Key Points
Rac1 and Cdc42 possess nonredundant roles in preventing apoptosis of NPM-ALK lymphoma cells.
Simultaneous deletions of both Rac1 and Cdc42 prevents NPM-ALK lymphoma dissemination in vivo.
Abstract
Increasing evidence suggests that Rho family GTPases could have a critical role in the biology of T-cell lymphoma. In ALK-rearranged anaplastic large cell lymphoma (ALCL), a specific subtype of T-cell lymphoma, the Rho family GTPases Cdc42 and Rac1 are activated by the ALK oncogenic activity. In vitro studies have shown that Cdc42 and Rac1 control rather similar phenotypes of ALCL biology such as the proliferation, survival, and migration of lymphoma cells. However, their role and possible redundancy in ALK-driven lymphoma development in vivo are still undetermined. We genetically deleted Cdc42 or Rac1 in a mouse model of ALK-rearranged ALCL to show that either Cdc42 or Rac1 deletion impaired lymphoma development, modified lymphoma morphology, actin filament distribution, and migration properties of lymphoma cells. Cdc42 or Rac1 deletion primarily affected survival rather than proliferation of lymphoma cells. Apoptosis of lymphoma cells was equally induced following Cdc42 or Rac1 deletion, was associated with upregulation of the proapoptotic molecule Bid, and was blocked by Bcl2 overexpression. Remarkably, Cdc42/Rac1 double deletion, but not Cdc42 or Rac1 single deletions, completely prevented NPM-ALK lymphoma dissemination in vivo. Thus, Cdc42 and Rac1 have nonredundant roles in controlling ALK-rearranged lymphoma survival and morphology but are redundant for lymphoma dissemination, suggesting that targeting both GTPases could represent a preferable therapeutic option for ALCL treatment.
Introduction
The Rho GTPases family members Cdc42 and Rac1 are thought to act as oncogenes in several cancer types by regulating proliferation, survival, migration and invasion.1-4 Although initial studies suggested that GTPases could act as oncogenes in cancer, recent studies unveiled a tumor suppressor rather than oncogenic function for Cdc42 in some tumors.1 In lymphoma and leukemia, Rho GTPases are more commonly activated by indirect mechanisms, such as increased Rho guanine nucleotide exchange factor (GEF) and/or decreased Rho GTPase-activating protein (GAP) activity,4,5 or inactivated by mutations in T-cell lymphoma.6-8 ALK-rearranged anaplastic large cell lymphoma (ALCL) is a subtype of T-cell lymphoma where the oncogenic translocation t(2;5) generates the constitutively active tyrosine kinase NPM-ALK.9,10 NPM-ALK increases the activity of Cdc42 and Rac1 by activating the RhoGEFs Vav1 and Vav3, respectively,11,12 thus suggesting that Cdc42 and Rac1 could act as oncogenes in ALCL. Indeed, previous in vitro studies in ALCL demonstrated that Cdc42 is essential for cell proliferation and survival,9 whereas Rac1 is implicated in cell migration of ALK-transformed cells.10 In accordance with these roles, the blockade of Cdc42 activity by short hairpin RNA knockdown or by pharmacologycal inhibition with secramine induced cell cycle arrest and apoptosis of ALCL cells,9 whereas Rac1 inhibition by the NSC23766 inhibitor abrogated NPM-ALK–elicited disease progression and metastasis in mice.13
Despite this evidence, however, the specific roles of Cdc42 and Rac1 in ALK-rearranged lymphoma development and dissemination in vivo have never been investigated at the genetic level. In the present work, we genetically ablated Cdc42 or Rac1 in a mouse model of NPM-ALK-driven lymphoma. By this approach, we show that Cdc42 or Rac1 are equally essential for ALCL development in vivo, because the deletion of either of them delays NPM-ALK lymphoma development by reducing the survival of lymphoma cells. Unexpectedly, Cdc42 or Rac1 single deletions have no effect on the dissemination potentials of NPM-ALK lymphoma cells in vivo. In contrast, Cdc42/Rac1 double deletions further impair lymphoma development and completely abrogate lymphoma dissemination in vivo. Thus, we demonstrate essential but nonredundant roles for Cdc42 and Rac1 in NPM-ALK lymphoma development and dissemination and suggest that effective therapies to target these GTPases in lymphoma should aim at inhibiting both Cdc42 and Rac1 simultaneously to achieve a maximal therapeutic effect.
Methods
Mice and immortalized cell lines
CD4-NPM-ALK, CD4Cre,13 Cdc42fl/fl14 , and Rac1fl/fl15 alleles have been previously described. Briefly, mice containing the Rac1 and/or Cdc42 gene flanked by loxP sites (Rac1fl/Cdc42fl) were crossed with NPM-ALK transgenic mice carrying the CD4Cre transgene. All animal experiments were approved by the ethical committees of the University of Torino as appropriate and performed in accordance with the guidelines stated in the International Guiding Principles for Biomedical Research Involving Animals, developed by the Council for International Organizations of Medical Sciences. Cdc42fl/fl, Rac1fl/fl, or Cdc42fl/fl/Rac1fl/fl cell lines were established from genetically corresponding tumors that developed in NPM-ALK transgenic mice by serial passaging as previously described.13 Cell lines were maintained in RPMI 1640 (Lonza) with 10% fetal bovine serum, 2% penicillin, streptomycin 5 mg/mL (Gibco), and 1% glutamine (Gibco). Cell lines were grown at 37°C in humidified atmosphere with 5% CO2.
In vivo experiments
NOD scid gamma (NSG) immunocompromised mice were purchased from Charles River Laboratories. For tumor dissemination analysis mice were inoculated intravenously (i.v.) with 5 × 106 lymphoma cell lines in 0.2 mL phosphate-buffered saline (PBS). The lymphoma cell lines used for the in vivo experiments were NPM-ALK, NPM-ALK-CD4Cre-Cdc42fl/fl, and NPM-ALK-CD4Cre-Rac1fl/fl, either uninfected or transduced with CreERT2/Bcl2 retroviruses. Mice were euthanized under anesthesia 15 days after the inoculation. All organs were isolated and immediately fixed in formalin solution for histopathological examination (hematoxylin and eosin [H&E] staining).
Retrovirus preparation and cell transduction
Retroviruses were generated by transient transfection of pWZLblast vector expressing CreERT2 or MSCV-EGFP vector carrying Bcl2 in 293 Phoenix packaging cells. After 24 hours of incubation at 37°C, supernatants containing viral particles were collected and used for further transduction. For the retroviral transduction, 300 µL supernatant was used to infect 5 × 104 lymphoma cells as previously described.13 CreERT2 transduced cells were selected using blasticidin (Calbiochem, San Diego, CA) at 25 µg/mL for 6 days. Bcl2 transduced cells were analyzed for GFP expression using a FACSCalibur flow cytometer (Becton Dickinson). CELLQuest software was used for data acquisition and analysis.
Flow cytometry
Pretumoral thymuses characterization.
Mice were euthanized at 6 weeks of age, and pretumoral thymuses were resected and used for flow cytometry analysis. Single-cell suspensions were prepared from fresh pretumoral thymuses with mechanic disaggregation and isolated with a 40-μm nylon cell strainer (BD Biosystems, San Jose, CA). Cells were resuspended in PBS and stained with rat anti–mouse CD4-PE (clone GK1.5; Miltenyi Biotec) and anti–mouse CD8a-PerCP (clone 53-6.7; BioLegend) antibodies.
Immunophenotype.
Immortalized cell lines and freshly isolated cells obtained from tumors of NPM-ALK, NPM-ALK-CD4Cre-Cdc42fl/fl, and NPM-ALK-CD4Cre-Rac1fl/fl transgenic mice were incubated and stained with the following antibodies: CD3-FITC, CD4-PE, CD25-APC, CD45R(B220)-PE, CD90-PE, NKp46-FITC (all from Milteny Biotec), and CD8a-PerCP (BioLegend). Cells were analyzed in a FACSCalibur flow cytometer (BD Bioscience) using FlowJo software (Treestar, Inc.).
Immunohistochemical analyses
Immunohistochemical studies were conducted on formalin-fixed (10%) paraffin-embedded tissues. Paraffin sections (2 μm thick) were processed for either H&E staining or immunohistochemistry. Images were acquired with an Olympus BX41 microscope equipped with ×2 and ×40 objectives. For immunostainings, anti-ALK (clone 18-0266, Zymed), anti-Ki-67 (clone Sp6, Abcam), and activated caspase-3 antibodies (clone 5A1E, Cell Signaling) were used. Spleen sections with reactive lymphoid hyperplasia were employed for positive and negative controls.
Immunofluorescence staining
Cells were grown for 12 hours on glass coverslips pretreated with fibronectine (10 μg/mL PBS) at 37°C for 1 hour to facilitate cell adhesion. Samples were fixed in PBS containing 4% paraformaldehyde at room temperature for 10 minutes and permeabilized with PBS containing 0.3% Triton X-100 for 5 minutes. Coverslips were incubated with PBS containing 3% bovine serum albumin (BSA) for 1 hour at room temperature and then stained with phycoerythrin (PE)–conjugated phalloidin (1:200 PBS; Sigma-Aldrich) for 40 minutes in order to stain actin filaments. Nuclei were stained 10 minutes at room temperature with HOECHST (300 ng/mL; Sigma). Coverslips were mounted in antifading solution and viewed using a Leica photomicroscope. Images were acquired at room temperature by means of a ×100/1.40 oil PL APO objective (Leica, Heidelberg, Germany) and analyzed using DM LM Leica software.
Apoptosis assay and cell cycle analysis
Cells (5 × 105 cells/mL) were grown in 6-well plates after treatment with 10 nM 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich) for 4 hours. 1 × 105 cells were then washed with PBS and incubated for 15 minutes at 37°C in Annexin V binding buffer containing 200 nM TMRM (tetrametylrodamine methyl-ester). Percentage of apoptotic cells was analyzed by FACSCalibur flow cytometer (BD Bioscience) using the CellQuest Program. For DNA content determination, cells were washed with PBS, resuspended in citric acid buffer (0.05 M Na2HPO4, 25 mM sodium citrate, and 0.1% Triton X-100 [pH 7.3]), treated with RNase (0.25 mg/mL), and then stained with propidium iodide (50 μg/mL). The S-phase fraction was calculated using the Modfit program from Becton Dickinson.
Immunoblotting
Approximately 40 μg protein extract was obtained from cell lysates using GST-FISH buffer (10 mM MgCl2, 150 mM NaCl, 1% NP40, 2% glycerol, 1 mM EDTA, and 25 mM HEPES [pH 7.5]) supplemented with 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4 (Sigma), and a cocktail of protease inhibitors (Roche) and then separated on SDS-PAGE (Bio-Rad), transferred to a nitrocellulose membrane, and blotted with primary antibodies raised against NPM-ALK (1:3000, Invitrogen), actin (1:4000, Sigma-Aldrich), Cdc42 (1:3000, BD Transduction Laboratories), Rac1 (1:4000, Upstate), Bcl2 (1:4000, Santa Cruz Biotechnology), and Bid, Bax, Bik, Bad, Bak, Bim, Puma, and cleaved caspase-3 (1:1000, Cell Signaling Technology). Secondary anti-mouse or anti-rabbit antibodies were purchased from Amersham.
Migration assays
The migration assay was performed using Transwell Permeable Supports (0.8 μm pore size; Corning) on a 24-well multiplate. Cells were serum starved in RPMI 1640 containing 0.1% fetal bovine serum and then placed in the upper chamber of the transwell at a concentration of 1 × 106 cells/well in 100 μL medium. Stromal-derived factor-1 α (SDF-1α; R&D Systems, Minneapolis, MN) was added at the bottom of the chamber as a chemoattractant at a concentration of 100 ng/mL in serum-free medium. After 3 hours of incubation at 37°C in a 5% CO2 atmosphere, the transwells were removed (with nonmigrating cells) and the number of the migrated cells was counted. The percentage of migrating cells was calculated as ratio to the controls treated with serum-free medium.
Statistical analysis
Statistical significance was calculated with the Student t test, and only values lower than 0.05 were considered significant. Tumor-free survival distribution was estimated by the nonparametric Kaplan-Meier method. Unless otherwise indicated, data are presented as mean ± standard deviation (SD).
Results
Rac1 and Cdc42 possess nonredundant roles in NPM-ALK–mediated lymphomagenesis
To study the functions of Cdc42 and Rac1 in NPM-ALK rearranged lymphoma in vivo, we deleted Cdc42 or Rac1 genes in NPM-ALK transgenic mice that develop ALK-driven T-cell lymphoma with high penetrance.14 Constitutive deletions of either Cdc4215 or Rac116 are embryonic lethal, whereas T-cell–specific deletions of Cdc42 or Rac1 are associated with modest impairment of T-cell development and functions.17-19 To achieve a T-cell–restricted deletion, we crossed conditional Cdc42fl/fl or Rac1fl/fl mice with CD4Cre mice that express the Cre recombinase in both CD4 and CD8 T cells under the control of the CD4 minimal promoter.20 By using this approach, we aimed at deleting Cdc42 or Rac1 simultaneously with the induction of NPM-ALK expression, because NPM-ALK expression is under the control of the same CD4 promoter.14 Indeed, we obtained an efficient deletion of Cdc42 or Rac1 in the large majority of thymocytes as demonstrated by an almost complete loss of protein expression by western blot (Figure 1A). Deletion of Cdc42 or Rac1 in the thymus resulted only in a slight impairment of T-cell maturation with decreased numbers of single-positive CD4+ or CD8+ T cells, a phenotype that was more pronounced in CD4Cre-Cdc42fl/fl mice (see supplemental Figure 1 and supplemental Table 1, available on the Blood Web site). These results are comparable to previous studies where Rac1 or Cdc42 deletions were mediated by Lck-Cre17,19 or hCD2-Cre.18 In contrast, double deletions of Cdc42 and Rac1 have never been described to our knowledge. By crossing CD4Cre mice with Cdc42fl/fl/Rac1fl/fl mice, we achieved an efficient double deletion of both Cdc42 and Rac1 (Figure 1A). In these mice, we observed a significant reduction in thymocyte numbers with a relative decrease in the CD4+/CD8+ double-positive population (supplemental Figure 1 and supplemental Table 1).
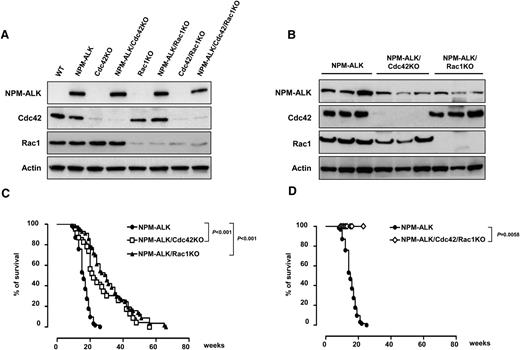
Deletion of Cdc42 or Rac1 impair NPM-ALK lymphoma development in vivo. (A) Representative western blots of Cdc42 and Rac1 deletion in pretumoral thymuses obtained from wild-type (WT) or NPM-ALK transgenic mice. Mice were euthanized at 4 weeks of age when lymphoma was not phenotypically and morphologically detectable as previously described.12 Cell lysates were extracted and membranes were blotted with the indicated antibodies. (B) Representative western blots for Cdc42 and Rac1 deletion in primary tumors from mice with the indicated genotype. Three tumors out of 10 analyzed for each genotype are shown. (C-D) Kaplan-Meier survival curves of NPM-ALK transgenic mice in the presence or absence of Cdc42 and/or Rac1. NPM-ALK mice were crossed with mice carrying the indicated conditional knocked-in alleles. All mice were analyzed by necropsy to demonstrate the presence of lymphoma: NPM-ALK (n = 70), NPM-ALK;CD4Cre;Cdc42fl/fl (NPM-ALK/Cdc42KO; n = 25), NPM-ALK;CD4Cre;Rac1fl/fl (NPM-ALK/Rac1KO; n = 58), and NPM-ALK;CD4Cre;Cdc42fl/fl;Rac1fl/fl (NPM-ALK/Cdc42/Rac1KO; n = 17).
Deletion of Cdc42 or Rac1 impair NPM-ALK lymphoma development in vivo. (A) Representative western blots of Cdc42 and Rac1 deletion in pretumoral thymuses obtained from wild-type (WT) or NPM-ALK transgenic mice. Mice were euthanized at 4 weeks of age when lymphoma was not phenotypically and morphologically detectable as previously described.12 Cell lysates were extracted and membranes were blotted with the indicated antibodies. (B) Representative western blots for Cdc42 and Rac1 deletion in primary tumors from mice with the indicated genotype. Three tumors out of 10 analyzed for each genotype are shown. (C-D) Kaplan-Meier survival curves of NPM-ALK transgenic mice in the presence or absence of Cdc42 and/or Rac1. NPM-ALK mice were crossed with mice carrying the indicated conditional knocked-in alleles. All mice were analyzed by necropsy to demonstrate the presence of lymphoma: NPM-ALK (n = 70), NPM-ALK;CD4Cre;Cdc42fl/fl (NPM-ALK/Cdc42KO; n = 25), NPM-ALK;CD4Cre;Rac1fl/fl (NPM-ALK/Rac1KO; n = 58), and NPM-ALK;CD4Cre;Cdc42fl/fl;Rac1fl/fl (NPM-ALK/Cdc42/Rac1KO; n = 17).
To study the effects of Cdc42 or Rac1 on NPM-ALK–mediated lymphomagenesis, we crossed NPM-ALK transgenic mice with CD4Cre-Cdc42fl/fl or CD4Cre-Rac1fl/fl mice. Deletion of Cdc42 or Rac1 was efficient and T-cell development was comparable to NPM-ALK–negative mice (Figure 1A, supplemental Figure 1, and supplemental Table 1), with a slight increase in mature CD4+ or CD8+ T cells in NPM-ALK transgenic mice as we previously described.14 Expression of NPM-ALK in T cells acts as a strong driver oncogene, as 100% of mice develop lymphoma, with a mean survival of 15 weeks.14 NPM-ALK lymphomas arose in both NPM-ALK/CD4Cre/Cdc42fl/fl and NPM-ALK/CD4Cre/Rac1fl/fl mice despite the efficient deletion of Cdc42 or Rac1 in all tumors tested (10 NPM-ALK/CD4Cre/Cdc42fl/fl and 10 NPM-ALK/CD4Cre/Rac1fl/fl) (Figure 1B and supplemental Table 2). Strikingly, however, deletion of either Cdc42 or Rac1 significantly delayed lymphoma onset and extended survival (Figure 1C). NPM-ALK/CD4Cre/Cdc42fl/fl/Rac1fl/fl mice (n = 17) did not develop lymphoma up to 28 weeks (Figure 1D). Unfortunately, longer follow-up was not reached as Cdc42/Rac1 double-knockout (DKO) mice died prematurely because of multiorgan failure, which is currently under investigation.
In vivo deletion of Cdc42 or Rac1 increases apoptosis of NPM-ALK lymphoma cells
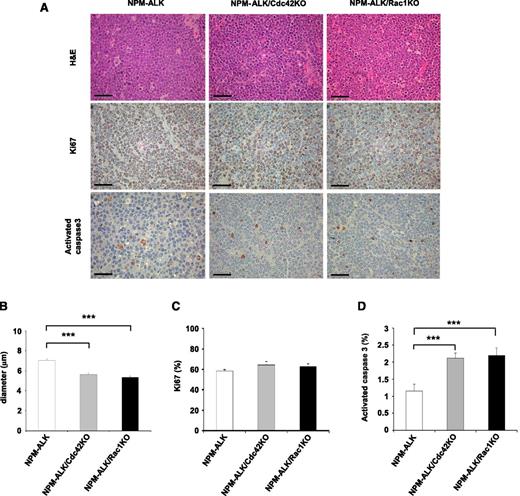
Deletions of Cdc42 or Rac1 did not substantially change the frequency of tumor subtypes or the phenotype of lymphoma developed in NPM-ALK transgenic mice alone. T-cell lymphoma was by far the predominant tumor subtype developed in each genetic strain (supplemental Table 2), in accordance with our previous report.14 The phenotype of NPM-ALK lymphoma was mostly negative for both CD4 and CD8 or weakly positive for CD4, corresponding to an early stage of T-cell development when the CD4 transgene is activated to expressed NPM-ALK (supplemental Figure 2 and supplemental Table 3), as we previously described.14 In terms of morphology, NPM-ALK/CD4Cre/Cdc42fl/fl and NPM-ALK/CD4Cre/Rac1fl/fl lymphomas were comparable to NPM-ALK lymphomas, with lymphoma cells showing smaller size and rounder morphology (Figure 2A-B). Interestingly, NPM-ALK/CD4Cre/Cdc42fl/fl and NPM-ALK/CD4Cre/Rac1fl/fl tumors showed a comparable proliferation index but significantly higher apoptotic rate than NPM-ALK tumors (Figure 2C-D). Overall, these data indicated that Cdc42 or Rac1 deletion changed the morphology and decreased cell viability, but not the proliferation, of NPM-ALK–transformed lymphoma cells.
Apoptosis induced by Cdc42 or Rac1 deletion impairs NPM-ALK lymphoma development in vivo. (A) Representative histology of lymphoma arising in NPM-ALK, NPM-ALK;CD4Cre;Cdc42fl/fl (NPM-ALK/Cdc42KO), or NPM-ALK;CD4Cre;Rac1fl/fl (NPM-ALK/Rac1KO) mice (top). Immunostainings for Ki-67 (middle) and activated caspase-3 (bottom) in tumors of the indicated genotypes are shown. Scale bar, 50 μm. (B) Histograms represent the average diameter quantified by counting at least 100 cells for each genotype. Error bars indicated SEM. ***P < .001. (C) Quantification of the percentages of proliferating cells based on Ki-67 counts on sections stained by immunohistochemistry. Data were obtained from 10 different areas in 3 independent mice for each genotype. (D) Quantification of the percentages of apoptotic cells based on activated caspase-3 counts on sections stained by immunohistochemistry. Data were obtained from 10 different areas in 3 independent mice for each genotype. Error bars indicated SEM. ***P < .001.
Apoptosis induced by Cdc42 or Rac1 deletion impairs NPM-ALK lymphoma development in vivo. (A) Representative histology of lymphoma arising in NPM-ALK, NPM-ALK;CD4Cre;Cdc42fl/fl (NPM-ALK/Cdc42KO), or NPM-ALK;CD4Cre;Rac1fl/fl (NPM-ALK/Rac1KO) mice (top). Immunostainings for Ki-67 (middle) and activated caspase-3 (bottom) in tumors of the indicated genotypes are shown. Scale bar, 50 μm. (B) Histograms represent the average diameter quantified by counting at least 100 cells for each genotype. Error bars indicated SEM. ***P < .001. (C) Quantification of the percentages of proliferating cells based on Ki-67 counts on sections stained by immunohistochemistry. Data were obtained from 10 different areas in 3 independent mice for each genotype. (D) Quantification of the percentages of apoptotic cells based on activated caspase-3 counts on sections stained by immunohistochemistry. Data were obtained from 10 different areas in 3 independent mice for each genotype. Error bars indicated SEM. ***P < .001.
Deletion of Cdc42 or Rac1 induces a Bcl2-dependent apoptosis in NPM-ALK lymphoma cells
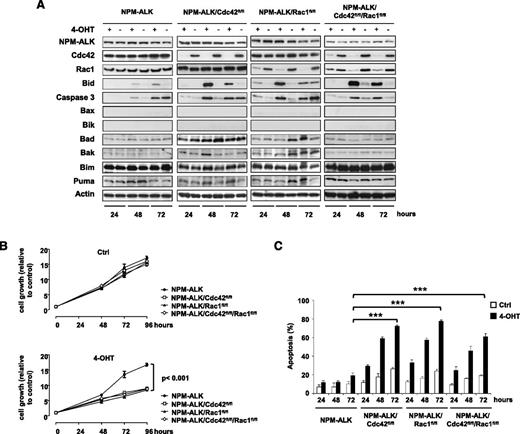
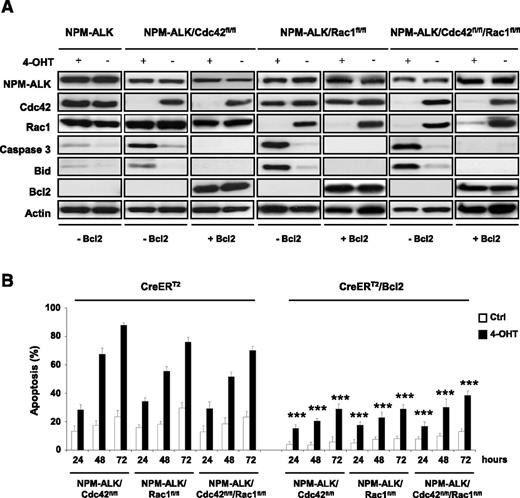
To better characterize the mechanistic role of Rac1 and Cdc42 in NPM-ALK–mediated lymphomagenesis, we generated NPM-ALK lymphoma cell lines where genetic Cdc42 or Rac1 deletions could be induced at will. Immortalized lymphoma cell lines with different genotypes (NPM-ALK, NPM-ALK/Cdc42fl/fl, NPM-ALK/Rac1fl/fl, and NPM-ALK/Rac1fl/fl/Cdc42fl/fl) were transduced with a retrovirus expressing an inducible CreERT2 construct that allows the activation of the Cre-recombinase upon treatment with 4-hydroxytamoxifen (4-OHT). Using this system, efficient deletions were achieved within 48 hours of initial treatment with 4-OHT (10 nM for 4 hours) in single Cdc42fl/fl or Rac1fl/fl as well as double Cdc42fl/fl/Rac1fl/fl lymphoma cells (Figure 3A). Single deletions of Cdc42 or Rac1 significantly impaired cell growth of NPM-ALK lymphoma cells compared with cells with functional Cdc42 and Rac1 (Figure 3B). A similar growth reduction was observed in Cdc42/Rac1 DKO lymphoma cells (Figure 3B). Consistent with the in vivo findings, Cdc42 or Rac1 deletions induced only a slight reduction in the proliferation of lymphoma cells as measured by S-phase and cell cycle (supplemental Figure 3). In contrast, single deletions of Cdc42 or Rac1 were associated with a potent induction of cell apoptosis (Figure 3C and supplemental Figure 4), which was consistent with the increased apoptosis observed in primary tumors (Figure 2D). Surprisingly, Cdc42/Rac1 DKO showed a phenotype comparable to single Cdc42 or Rac1 KOs without additional effects on the cell cycle or apoptosis (Figure 3C and supplemental Figures 3 and 4). Next, we further investigated the mechanisms of apoptosis induced by Cdc42 or Rac1 deletions. To this end, we exploited our system of inducible Cdc42 or Rac1 deletions in NPM-ALK lymphoma cells to perform a screening for key factors involved in apoptosis. We found a strong upregulation of the BH3 interacting-domain death agonist Bid in single Cdc42 or Rac1 knockouts as well as in Cdc42/Rac1 DKOs, whereas no changes were observed in the proapoptotic NPM-ALK target Bim21,22 or in other key mediators of apoptosis (Figure 3A). Bid interacts with Bax to induce apoptosis by increasing the permeability of mitochondria and can be counteracted by Bcl2.23 Therefore, we reasoned that overexpression of Bcl2 would prevent Bid-mediated apoptosis in Cdc42- or Rac1-deleted as well as in Cdc42/Rac1 DKO lymphoma cells. To test this hypothesis, we overexpressed Bcl2 in NPM-ALK lymphoma cells (Figure 4A). Bcl2 overexpression completely blocked Bid upregulation and caspase-3 activation upon deletion of Cdc42 or Rac1 and in DKO cells (Figure 4A). Consistently, apoptosis of NPM-ALK lymphoma cells induced by Cdc42 or Rac1 deletions was markedly reduced (Figure 4B and supplemental Figure 4).
Deletion of Cdc42 or Rac1 increases apoptosis mediated by Bid upregulation. (A) Representative western blots of immortalized NPM-ALK lymphoma cell lines obtained from mice with the indicated genotypes. Cells were transduced by retroviruses expressing an inducible CreERT2 recombinase system and selected using 25 μg/mL blasticidin for 6 days. Cre recombinase was shortly activated by treatment with 10 nM 4-hydroxytamoxifen (4-OHT) for 4 hours to induce deletion of the floxed genes. Cells were collected 24, 48, and 72 hours after 4-OHT induction, lysed, and blotted with the indicated antibodies. Data are from 1 representative cell line for each genotype out of 3 NPM-ALK, 5 NPM-ALK;CreERT2;Cdc42fl/fl, 4 NPM-ALK;CreERT2;Rac1fl/fl, and 3 NPM-ALK;CreERT2;Cdc42fl/fl;Rac1fl/fl cell lines transduced with CreERT2 recombinase. (B) NPM-ALK lymphoma cell lines obtained from mice with the genotypes as in panel A and expressing CreERT2 were conditionally deleted of the indicated floxed genes by treatment with 10 nM 4-OHT for 4 hours as described above. Cell growth/viability were measured by CellTiter-Glo at the indicated time points. Data are presented as mean ± SD of triplicate experiments, each performed with 3 independent cell lines for each genotype. (C) Percentages of apoptotic cells measured by TMRM staining at the indicated time points in NPM-ALK immortalized lymphoma cells from mice with the indicated genotypes after CreERT2 induction of the floxed genes as described above. Data are indicated as means ± SD of triplicate experiments, each performed with 3 independent cell lines for each genotype. ***P < .001.
Deletion of Cdc42 or Rac1 increases apoptosis mediated by Bid upregulation. (A) Representative western blots of immortalized NPM-ALK lymphoma cell lines obtained from mice with the indicated genotypes. Cells were transduced by retroviruses expressing an inducible CreERT2 recombinase system and selected using 25 μg/mL blasticidin for 6 days. Cre recombinase was shortly activated by treatment with 10 nM 4-hydroxytamoxifen (4-OHT) for 4 hours to induce deletion of the floxed genes. Cells were collected 24, 48, and 72 hours after 4-OHT induction, lysed, and blotted with the indicated antibodies. Data are from 1 representative cell line for each genotype out of 3 NPM-ALK, 5 NPM-ALK;CreERT2;Cdc42fl/fl, 4 NPM-ALK;CreERT2;Rac1fl/fl, and 3 NPM-ALK;CreERT2;Cdc42fl/fl;Rac1fl/fl cell lines transduced with CreERT2 recombinase. (B) NPM-ALK lymphoma cell lines obtained from mice with the genotypes as in panel A and expressing CreERT2 were conditionally deleted of the indicated floxed genes by treatment with 10 nM 4-OHT for 4 hours as described above. Cell growth/viability were measured by CellTiter-Glo at the indicated time points. Data are presented as mean ± SD of triplicate experiments, each performed with 3 independent cell lines for each genotype. (C) Percentages of apoptotic cells measured by TMRM staining at the indicated time points in NPM-ALK immortalized lymphoma cells from mice with the indicated genotypes after CreERT2 induction of the floxed genes as described above. Data are indicated as means ± SD of triplicate experiments, each performed with 3 independent cell lines for each genotype. ***P < .001.
Bcl2 overexpression blocks apoptosis, Bid upregulation, and caspase-3 activation associated with Cdc42 and Rac1 deletion in NPM-ALK lymphoma. Three independent CreERT2 lymphoma cell lines for each indicated genotype as in Figure 2 were transduced with a retrovirus expressing Bcl2 and GFP as reporter. Percentages of transduced cells were calculated by GFP positivity in flow cytometry and were above 90% in all cell lines. Cdc42 and Rac1 deletions were induced by treatment with 10 nM 4-hydroxytamoxifen for 4 hours. (A) Cells were collected at the indicated time points and lysed, and western blots were performed with the indicated antibodies. (B) Overexpression of Bcl2 rescues apoptosis induced by Cdc42 or Rac1 deletion. Three independent CreERT2 lymphoma cell lines for each indicated genotype as in panel A were transduced with a retrovirus expressing Bcl2 and GFP. Cdc42 or Rac1 deletion was induced by 4-OHT treatment as described above. Analysis of apoptosis was carried out by TMRM staining and flow cytometry at the indicated time points. Data are indicated as mean ± SD of triplicate experiments, each performed with 3 independent cell lines for each genotype. P values are calculated by comparing control (Ctrl) vs 4-OHT–induced cells at each indicated time point (***P < .001).
Bcl2 overexpression blocks apoptosis, Bid upregulation, and caspase-3 activation associated with Cdc42 and Rac1 deletion in NPM-ALK lymphoma. Three independent CreERT2 lymphoma cell lines for each indicated genotype as in Figure 2 were transduced with a retrovirus expressing Bcl2 and GFP as reporter. Percentages of transduced cells were calculated by GFP positivity in flow cytometry and were above 90% in all cell lines. Cdc42 and Rac1 deletions were induced by treatment with 10 nM 4-hydroxytamoxifen for 4 hours. (A) Cells were collected at the indicated time points and lysed, and western blots were performed with the indicated antibodies. (B) Overexpression of Bcl2 rescues apoptosis induced by Cdc42 or Rac1 deletion. Three independent CreERT2 lymphoma cell lines for each indicated genotype as in panel A were transduced with a retrovirus expressing Bcl2 and GFP. Cdc42 or Rac1 deletion was induced by 4-OHT treatment as described above. Analysis of apoptosis was carried out by TMRM staining and flow cytometry at the indicated time points. Data are indicated as mean ± SD of triplicate experiments, each performed with 3 independent cell lines for each genotype. P values are calculated by comparing control (Ctrl) vs 4-OHT–induced cells at each indicated time point (***P < .001).
Redundant roles of Cdc42 and Rac1 in NPM-ALK lymphoma dissemination
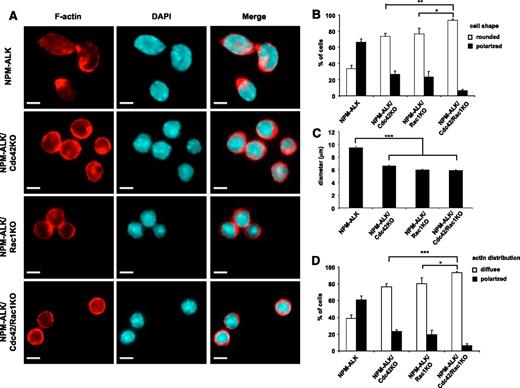
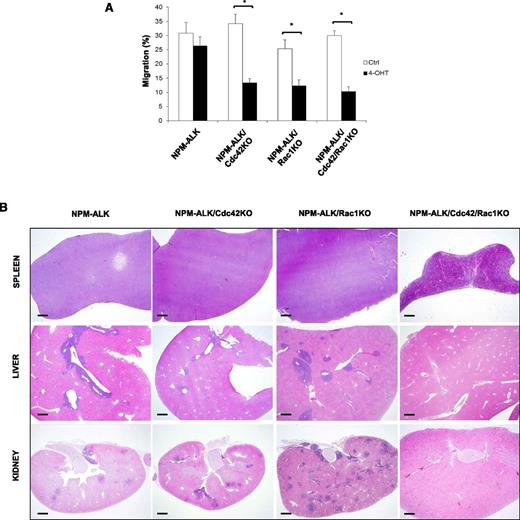
We previously reported that Cdc42 activation directly governs the NPM-ALK lymphoma cell shape (ie, the anaplastic morphology),11 whereas a similar effect for Rac1 activation has not been studied yet. Thus, we decided to precisely address this question by taking advantage of our system of inducible genetic deletion of Cdc42 or Rac1. Cell morphology can be properly evaluated by analyzing the distribution of F-actin filaments within the cytoplasm of anaplastic lymphoma cells, where F-actin is unevenly distributed and polarized. The pattern of actin polarization directly reflects the activity of NPM-ALK on the cytoskeleton and is mediated by Cdc42.11 As expected, deletion of Cdc42 significantly reduced the irregular shape and actin polarization of NPM-ALK lymphoma cells (Figure 5A), as cells became more regularly rounded (Figure 5B), smaller (Figure 5C), and lost F-actin polarization (Figure 5D). Remarkably, Rac1 deletion induced comparable changes in NPM-ALK lymphoma cell morphology, thus indicating that both Cdc42 and Rac1 control lymphoma cell morphology (Figure 5). Simultaneous deletions of both Cdc42 and Rac1 further increased the phenotype of loss of polarization (Figure 5 B,D), indicating similar but nonredundant roles in regulating NPM-ALK lymphoma cell shape. Finally, we asked whether Cdc42 or Rac1 were critical for NPM-ALK lymphoma cell migration and dissemination in vivo. First, we tested lymphoma migration in vitro to confirm that Cdc42 or Rac1 deletions resulted in reduced cell migration, as expected from previous studies.11,12 Cdc42/Rac1 DKO lymphoma cells showed a comparable impairment of migration (Figure 6A). Next, we studied in vivo dissemination of NPM-ALK lymphoma. For this experiment, we generated cell lines with stable deletions of Cdc42 or Rac1 after 4-OHT induction of CreERT2 recombinase by expressing Bcl2 to block apoptosis as shown above. Indeed, cell lines with stable CreERT2-induced Cdc42 or Rac1 deletion showed comparable deletion efficacy and growth rates similar to cell lines immortalized from transgenic mice that express CD4Cre (supplemental Figure 5A-B). Importantly, this approach allowed us to generate stable Cdc42/Rac1 DKO cell lines, as DKO lymphomas were never obtained in transgenic mice (Figure 1D). Cdc42/Rac1 DKO NPM-ALK lymphoma cell lines showed comparable growth rates to singly Cdc42- or Rac1-deleted lymphoma cell lines (supplemental Figure 5). Equal numbers of NPM-ALK cells were injected i.v. in recipient mice, and lymphoma dissemination was analyzed in various organs 2 weeks after cell injection. Control NPM-ALK lymphoma cells completely colonized lymphoid organs, such as spleen and lymph nodes, as well as several other organs including liver, kidneys, and lungs (Figure 6 and Table 1). The presence and dissemination of NPM-ALK lymphoma cells was confirmed by ALK immunostaining (supplemental Figure 6). NPM-ALK lymphoma cells with single deletion of Cdc42 or Rac1, by either CD4Cre or CreERT2, equally disseminated to the same organs, whereas lymphoma cells with Cdc42/Rac1 DKO were almost completely unable to disseminate and colonize lymphoid and nonlymphoid organs (Figure 6, supplemental Figure 6, and Table 1). Overall, these results show that Cdc42 and Rac1 control cell shape and migration of NPM-ALK lymphoma cells. However, only a simultaneous elimination of Cdc42 and Rac1 activities can prevent lymphoma dissemination in vivo.
Control of NPM-ALK lymphoma cell shape by Cdc42 or Rac1. NPM-ALK, NPM-ALK;Cdc42fl/fl, NPM-ALK;Rac1fl/fl, and NPM-ALK;Cdc42fl/fl;Rac1fl/fl lymphoma cell lines were immortalized from primary tumors arising in mice with the indicated genotype. Cells were transduced with CreERT2 and Bcl2 retroviruses and then conditionally deleted for the indicated floxed genes by treatment with 10 nM 4-hydroxytamoxifen for 4 hours as described above. Stable cell lines were obtained by culturing deleted cells for at least 3 weeks. (A) Cell morphology and cell shape were evaluated by immunofluorescence using phycoerythrin-conjugated phalloidin staining to detect actin filaments. Scale bar, 5 μm. NPM-ALK;CreERT2 (NPM-ALK), NPM-ALK;CreERT2;Cdc42fl/fl;Bcl2 (NPM-ALK/Cdc42KO), NPM-ALK;CreERT2;Rac1fl/fl;Bcl2 (NPM-ALK/Rac1KO), NPM-ALK;CreERT2;Cdc42fl/fl;Rac1fl/fl;Bcl2 (NPM-ALK/Cdc42/Rac1KO). (B-D) Histograms represent the percentage of round versus polarized cells expressed as cell shape (B), average diameter (C), and actin distribution around the membrane or in the lamellipodial membrane protrusion (D). Each quantification was obtained by counting at least 100 cells for each condition. Two independent lymphoma cell lines for each genotype were studied in triplicate experiments. Error bars indicate SEM. ***P < .001; **P < .002; *P < .05.
Control of NPM-ALK lymphoma cell shape by Cdc42 or Rac1. NPM-ALK, NPM-ALK;Cdc42fl/fl, NPM-ALK;Rac1fl/fl, and NPM-ALK;Cdc42fl/fl;Rac1fl/fl lymphoma cell lines were immortalized from primary tumors arising in mice with the indicated genotype. Cells were transduced with CreERT2 and Bcl2 retroviruses and then conditionally deleted for the indicated floxed genes by treatment with 10 nM 4-hydroxytamoxifen for 4 hours as described above. Stable cell lines were obtained by culturing deleted cells for at least 3 weeks. (A) Cell morphology and cell shape were evaluated by immunofluorescence using phycoerythrin-conjugated phalloidin staining to detect actin filaments. Scale bar, 5 μm. NPM-ALK;CreERT2 (NPM-ALK), NPM-ALK;CreERT2;Cdc42fl/fl;Bcl2 (NPM-ALK/Cdc42KO), NPM-ALK;CreERT2;Rac1fl/fl;Bcl2 (NPM-ALK/Rac1KO), NPM-ALK;CreERT2;Cdc42fl/fl;Rac1fl/fl;Bcl2 (NPM-ALK/Cdc42/Rac1KO). (B-D) Histograms represent the percentage of round versus polarized cells expressed as cell shape (B), average diameter (C), and actin distribution around the membrane or in the lamellipodial membrane protrusion (D). Each quantification was obtained by counting at least 100 cells for each condition. Two independent lymphoma cell lines for each genotype were studied in triplicate experiments. Error bars indicate SEM. ***P < .001; **P < .002; *P < .05.
Effects of Cdc42 and Rac1 deletions on in vitro migration and in vivo dissemination of NPM-ALK lymphoma cells. (A) Three independent CreERT2 lymphoma cell lines for each genotype were transduced with a retrovirus expressing Bcl2 to protect cells from apoptosis as in Figure 5. Cdc42 and Rac1 deletions were induced by treatment with 10 nM 4-hydroxytamoxifen for 4 hours. Cell were then seeded into the upper chamber of transwells (0.8 µm pore size) and allowed to migrate toward a gradient of SDF-1α (100 ng/mL) that was plated in the lower chamber for 4 hours in CO2 incubator. Migrated cells in the bottom chamber were counted. The histograms indicate means ± SD from 3 independent cell lines for each genotype using triplicate wells for experimental point. The Student t test was used to calculate statistical significance (*P < .001). (B) Immortalized NPM-ALK lymphoma cell lines with the genotypes as in panel A were inoculated i.v. (5 × 106) in NOD scid γ (NSG) mice. After 15 days, mice were euthanized and all organs were isolated and fixed in formalin solution for H&E staining. Figure shows representative histology of spleen, liver, and kidney. NPM-ALK;CreERT2 (NPM-ALK), NPM-ALK;CreERT2;Cdc42fl/fl;Bcl2 (NPM-ALK/Cdc42KO), NPM-ALK;CreERT2;Rac1fl/fl;Bcl2 (NPM-ALK/Rac1KO), NPM-ALK;CreERT2;Cdc42fl/fl;Rac1fl/fl;Bcl2 (NPM-ALK/Cdc42/Rac1KO). Scale bar, 1 mm.
Effects of Cdc42 and Rac1 deletions on in vitro migration and in vivo dissemination of NPM-ALK lymphoma cells. (A) Three independent CreERT2 lymphoma cell lines for each genotype were transduced with a retrovirus expressing Bcl2 to protect cells from apoptosis as in Figure 5. Cdc42 and Rac1 deletions were induced by treatment with 10 nM 4-hydroxytamoxifen for 4 hours. Cell were then seeded into the upper chamber of transwells (0.8 µm pore size) and allowed to migrate toward a gradient of SDF-1α (100 ng/mL) that was plated in the lower chamber for 4 hours in CO2 incubator. Migrated cells in the bottom chamber were counted. The histograms indicate means ± SD from 3 independent cell lines for each genotype using triplicate wells for experimental point. The Student t test was used to calculate statistical significance (*P < .001). (B) Immortalized NPM-ALK lymphoma cell lines with the genotypes as in panel A were inoculated i.v. (5 × 106) in NOD scid γ (NSG) mice. After 15 days, mice were euthanized and all organs were isolated and fixed in formalin solution for H&E staining. Figure shows representative histology of spleen, liver, and kidney. NPM-ALK;CreERT2 (NPM-ALK), NPM-ALK;CreERT2;Cdc42fl/fl;Bcl2 (NPM-ALK/Cdc42KO), NPM-ALK;CreERT2;Rac1fl/fl;Bcl2 (NPM-ALK/Rac1KO), NPM-ALK;CreERT2;Cdc42fl/fl;Rac1fl/fl;Bcl2 (NPM-ALK/Cdc42/Rac1KO). Scale bar, 1 mm.
In vivo NPM-ALK lymphoma dissemination
| Genotype . | Cre . | Spleen . | Liver . | Kidney . | Lung . | Bone marrow . | Brain . | Heart . |
|---|---|---|---|---|---|---|---|---|
| NPM-ALK | CD4Cre | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 0/4 | 0/4 |
| NPM-ALK;Cdc42fl/fl | CD4Cre | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 0/3 | 0/3 |
| NPM-ALK;Rac1fl/fl | CD4Cre | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 0/3 | 0/3 |
| NPM-ALK;Cdc42fl/fl | CreERT2 | 3/3 | 3/3 | 3/3 | 0/3 | 3/3 | 0/3 | 0/3 |
| NPM-ALK;Rac1fl/fl | CreERT2 | 3/3 | 3/3 | 3/3 | 2/3 | 3/3 | 0/3 | 0/3 |
| NPM-ALK;Cdc42fl/fl;Rac1fl/fl | CreERT2 | 0/6 | 0/6 | 0/6 | 1/6 | 0/6 | 0/6 | 0/6 |
| Genotype . | Cre . | Spleen . | Liver . | Kidney . | Lung . | Bone marrow . | Brain . | Heart . |
|---|---|---|---|---|---|---|---|---|
| NPM-ALK | CD4Cre | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 0/4 | 0/4 |
| NPM-ALK;Cdc42fl/fl | CD4Cre | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 0/3 | 0/3 |
| NPM-ALK;Rac1fl/fl | CD4Cre | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 0/3 | 0/3 |
| NPM-ALK;Cdc42fl/fl | CreERT2 | 3/3 | 3/3 | 3/3 | 0/3 | 3/3 | 0/3 | 0/3 |
| NPM-ALK;Rac1fl/fl | CreERT2 | 3/3 | 3/3 | 3/3 | 2/3 | 3/3 | 0/3 | 0/3 |
| NPM-ALK;Cdc42fl/fl;Rac1fl/fl | CreERT2 | 0/6 | 0/6 | 0/6 | 1/6 | 0/6 | 0/6 | 0/6 |
Discussion
ALCL has peculiar morphologic features compared with most T-cell and non–T-cell lymphomas. They have a highly atypical morphology and peculiar patterns of dissemination.9,24 Previous reports showed that in ALK-rearranged ALCL the oncogenic NPM-ALK directly controls cell shape and migration of lymphoma cells by activating the Rho GTPases Cdc42 and Rac1.11-13 Mechanistically, NPM-ALK directly phosphorylates the RhoGEFs Vav1 and Vav3 to activate in turn Cdc42 and Rac1, respectively.11,12 Recent work showed that NPM-ALK also exploits the Tiam1 RhoGEF to activate Rac1 via phosphatidylinositol 5-phosphate (PtdIns5P) produced by the phosphatidylinositol 5-kinase PIKfyve.25 NPM-ALK not only activates Cdc42 and Rac1 but also appears to regulate a larger program of GTPase activity regulation that also involves RhoA inhibition,11 thus suggesting that in NPM-ALK–driven lymphoma, Cdc42 and Rac1 could act as oncogenes whereas RhoA could represent a tumor suppressor. This view is corroborated by recent studies that found RhoA frequently inactivated by mutations in peripheral T-cell lymphoma6-8 and Burkitt lymphoma.26
In this work, we developed a genetic approach to specifically delete Cdc42 or Rac1 in T cells transformed by NPM-ALK oncogenic activity. In addition, we also simultaneously deleted Cdc42 and Rac1 in the same lymphoma cells, a feat never accomplished before. Using this approach, we showed that Cdc42 or Rac1 deletions delayed to a comparable extent the onset of NPM-ALK–driven lymphoma in mice. Cdc42/Rac1 double-deleted NPM-ALK mice never developed lymphoma, but the follow-up was relatively short due to the early lethality in these mice. The causes of this early lethality are not yet understood and currently under investigation in our laboratory. In necropsy studies carried out in most of the mice, we could not detect any evidence of lymphoma or plasma cell tumors, but rather detected signs of multiorgan failure that involved the liver, kidneys, and lungs.
In vivo deletion of Cdc42 or Rac1, as well as in vitro through an inducible system, was associated mostly with an increased apoptotic rate of lymphoma cells, with relatively mild effects on the cell cycle and proliferation. Thus, by this genetic approach, we could determine that Cdc42 and Rac1 control survival rather than proliferation of lymphoma cells, a conclusion partially in contrast with previous studies based on short hairpin RNA knockdowns or not-specific inhibitors such as secramine for Cdc4211 or NSC23766 for Rac1.13 In our genetically deleted models, a predominant role for Cdc42 or Rac1 on survival was further supported by results in lymphoma cell lines where apoptosis was blocked by overexpression of Bcl2. Indeed, in the presence of high levels of Bcl2, Cdc42- or Rac1-deleted NPM-ALK cell lines showed a similar proliferation rate to lymphoma cells with normal expression of Cdc42 or Rac1, thus further supporting the conclusion that lymphoma proliferation is only marginally dependent on the activity of Cdc42 or Rac1 in NPM-ALK lymphoma. The signaling network activated by Cdc42 and Rac1 is extremely complex. Antiapoptotic and survival signals activated by Cdc42 or Rac1 include nuclear factor κB, p38/MAPK, mTOR, as well as other molecules that greatly overlap with targets activated by NPM-ALK itself.2,9,10 In addition to an involvement in cell survival or proliferation, Cdc42 or Rac1 have well-established roles to govern cell movements, migration, and metastasis formation.1-3 As expected by previous studies from our group and others,11,12 we found that either Cdc42 or Rac1 deletion was associated to a reduction of cell migration in vitro induced by the chemokine SDF-1. Surprisingly, however, neither Cdc42 nor Rac1 single deletions impaired lymphoma migration and dissemination in vivo. NPM-ALK lymphoma cells deleted for Cdc42 or Rac1 efficiently colonized not only lymphoid organs, such as the spleen and the bone marrow, but also nonlymphoid organs, such as liver, kidneys, and lungs. In contrast, when both Cdc42 and Rac1 were simultaneously deleted, NPM-ALK lymphoma cells lost completely their capacity of in vivo dissemination despite retaining a similar proliferation rate (Figure 5 and supplemental Figures 5 and 6). These results indicate that for in vivo dissemination, NPM-ALK lymphoma cells can compensate for the loss of function of one GTPase, but not both, thus indicating that Cdc42 and Rac1 have nonredundant roles in NPM-ALK–driven lymphoma growth but are redundant for lymphoma dissemination. As an alternative to redundancy, it is possible that double Cdc42 and Rac1 deletion could act with a threshold mechanism, because we observed more dramatic changes in cell morphology and F-actin filament distribution in Cdc42/Rac1 double-deleted lymphoma cells compared with single deletions. In this view, only a more profound loss of GTPase activity, as in double Cdc42- and Rac1-deleted cells, would result in an impairment of cell migration in vivo.
Overall, these results could have substantial therapeutic implications as the interest in developing Cdc42 or Rac1 inhibitors for clinical use is constantly growing.2,27 Our data indicate that a simultaneous inhibition of both Cdc42 and Rac1 would likely have a more profound effect on lymphoma growth and dissemination. As dual Cdc42/Rac1 inhibitors have been recently developed and are currently tested in preclinical mouse models,28 this approach could represent a potential additional therapeutic strategy for NPM-ALK lymphoma.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Maria Stella Scalzo and Daniele Corino for their precious technical assistance.
This work was supported by grant FP7 ERC-2009-StG (proposal 242965, “Lunely”), Associazione Italiana per la Ricerca sul Cancro (AIRC) grant IG-12023, International Association for Cancer Research grant 12-0216 (R. Chiarle), My First AIRC Grant (MFAG) (C.A. and M.C.), the Bando Giovani Ricercatori grant 2009-GR 1603126 (M.C.), the Ellison Foundation Boston, Grant for Oncology Innovation by Merck-Serono, and National Institutes of Health, National Cancer Institute grant R01 CA196703-01 (R. Chiarle). C.A. is recipient of a postdoctoral fellowship from the Spanish Association Against Cancer.
Authorship
Contribution: R. Choudhari, V.G.M., M.M., and R.P. performed research, analyzed data, and contributed to writing the paper; C.B. provided essential mouse strains; C.V. and M.C. supervised experiments and contributed key reagents; C.A. and R. Chiarle conceived the project, designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for R. Choudhari is Department of Biotechnology, Jnana Shakthi Campus, Torvi, Karnataka State Womens University, Vijayapur-586101, Karnataka, India.
Correspondence: Roberto Chiarle, Department of Pathology, Children’s Hospital Boston and Harvard Medical School, Enders 1116.1, 320 Longwood Ave, Boston, MA 02115; e-mail: roberto.chiarle@childrens.harvard.edu; Chiara Ambrogio, Molecular Oncology Program, Centro Nacional de Investigaciones Oncológicas (CNIO), C/ Melchor Fernández Almagro, 3. 28029 Madrid, Spain; e-mail: cambrogio@cnio.es; and Claudia Voena, Department of Molecular Biotechnology and Health Sciences, University of Torino, Italy, Via Santena 7, 10126 Torino, Italy; e-mail: claudia.voena@unito.it.
References
Author notes
R. Choudhari and V.G.M. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal