliper, Hopkinton, MA) system attached to a Nikon E800 fluorescent microscope.
Bone marrow chimeras
Donor bone marrow cells were isolated from femurs of wild-type, cRel−/−, Relb−/−, or Relb−/−cRel−/− mice, mixed in a 50:50 ratio, and injected (∼5 × 106 cells intravenously per mouse) into 2- to 3-month-old C57BL/6 or C57BL/6 x B6.SJL-Ptprca F1 recipients. Before injection, recipient mice were lethally irradiated (1000 rads). Chimeric mice were analyzed 8 weeks after bone marrow reconstitution. Origin and composition of lymphoid cells was determined by Ly5.1 and Ly5.2 markers.
In vitro B-cell differentiation
In vitro differentiation was performed as previously described.29 Briefly, bone marrow cells were cultured in complete Iscove modified Dulbecco medium with 20 ng/mL interleukin (IL) 7 for 3 to 4 days to enrich for IgM+ transitional B cells. Cells were then washed twice with PBS to remove IL-7 and plated with 50 ng/mL recombinant murine BAFF. On subsequent days, cells were stained with anti-B220, IgM, and IgD antibodies to determine maturation.
Biochemical analysis
Electrophoretic mobility shift assays (EMSAs) were conducted as described previously22,30 and modified to include antibodies specific for RelA (Santa Cruz SC372), RelB (SC226), cRel (SC71), or a combination thereof. For EMSAs focusing on cRel-DNA binding activity, nuclear extracts were preincubated with RelA and RelB antibodies for 15 minutes on ice prior to addition of radiolabeled probe to ablate their specific DNA binding activities. Similarly, nuclear extracts were preincubated with RelA and cRel antibodies or RelB and cRel antibodies when RelB- or RelA-DNA binding activity was the focus, respectively (see Figure 1B). Other antibodies used for immunoblotting were Akt (Cell Signaling Technology, CST#4691), phospho-Akt (Ser473) (CST#4060), nuclear factor of κ light polypeptide gene enhancer in B-cells inhibitor (IκB) α (SC371), IκBε (SC7155), α-tubulin (SC26), NFkB1/p50 and NFkB2/p52 (Dr Nancy Rice), and Bclxl (Abcam, #ab256).
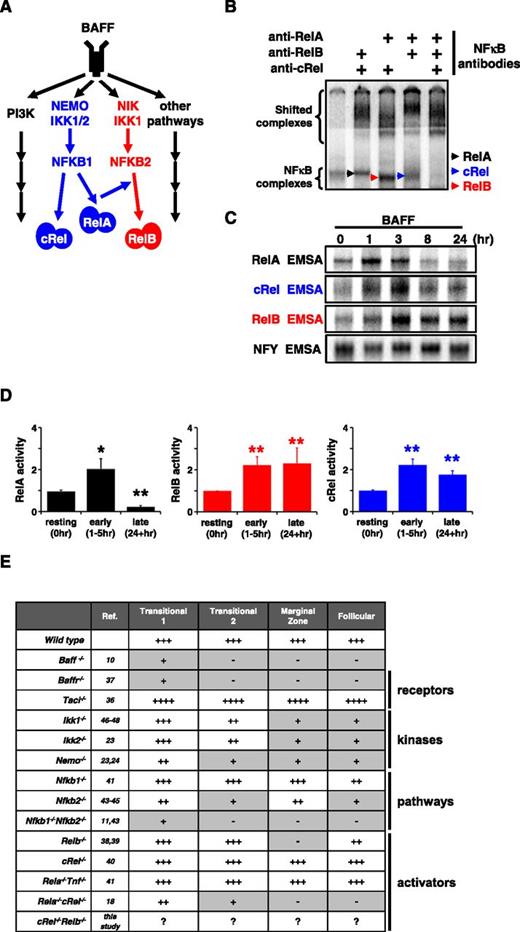
BAFF-R stimulation triggers persistent RelB and cRel activity to control in vitro survival functions. (A) BAFF, which binds TACI and BAFF-R, is a key regulator of peripheral B-cell maintenance. BAFF-R signaling functions occur primarily in an NIK-dependent, noncanonical NF-κB manner, mediated through RelB activity, whereas TACI triggers NEMO-dependent canonical NF-κB. (B) Identification of activated NF-κB species in B cells stimulated with BAFF for 24 hours by supershift analysis. (C) RelA, RelB, and cRel EMSA (see “Materials and methods”) of B cells are used to reveal their respective activity kinetics following BAFF-R perturbation. (D) Quantification of RelA, RelB, and cRel activities of BAFF-stimulated primary B cells derived from EMSA. Naive is resting, unstimulated B cells; early and late activity correspond to activities following 1 to 5 hours and 24+ hours of BAFF stimulation, respectively. Representative of ≥4 experiments. *P < .05; **P < .01. (E) Summary table of known NF-κB–deficient mouse models and the effect on B-cell populations. Gray boxes denote a B-cell developmental defect. +++, cell numbers equate to typical wild-type level; ++++, greater than wild-type; ++, intermediate level; +, low; -, not present. EMSA for panels B-C representative of at least 3 experiments.
BAFF-R stimulation triggers persistent RelB and cRel activity to control in vitro survival functions. (A) BAFF, which binds TACI and BAFF-R, is a key regulator of peripheral B-cell maintenance. BAFF-R signaling functions occur primarily in an NIK-dependent, noncanonical NF-κB manner, mediated through RelB activity, whereas TACI triggers NEMO-dependent canonical NF-κB. (B) Identification of activated NF-κB species in B cells stimulated with BAFF for 24 hours by supershift analysis. (C) RelA, RelB, and cRel EMSA (see “Materials and methods”) of B cells are used to reveal their respective activity kinetics following BAFF-R perturbation. (D) Quantification of RelA, RelB, and cRel activities of BAFF-stimulated primary B cells derived from EMSA. Naive is resting, unstimulated B cells; early and late activity correspond to activities following 1 to 5 hours and 24+ hours of BAFF stimulation, respectively. Representative of ≥4 experiments. *P < .05; **P < .01. (E) Summary table of known NF-κB–deficient mouse models and the effect on B-cell populations. Gray boxes denote a B-cell developmental defect. +++, cell numbers equate to typical wild-type level; ++++, greater than wild-type; ++, intermediate level; +, low; -, not present. EMSA for panels B-C representative of at least 3 experiments.
Transcriptome analysis
Total RNA was extracted from 50 ng/mL BAFF-stimulated wild-type B cells. Messenger RNA was isolated from 2 μg total RNA using oligo (dT) magnetic beads and fragmented at high temperature using divalent cations. Complementary DNA libraries were generated using the Illumina Truseq kits, and quantitation was performed using the Roche Light Cycler 480. Sequencing was performed on Illumina's HiSequation 2000, according to the manufacturer’s recommendations and prepared for RNA-sequencing (RNA-seq) analysis by the Broad Stem Cell Research Center core facility at the University of California, Los Angeles. Reads were aligned to the mouse mm10 genome and RefSeq genes31,32 with Tophat.33 Cufflinks CummRbund34 was used to ascertain differential expression of genes. Gene differential FPKMs were obtained from the cuffdiff program in the Tuxedo RNA-seq analysis suite. RNA-seq data are deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database (accession number GSE62559).
Results
BAFF induces persistent NF-κB RelB and cRel activities that have complementary roles in mediating B-cell survival
BAFF is known to engage 3 BLyS family receptors, namely, B-cell maturation antigen (BCMA), transmembrane activator and calcium-modulating cyclophilin ligand-interactor protein (TACI), and BAFF-R (also known as BR3 or BLYS receptor),35 and to activate several signaling pathways including the canonical and noncanonical NF-κB pathways, whose transcriptional effectors are RelA/cRel and RelB, respectively (Figure 1A).
We examined which NF-κB transcription factors are activated by BAFF in murine splenic B cells. We used a modified EMSA procedure in which antibodies are used to ablate 2 of 3 activation-domain-containing NF-κB family members (Figure 1B). Stimulation with BAFF ligand resulted in activation of RelA-, cRel-, and RelB-containing dimers, but the time courses reveal that only cRel and RelB show persistent activation: RelA activity peaks at 1 hour and returns to basal levels within 8 hours. In contrast, cRel and RelB activities are not short lived, peaking at ∼3 hours and persisting beyond 24 hours (Figure 1C). Repeated experimental analysis showed that these conclusions were statistically significant (Figure 1D).
Numerous studies have investigated which signaling pathways mediate BAFF’s physiological functions, and these genetic knockout studies are summarized in Figure 1E. Of the 3 known BAFF-binding receptors, expression of BCMA is not observed in naive B-cell populations, and TACI deletion actually results in enhanced B-cell numbers and splenomegaly, whereas BAFF-R deletion phenocopies BAFF deletion in resulting in a near loss of peripheral B cells.36 BAFF activates both canonical and noncanonical signaling branches of the NF-κB system, yet deficiencies in only canonical components (eg, NEMO, RelA/cRel) have been reported to result in defective B-cell development (Figure 1E).37-48 However, the canonical pathway, and RelA in particular, is known to enable noncanonical signaling via NFkB2 and Relb expression20,21 (Figure 1A), suggesting that phenotypes observed in severe canonical deficiencies might be mediated by defects in noncanonical signaling.
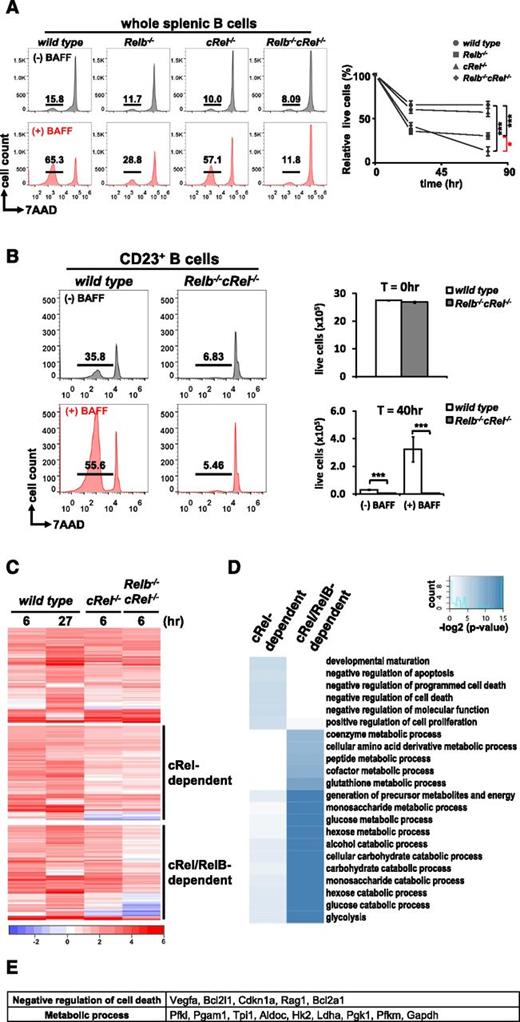
As both RelB and cRel dimers are activated persistently by BAFF, we focused on examining their roles in mediating its key function, survival. Whole splenic B cells were isolated from Relb−/−, cRel−/−, and Relb−/−cRel−/− mice and stimulated with BAFF for up to 4 days. In the in vitro survival assay, RelB-deficient B cells respond poorly to 70 hours of BAFF stimulation, and cRel deletion also displays a minor defect in the BAFF response; however, compound deletion of both cRel and RelB resulted in a statistically more severe phenotype (Figure 2A) than single deletion compared with wild-type. We observe a similarly severe survival defect in a homogeneous population of FO (CD23+) B cells isolated from the spleen of wild-type and Relb−/−cRel−/− mice (Figure 2B). These data suggest that BAFF signaling coordinates the functions of both canonical and noncanonical NF-κB transcription factors to promote survival.
RelB and cRel coordinate together to provide proper BAFF-mediated survival signals in vitro. (A) FACS plots of in vitro survival assay of whole splenic wild-type, Relb−/−, cRel−/−, and Relb−/−cRel−/− B cells stimulated with BAFF ligand for 70 hours. Numbers represent the percentage of live cells (7AAD−) found in culture. Graphical representation of the FACS plots (right), n = 3. (B) FACS plots of in vitro survival assay of FO (CD23+) wild-type and Relb−/−cRel−/− B cells stimulated with BAFF for 40 hours. (C) RNA-seq analysis from BAFF-stimulated CD23+wild-type, cRel−/−, and Relb−/−cRel−/− B cells at the indicated time points. Genes (517) were upregulated in BAFF-stimulated follicular B cells; 289 of these showed substantial expression defect in B cells lacking both cRel and RelB (middle and bottom panels). Of these, 127 showed expression defects even in the single cRel knockout (middle panel); 162 showed expression defects only in the Relb−/−cRel−/− double knockout (bottom panel). (D) cRel-dependent genes protect cells against cell death. Gene ontology analysis identifies distinct process terms for cRel-dependent vs RelB/cRel-dependent gene clusters. Whereas RelB/cRel-dependent clusters are significantly associated with terms describing metabolic processes, the cRel-dependent cluster shows overrepresentation of negative regulation of cell death/apoptosis. *P < .05; **P < .005; ***P < .001. Also see supplemental Figure 1. (E) List of representative cRel-dependent and RelB/cRel-dependent genes identified as “negative regulators of cell death” and “metabolic process,” respectively, by gene ontology analysis.
RelB and cRel coordinate together to provide proper BAFF-mediated survival signals in vitro. (A) FACS plots of in vitro survival assay of whole splenic wild-type, Relb−/−, cRel−/−, and Relb−/−cRel−/− B cells stimulated with BAFF ligand for 70 hours. Numbers represent the percentage of live cells (7AAD−) found in culture. Graphical representation of the FACS plots (right), n = 3. (B) FACS plots of in vitro survival assay of FO (CD23+) wild-type and Relb−/−cRel−/− B cells stimulated with BAFF for 40 hours. (C) RNA-seq analysis from BAFF-stimulated CD23+wild-type, cRel−/−, and Relb−/−cRel−/− B cells at the indicated time points. Genes (517) were upregulated in BAFF-stimulated follicular B cells; 289 of these showed substantial expression defect in B cells lacking both cRel and RelB (middle and bottom panels). Of these, 127 showed expression defects even in the single cRel knockout (middle panel); 162 showed expression defects only in the Relb−/−cRel−/− double knockout (bottom panel). (D) cRel-dependent genes protect cells against cell death. Gene ontology analysis identifies distinct process terms for cRel-dependent vs RelB/cRel-dependent gene clusters. Whereas RelB/cRel-dependent clusters are significantly associated with terms describing metabolic processes, the cRel-dependent cluster shows overrepresentation of negative regulation of cell death/apoptosis. *P < .05; **P < .005; ***P < .001. Also see supplemental Figure 1. (E) List of representative cRel-dependent and RelB/cRel-dependent genes identified as “negative regulators of cell death” and “metabolic process,” respectively, by gene ontology analysis.
Next we asked whether the cRel and RelB mediated their complementary functions by inducing the same or different sets of survival genes in B cells. We undertook RNA-seq of BAFF-stimulated FO (CD23+) wild-type, cRel−/−, and Relb−/−cRel−/− B cells. Here, we identified 517 genes upregulated in BAFF-stimulated wild-type B cells, 289 genes of which were substantially reduced in Relb−/−cRel−/− B cells (Figure 2C). Of these, 127 genes were largely cRel dependent, as they showed lower BAFF-induced expression in both cRel−/− or Relb−/−cRel−/− B cells, and 162 were identified as RelB dependent in the context of cRel deficiency and may thus be described as potential RelB target genes. Interestingly, gene ontology analysis focusing on biological process terms identified a large number of receptor-associated and mitochondrial antiapoptotic regulators within the cRel-dependent cluster, whereas metabolic regulators were primarily identified in the RelB/cRel-dependent cluster (Figure 2D-E). These data suggest that coordination of cRel and RelB activities by BAFF serves the expression of complementary sets of genes that ensure B-cell survival. Confirmatory reverse transcription–quantitative polymerase chain reaction showed that Bcl2 family members A1 and Bcl-xL required only cRel but not RelB for their expression, and the reduction is messenger RNA expression also resulted in a reduction in Bcl-xL protein (supplemental Figure 2).
Compound deletion of RelB and cRel results in severe peripheral B-cell developmental defects
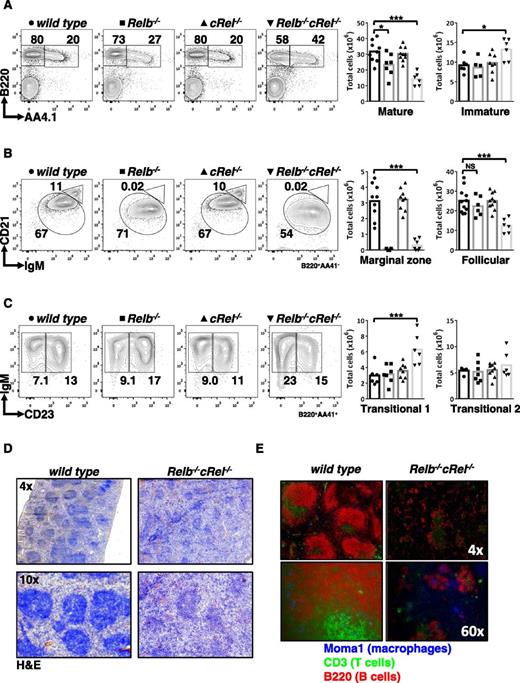
As RelB and cRel are critical for BAFF-mediated survival functions in vitro, we examined B-cell development in vivo in mice lacking RelB, cRel, or both. Using an established FACS strategy,49 we confirmed that there is no defect in peripheral B-cell populations in cRel−/− mice and only a modest decrease in Relb−/− mice. However, we found a significant loss of mature B cells and an increase in the transitional compartment in double knockout mice (Figure 3A), whereas T-cell numbers were normal (supplemental Figure 3). Distinguishing mature B-cell subsets, we confirmed previous reports that Relb−/− spleens were deficient in the MZ B-cell population38 but now found that the double knockout mice generate significantly fewer FO B cells (Figure 3B; Table 1).
Compound deletion of RelB and cRel results in developmental block at the T1 stage. (A-C) FACS analysis of peripheral B-cell development in whole splenic extracts from wild-type, Relb−/−, cRel−/−, and Relb−/−cRel−/− mice. Identification of mature (B220+AA4.1−) and transitional B cells (B220+AA4.1+). Mature B cells (B220+AA4.1−) are further classified into FO B cells (CD21+IgM+) and mature MZ B cells (CD21highIgM+). Transitional B cells (B220+AA4.1+) are subdivided into T1 B cells (CD23−IgM+) and T2 B cells (CD23+IgM+). Scatter plots are graphical representation of FACS plots: wild-type (●), Relb−/− (▪), cRel−/− (▲), and Relb−/−cRel−/− (▼). (D) Histologic analysis of splenic sections taken from wild-type and Relb−/−cRel−/− mouse using hematoxylin and eosin stain (H&E). (E) In vivo analysis of mature B-cell development in double knockout mouse using immunofluorescence of frozen splenic sections stained with anti-CD3 fluorescein isothiocyanate, anti-B220 allophycocyanin, and anti-MOMA-1 Alexa Fluor 405. *P < .05; **P < .005; ***P < .001. Also see supplemental Figure 2.
Compound deletion of RelB and cRel results in developmental block at the T1 stage. (A-C) FACS analysis of peripheral B-cell development in whole splenic extracts from wild-type, Relb−/−, cRel−/−, and Relb−/−cRel−/− mice. Identification of mature (B220+AA4.1−) and transitional B cells (B220+AA4.1+). Mature B cells (B220+AA4.1−) are further classified into FO B cells (CD21+IgM+) and mature MZ B cells (CD21highIgM+). Transitional B cells (B220+AA4.1+) are subdivided into T1 B cells (CD23−IgM+) and T2 B cells (CD23+IgM+). Scatter plots are graphical representation of FACS plots: wild-type (●), Relb−/− (▪), cRel−/− (▲), and Relb−/−cRel−/− (▼). (D) Histologic analysis of splenic sections taken from wild-type and Relb−/−cRel−/− mouse using hematoxylin and eosin stain (H&E). (E) In vivo analysis of mature B-cell development in double knockout mouse using immunofluorescence of frozen splenic sections stained with anti-CD3 fluorescein isothiocyanate, anti-B220 allophycocyanin, and anti-MOMA-1 Alexa Fluor 405. *P < .05; **P < .005; ***P < .001. Also see supplemental Figure 2.
Summary of peripheral B-cell development in wild-type, Relb−/−, cRel−/−, and Relb−/−cRel−/− mice
| Phenotype . | Total B cells (×106) . | |||
|---|---|---|---|---|
| wild-type . | Relb−/− . | cRel−/− . | Relb−/−cRel−/− . | |
| Total cells | 75.4 ± 17 | 131 ± 15** | 76.1 ± 8.3 | 104 ± 25* |
| Total B cells | 51.3 ± 6.9 | 39.8 ± 8.6* | 41.6 ± 4.9 | 29.1 ± 7.8*** |
| Mature | 32.8 ± 6.7 | 23.3 ± 7.1* | 30.1 ± 4.4 | 14.3 ± 4.2* |
| Transitional | 9.28 ± 1.9 | 7.85 ± 1.7 | 9.86 ± 2.5 | 12.7 ± 2.7* |
| T1 | 3.00 ± 1.2 | 3.17 ± 1.2 | 3.58 ± 0.94 | 6.30 ± 2.0** |
| T2 | 5.42 ± 0.64 | 5.21 ± 2.0 | 5.54 ± 1.3 | 6.48 ± 2.5 |
| MZ | 3.1 ± 1.1 | 0.067 ± 0.052**** | 3.2 ± 0.071 | 0.31 ± 0.34**** |
| Follicular | 25.3 ± 6.3 | 22.4 ± 5.1 | 24.1 ± 4.4 | 12.5 ± 3.6*** |
| Phenotype . | Total B cells (×106) . | |||
|---|---|---|---|---|
| wild-type . | Relb−/− . | cRel−/− . | Relb−/−cRel−/− . | |
| Total cells | 75.4 ± 17 | 131 ± 15** | 76.1 ± 8.3 | 104 ± 25* |
| Total B cells | 51.3 ± 6.9 | 39.8 ± 8.6* | 41.6 ± 4.9 | 29.1 ± 7.8*** |
| Mature | 32.8 ± 6.7 | 23.3 ± 7.1* | 30.1 ± 4.4 | 14.3 ± 4.2* |
| Transitional | 9.28 ± 1.9 | 7.85 ± 1.7 | 9.86 ± 2.5 | 12.7 ± 2.7* |
| T1 | 3.00 ± 1.2 | 3.17 ± 1.2 | 3.58 ± 0.94 | 6.30 ± 2.0** |
| T2 | 5.42 ± 0.64 | 5.21 ± 2.0 | 5.54 ± 1.3 | 6.48 ± 2.5 |
| MZ | 3.1 ± 1.1 | 0.067 ± 0.052**** | 3.2 ± 0.071 | 0.31 ± 0.34**** |
| Follicular | 25.3 ± 6.3 | 22.4 ± 5.1 | 24.1 ± 4.4 | 12.5 ± 3.6*** |
****P < .0001, ***P < .001, **P < .01, *P < .05.
Dissection of transitional B-cell populations revealed an increased proportion of transitional T1 but not T2 cells (Figure 3C). T1 B cells are the initial class of B cells that give rise to the mature populations.50 In addition, we examined the B-cell maturation in the bone marrow of the double knockout and found no discernible defect compared with wild-type controls (supplemental Figure 4). The increase in T1 B cells found in the double knockout mouse suggested to us that RelB and cRel are required not only for the survival of developing B cells but also for peripheral B-cell development per se. Although the Relb−/−cRel−/− developmental phenotype is not as severe as that caused by BAFF deficiency, the block at the T1 stage is reminiscent.37 Further, histologic analysis corroborated these findings: in the Relb−/−cRel−/− spleen, the white pulp, although present, was not as dense (Figure 3D). The reduction in white pulp may be caused by reduced numbers of B cells in the spleen, as our FACS analysis suggested, or it may be a consequence of improper homing of the lymphocytes causing them to be dispersed throughout the spleen. To address this, we investigated the organization of B cells using immunofluorescence. Frozen sections of splenic tissue were stained with anti-CD3 and anti-B220 to identify the T-cell zone and B-cell zone, respectively. MOMA-1 (which stains MZ metallophilic macrophages) outlined the follicular rim surrounding the B-cell–rich zone. In agreement with our FACS data, the presence of B cells was greatly diminished in the Relb−/−cRel−/− spleen, as shown by the staining of B220 (red; Figure 3E). We also observed that the characteristic ringlike structure of MOMA-1 staining (blue) is consistently less organized in the double knockout mouse, suggestive of a defective splenic architecture, which may51,52 or may not contribute to the B-cell development phenotype in the mutant mice.
B-cell intrinsic developmental defect in vivo
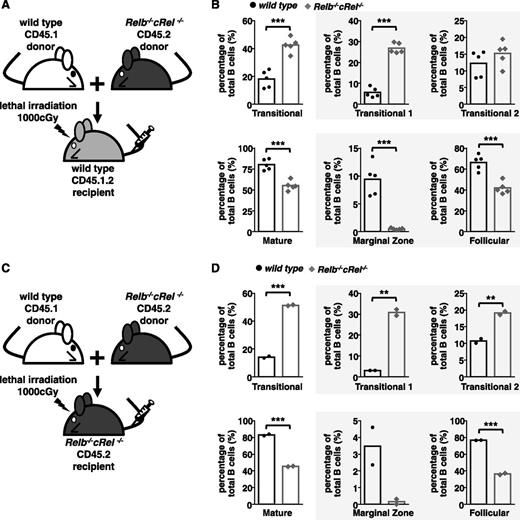
We studied whether the B-cell developmental phenotype is in fact B-cell intrinsic by generating mixed bone marrow chimeras, where bone marrow–derived hematopoietic stem cells from both wild-type and mutant donor mice were injected into a lethally irradiated wild-type or mutant recipient mice. To distinguish cells derived from either donor, we used the CD45 congenic marker system, in which donor cells expressed the CD45.1 or CD45.2 variant and recipient mice were CD45.1.2 (supplemental Figure 5A-B).
Within this experimental system, we found that Relb−/−cRel−/− (CD45.2) hematopoietic stem cells were compromised in their development even when wild-type recipients provided a normal milieu of architecture and soluble factors (Figure 4A-B) akin to observations made in cRel- and RelB-deficient animals (Figure 3A-C). Again, the transitional B-cell populations are higher than the wild-type controls, whereas FO populations were severely decreased and MZ B cells were undetectable (Figure 4A-B). As a reference, we also generated chimeras with RelB and cRel single knockout bone marrow cells, with no observable phenotype (supplemental Figure 5C-D). When wild-type and Relb−/−cRel−/− donor bone marrow stem cells were injected into lethally irradiated Relb−/−cRel−/− recipient mice, wild-type donor stem cells were able to mature normally from T1 to T2, FO, and MZ B cells, whereas Relb−/−cRel−/− donor stem cells matured poorly (similar to when they were in wild-type host) (Figure 4C-D). These data provide strong evidence that the B-cell developmental phenotype of Relb−/−cRel−/− mice is in fact cell intrinsic, and not caused by improper splenic architecture or cytokine milieu.
B-cell developmental defect displayed in Relb−/−cRel−/− mouse is B-cell intrinsic. (A and C) Schematic for the generation of CD45.1 wild-type and CD45.2 Relb−/−cRel−/− mixed bone marrow chimeras. Bone marrow stem cells from CD45.1 wild-type and CD45.2 Relb−/−cRel−/− are mixed in a 50:50 ratio and then injected into a lethally irradiated CD45.1.2 wild-type or Relb−/−cRel−/− mouse. (B and D) Graphical plots of percentages of specific subsets relative to total B cells in indicated mixed bone marrow chimeras, as described previously. The gating strategy used in the analysis is depicted in Figure 3. Representative FACS plots see supplemental Figure 3. For panel B, n = 5; for panel D, n = 2. *P < .05; **P < .005; ***P < .001.
B-cell developmental defect displayed in Relb−/−cRel−/− mouse is B-cell intrinsic. (A and C) Schematic for the generation of CD45.1 wild-type and CD45.2 Relb−/−cRel−/− mixed bone marrow chimeras. Bone marrow stem cells from CD45.1 wild-type and CD45.2 Relb−/−cRel−/− are mixed in a 50:50 ratio and then injected into a lethally irradiated CD45.1.2 wild-type or Relb−/−cRel−/− mouse. (B and D) Graphical plots of percentages of specific subsets relative to total B cells in indicated mixed bone marrow chimeras, as described previously. The gating strategy used in the analysis is depicted in Figure 3. Representative FACS plots see supplemental Figure 3. For panel B, n = 5; for panel D, n = 2. *P < .05; **P < .005; ***P < .001.
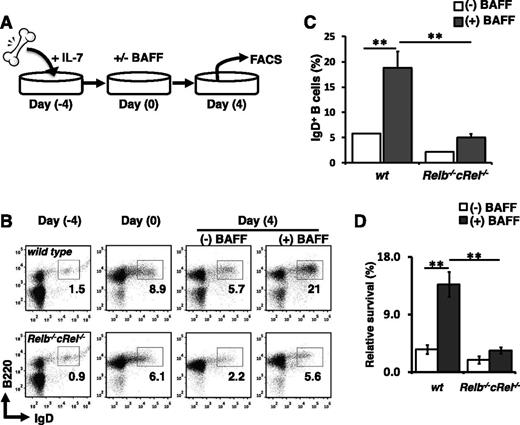
Relb−/−cRel−/− progenitors are defective in BAFF-mediated B-cell development in vitro
A true cell intrinsic phenotype is expected to be recapitulated in an in vitro differentiation assay in which all nonintrinsic components are held equal. We employed an in vitro B-cell development assay system described previously.11,53 In this system, transitional B cells are generated from bone marrow progenitors in the presence of IL-7. The cytokine is washed away, then recultured with BAFF ligand (Figure 5A). Expression of B-cell markers CD23 and CD21 has been reported to be regulated by BAFF.54 Further, FACS analysis suggested that RelB plays a role in CD21 expression (Figure 3B); thus we used IgD and B220 as our primary developmental markers in these analyses. Exposure of wild-type bone marrow cultures to BAFF for 4 days increased the fraction of IgD+ cells relative to untreated controls, demonstrating BAFF’s capacity to enhance B-cell development in vitro. In contrast, BAFF stimulation failed to increase the percentage of IgD+ cells present in Relb−/−cRel−/− bone marrow cultures (Figure 5B-C). Even in the absence of BAFF, the numbers of IgD+ cells were consistently lower in mutant cultures compared with wild-type. In fact, the reduction in B-cell numbers in this assay may be dominated by a BAFF-dependent survival defect (Figure 5D). Together, these experimental results further illustrate the necessity of RelB and cRel in BAFF-mediated B-cell maturation.
Relb−/−cRel−/− B-cell progenitors fail to respond to BAFF-mediated developmental signal in vitro. (A) Schematic of in vitro B-cell differentiation system. (B) Representative FACS plots of B220 and IgD expression in transitional B cells at the beginning (day 0) and end (day 4) of culture with or without BAFF. (C) Mean frequency of B220+IgD+ (T2-like) B cells following 4 days of culture as described in panel B. Numbers represent frequencies of live B220+ B cells in indicated gates. (D) Survival of in vitro cultured B cells. Live cells were identified by gating out 7AADHi population. Fold survival calculated from initial B-cell population (day 0). For FACS plots, n = 4. wt, wild-type.
Relb−/−cRel−/− B-cell progenitors fail to respond to BAFF-mediated developmental signal in vitro. (A) Schematic of in vitro B-cell differentiation system. (B) Representative FACS plots of B220 and IgD expression in transitional B cells at the beginning (day 0) and end (day 4) of culture with or without BAFF. (C) Mean frequency of B220+IgD+ (T2-like) B cells following 4 days of culture as described in panel B. Numbers represent frequencies of live B220+ B cells in indicated gates. (D) Survival of in vitro cultured B cells. Live cells were identified by gating out 7AADHi population. Fold survival calculated from initial B-cell population (day 0). For FACS plots, n = 4. wt, wild-type.
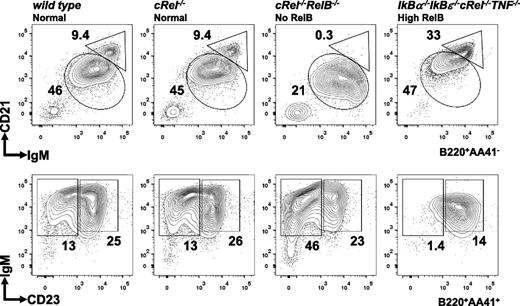
Deregulating NF-κB accelerates splenic B-cell development
Given that NF-κB RelB/cRel activities are required for B-cell maturation past the T1 stage (Figure 3), we asked whether elevated NF-κB might be sufficient to accelerate B-cell maturation. Compound deficiency of IκBα and IκBε leads to elevation of both constitutive and stimulus-responsive NF-κB activity,55 inflammatory gene expression, and neonatal death.56 We restored viability by introducing compound cRel and TNF deficiencies, leaving RelA and RelB deregulated, and examined the B-cell compartment. Although the total splenocytes count is lower in IκBα−/−IκBε−/−cRel−/−TNF−/− mice than wild-type controls, the proportion of B220+ cells is relatively normal (∼40% to 50%). Interestingly, in contrast to hypo-NF-κB activity in Relb−/−cRel−/− mice, hyper-NF-κB activity in IκBα−/−IκBε−/−cRel−/−TNF−/− mice leads to reduced T1 cell numbers and a preponderance of MZ B cells (Figure 6). Although the reduced B-cell numbers also indicate restrictions in bone marrow developmental events, these observations are indicative of accelerated splenic B-cell development.
Deregulated NF-κB activity in IκBα−/−IκBε−/−cRel−/−TNF−/− leads to reduction in T1 cells and increased MZ B-cell generation. FACS analysis of peripheral B-cell development in whole splenic extracts from wild-type, cRel−/−, Relb−/−cRel−/−, and IκBα−/−IκBε-/cRel−/−TNF−/− mice. The gating strategy used in the analysis is depicted in Figure 3. Numbers represent percentage of denoted subsets as proportion of total B cells. Representative FACS results of 3 experiments shown.
Deregulated NF-κB activity in IκBα−/−IκBε−/−cRel−/−TNF−/− leads to reduction in T1 cells and increased MZ B-cell generation. FACS analysis of peripheral B-cell development in whole splenic extracts from wild-type, cRel−/−, Relb−/−cRel−/−, and IκBα−/−IκBε-/cRel−/−TNF−/− mice. The gating strategy used in the analysis is depicted in Figure 3. Numbers represent percentage of denoted subsets as proportion of total B cells. Representative FACS results of 3 experiments shown.
Discussion
For more than a decade, numerous reports have focused on the characterization of noncanonical NF-κB signaling in response to the BAFF signaling pathway,17,24,57 yet only mutants of the canonical pathway have BAFF−/−-like phenotypes. In this study, we report that the canonical NF-κB effectors cRel and RelA are activated following BAFF stimulation. Further, although RelA activation is transient, cRel activation is persistent and thus mimics the activation profile of RelB, the well-known effector of the noncanonical pathway.
Indeed, both cRel and RelB play a role in BAFF functions ex vivo, as either singly deficient B cell showed reduced BAFF-responsive survival. Remarkably, however, these survival functions appear to be mediated by different gene expression programs, with cRel being required for the expression of known prosurvival genes A1 and Bcl-xL,16,58 but Relb−/− B-cells showing no defects in their expression. Whereas unbiased transcriptome analysis of RelB-deficient B cells is compromised by the confounding secondary effects of systemic inflammation in the Relb−/− mouse,39 Relb−/−cRel−/− mice, which do not show an inflammatory phenotype, allowed us to identify the RelB-dependent gene expression program in the context of cRel deficiency. The complementary nature of the cRel- and RelB-dependent expression programs manifests itself in the fact that double deletion leads to a more severe B-cell survival defect not only ex vivo (Figure 2) but also in vivo (Figure 3).
Further, we found that double deletion of RelB and cRel results in a block in splenic B-cell development at the T1 to T2 stage. These mutant mice are not only deficient in mature cells but possess a twofold increase in the percentage of transitional B cells and almost fourfold enhancement in the population of T1 cells. This developmental blockade at the T1 to T2 stage mimics the phenotype of BAFF knockout mice, although the defect is not quite as severe, suggesting a role for RelA, or an NF-κB-independent mechanism. This developmental defect is B-cell intrinsic, as Relb−/−cRel−/− bone marrow stem cells failed to develop normally in mixed bone marrow chimeras and wild-type hematopoietic stem cells are capable of proper B-cell development in lethally irradiated Relb−/−cRel−/− mice.
How does BAFF coordinately activate cRel and RelB? Three TNF receptor superfamily members bind BAFF, namely, BCMA, TACI, and BAFF-R (also known as BR3 or BLYS receptor),51 with BAFF-R and TACI being expressed on peripheral B-cell subsets.59 TACI activates canonical NF-κB signaling,60 but TACI-deficient mice show no defect on B-cell development and B cells are capable of degrading IκBα in response to BAFF.15 Our results are consistent with BAFF-R triggering both canonical, IκBα-mediated NF-κB signaling and also noncanonical, IκBδ-mediated activation of RelA and cRel. As a key survival receptor, BAFF-R signaling maintains the B-cell pool. In contrast, TACI acts as a negative regulator of B-cell homeostasis, indirectly regulating peripheral B-cell numbers by competing for the amount of BAFF available for signaling through BAFF-R.51,61
CD21 expression is reduced in the absence of RelB, and further decreased in the combined absence of RelB and cRel (Figure 3B). CD21 is known to be controlled by NF-κB-p52.62,63 Considering our data, CD21 is probably controlled by RelB:p52 complex during development, and cRel:p52 to a lesser extent (possibly providing compensation). Given that CD21 is essential in humoral immune response,64,65 it is likely RelB-deficient B cells will have impaired response to certain T-dependent antigens, especially complement-tagged antigens.66-68 Further functional studies are needed to address this.
Previous studies suggested that NF-κB regulates BAFF-R expression,69,70 although none of the NF-κB knockout studies cited in Figure 1E had examined BAFF-R expression. We examined BAFF-R expression in 2 ways: replicate RNA-seq did not show an expression defect in Relb−/−cRel−/− cells over wild-type controls (supplemental Figure 6A), thereby not corroborating transcriptional control by RelB/cRel. However, by FACS, BAFF-R staining was reduced (supplemental Figure 6B) and BAFF-responsive AKT phosphorylation (indicative of phosphatidylinositol 3-kinase activation) was also lower (supplemental Figure 6C). Together, this suggests that the phenotype of Relb−/−cRel−/− B cells may in part be reinforced by a reduction in BAFF-R surface expression. As BAFF-R is transcribed at the normal rate, this reduction is not a direct consequence of RelB/cRel deficiency, but probably rather a consequence of cell-biological deficiencies that hinder B-cell maturation from the T1 stage. Further, although the loss of cRel and RelB reduces the amount of p100, as observed elsewhere,71 BAFF-responsive noncanonical NFkB2/p100 degradation remained intact (supplemental Figure 6C).
The coordinate activation of cRel and RelB renders BAFF’s physiological responses relatively robust, as the single knockout shows little in vivo defect. At the same time, cell-context-specific control of the 2 branches of NF-κB signaling and activation of their respective gene expression programs may allow for fine tuning of BAFF’s broad physiological functions in controlling survival, B-cell maturation, cell growth, and even proliferation. Matching quantitative studies of signaling strength and dynamics to gene expression programs and cell biological level analyses may lead to a better understanding how BAFF functions in vivo and enable its pharmacologic manipulation in the clinical setting.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE62559).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank L. Delacruz for irradiating donor mice and University of California, San Diego histology core at Moores Cancer Center for sectioning of frozen spleens for immunofluorescence. The authors also thank V. Shih and S. Basak for experimental advice and S. Hedrick, C. Murre, and R. Rickert for critical discussions.
This work was supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (R01 AI083453), National Cancer Institute (R01 CA141722), and National Institute of General Medical Sciences (R01 GM071573, P50 GM085764) (A.H.); and a Cell and Molecular Genetics training grant (J.V.A.).
Authorship
Contribution: J.V.A., Y.C.L., A.H., M.A., M.D., and A.W.G. designed the research and interpreted data; J.V.A. and Y.C.L. performed all B-cell biochemistry; J.V.A., D.C.O., and M.D. performed immunofluorescence; J.V.A. and Y.C.L. performed in vitro B-cell development; J.V.A., H.B., and J.D.-T. carried out RNA-sequencing analysis; J.V.A., E.Y., M.A., and A.W.G. designed and made bone marrow chimeras; and J.V.A., Y.C.L., and A.H. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexander Hoffmann, University of California, Los Angeles, Los Angeles, CA 90095; e-mail: ahoffmann@ucla.edu.
References
Author notes
J.V.A. and Y.C.L. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal