Key Points
ATL involves genome-wide reprogramming of the H3K27me3 pattern that is distinct from other cell types.
Druggable epigenetic mechanisms are associated with ATL cell development and HTLV-1–mediated transformation.
Abstract
Adult T-cell leukemia-lymphoma (ATL) shows global gene expression alterations that confer cellular characteristics and unfavorable prognosis. However, molecular mechanisms of the sustained expression changes are largely unknown, because there is no study addressing the relationship between landscapes of the gene expression and epigenetic modifications. Here, we analyzed ATL epigenome and integrated it with transcriptome from primary ATL cells and those from corresponding normal CD4+ T cells to decipher ATL-specific “epigenetic code” that was critical for cell identity. We found that polycomb-repressive complex 2 (PRC2)-mediated trimethylation at histone H3Lys27 (H3K27me3) was significantly and frequently reprogrammed at half of genes in ATL cells. A large proportion of the abnormal gene downregulation was detected at the early stage of disease progression and was explained by H3K27me3 accumulation. The global H3K27me3 alterations involved ATL-specific gene expression changes that included several tumor suppressors, transcription factors, epigenetic modifiers, miRNAs, and developmental genes, suggesting diverse outcomes by the PRC2-dependent hierarchical regulation. Interestingly, a key enzyme, EZH2, was sensitive to promiscuous signaling network including the NF-κB pathway and was functionally affected by human T-cell leukemia virus type I (HTLV-1) Tax. The Tax-dependent immortalized cells showed H3K27me3 reprogramming that was significantly similar to that of ATL cells. Of note, a majority of the epigenetic silencing has occurred in leukemic cells from indolent ATL and also in HTLV-1–infected T cells from asymptomatic HTLV-1 carriers. Because pharmacologic inhibition of EZH2 reversed epigenetic disruption and selectively eliminated leukemic and HTLV-1–infected cells, targeting the epigenetic elements will hold great promise in treatment and prevention of the onset of ATL and HTLV-1–related diseases.
Introduction
Adult T-cell leukemia-lymphoma (ATL) is an aggressive T-cell leukemia/lymphoma refractory to currently available combination chemotherapies.1-6 The unfavorable prognosis results from an inadequate understanding of how diseases are caused and maintained in human T-cell leukemia virus type I (HTLV-1)–infected individuals. Thus far, direct comprehensive analyses of leukemic cells from ATL patients have identified the intrinsic molecular hallmarks of ATL, which comprise changes in gene expression, producing cytokines, genomic abnormalities, sustained signaling, microRNA (miRNA) deregulation, and epigenetic alterations.7-13 Because these organized principles serve the characteristics of ATL itself and are directly associated with its clinical traits, targeting them with mechanism-guided combination therapies may yield more effective and durable treatments for aggressive ATL with fewer adverse effects. There is also an urgent need to prevent the onset of the disease in HTLV-1–endemic areas in ways that selectively eliminate HTLV-1–infected cells.
The Roadmap Epigenomics Project14 has been successful in understanding the human epigenome and its biological impact; tumor development involves the chronic alteration of gene expression owing to epigenetic reprogramming, which includes pattern changes in DNA methylation and covalent histone modifications.15 The heritable chromatin architecture is maintained by the antagonistic actions of the transcriptional activators of the trithorax group (TrxG) proteins and repressors of the polycomb group (PcG) proteins.16 PcG-mediated dynamic control of H3K27 trimethylation (H3K27me3) is central to gene silencing in development, regeneration, stem cell pluripotency, and various cellular processes.17 T cells undergo dynamic PcG-mediated epigenetic changes at lineage-specific genes, such as transcription factors and cytokines.18 Accordingly, the deregulation of PcG generates a specific gene expression pattern responsible for the chronic proliferation, survival, peculiar development, and stemness in various human tumors, including non-Hodgkin lymphomas.19
A PcG-mediated epigenetic mechanism plays an important role for the characteristics of ATL. Enhancer of zeste homolog 2 (EZH2) expression level and the concomitant H3K27me3 mark are altered in ATL cells.11,20 The overexpression of factors associated with PcG induces and maintains the epigenetic silencing of multifunctional miR-31, which activates the NF-κB pathway through NF-κB–inducing kinase (NIK) induction and confers antiapoptotic features on T cells.11 However, no attempt has been made to determine the global epigenomic states explaining the deregulated gene expression specific to ATL. Furthermore, the determinants of PcG and EZH2 activities have not been identified. Mapping the regulatory network of PcG and characterizing its target genes may provide insight into its function in the development and pathogenesis of ATL. Importantly, epigenomic states that maintain and perpetuate tumor characteristics are potentially reversible cellular states that may be unlocked by epigenetic therapy, which is more clinically manageable than targeting irreversible genomic alterations.21
In this study, we performed integrative analyses of the epigenome (n = 3) and transcriptome (n = 58) of primary ATL cells and corresponding normal CD4+ T cells to delineate an overall perspective of epigenetic events and their roles in ATL pathogenesis. We found that PRC2-mediated epigenetic reprogramming is a fundamental characteristic of ATL. Global H3K27me3 gain is a basis of ATL-specific gene expression pattern that includes silencing of several tumor suppressors, transcription factors, epigenetic modifiers, and miRNAs. The epigenetic reprogramming occurs at an early stage of ATL development and seems to be associated with HTLV-1 infection.
Methods
Cell samples
Primary peripheral blood mononuclear cells (PBMCs) from ATL patients (supplemental Table 1, available on the Blood Web site), asymptomatic HTLV-1 carriers, and healthy volunteers were isolated by Ficoll separation and maintained in RPMI 1640 supplemented with 1% to 10% self-serum or fetal bovine serum (FBS), as described previously.9,11 The PBMCs were a part of those collected with informed consent as a collaborative project of the Joint Study on Prognostic Factors of ATL Development (JSPFAD).22 The study was approved by research ethics committee of the University of Tokyo (approval No. 14-155). Normal CD4+ T cells were purified by CD4+ T-cell isolation kit (Miltenyi Biotec) from PBMCs and maintained in RPMI 1640 with 10% FBS. T-cell activation was accomplished by treating the anti-CD3/CD28 antibodies (Miltenyi Biotec) for 2 days.
Microarray analysis
ChIP-on-chip experiments
Chromatin immunoprecipitation (ChIP) was performed according to the Agilent ChIP-on-chip (Coc) protocol (v11.0) with slight modification. Briefly, 1 × 107 cells were chemically crosslinked by 1% formaldehyde and lysed in lysis buffer. The lysates were digested by micrococcal nuclease and sonicated. After centrifugation, the supernatants were subjected to immunoprecipitation by specific antibodies conjugated with Dynabeads (Life Technologies, 11203D for rabbit antibody; 10004D for mouse antibody). After immunoprecipitation, obtained DNA fragments were amplified by ligation-mediated polymerase chain reaction (PCR) and then labeled with Cy3 (for input) or Cy5 (for immunoprecipitation) by SureTag DNA Labeling Kit (Agilent Technologies). The labeling efficiency was verified by nanodrop. Equal amounts of the Cy3- or Cy5-labeled DNA were hybridized to the Agilent G3 promoter array containing 400K probes. The promoter arrays were washed according to the manufacturer’s instructions and then scanned by a Scanner C (Agilent Technologies). The resulting fluorescence intensity was quantified by Feature Extraction 10.7 software and analyzed by Gene Spring 12.0 software (Agilent Technologies). The ChIP-on-chip data are available in the GEO database under the accession number GSE71450. For data analysis, entities were selected based on their signal intensity values (removing very low signals). The data were normalized by 75% centering of intensity distributions of all samples within a batch and by removing any variations based on standard deviation ([SD] <2).
Statistical analyses and bioinformatic analyses
Significant differences in gene expression, histone methylation, and other biological assays between the 2 groups were analyzed by Welch t test. Correlations between 2 groups were analyzed by Pearson’s correlation coefficients, and probabilities of overlap between gene sets were statistically tested, using R version 3.1.3 (http://www.R-project.org/). A value of P < .05 was considered statistically significant. Gene ontology (GO) analysis was performed by DAVID (http://david.abcc.ncifcrf.gov/).24,25 Motif analysis was performed by Human Single Site Analysis of oPOSSUM (http://opossum.cisreg.ca/oPOSSUM3/).26 Archival gene-expression data were obtained from Array Express (http://www.ebi.ac.uk/arrayexpress/).27 The mapping and peak calling of archival ChIP-seq data were processed with Bowtie2 and MACS using SraTailor.28
Establishment of Tax-cell
HTLV-1 Tax cDNA was cloned into a CSII-EF1α-IRES-Venus lentivirus vector.29,30 PHA-stimulated PBMCs were infected with lentivirus vector and cultured in RPMI 1640 with 10% FBS and 10 ng/mL recombinant human IL-2 (R&D Systems). Half the volume of media was changed every 3 days. Venus-positive cells were detected by FACSCalibur (Becton Dickinson).
Results
EZH2-dependent epigenetic abnormalities control ATL cell survival
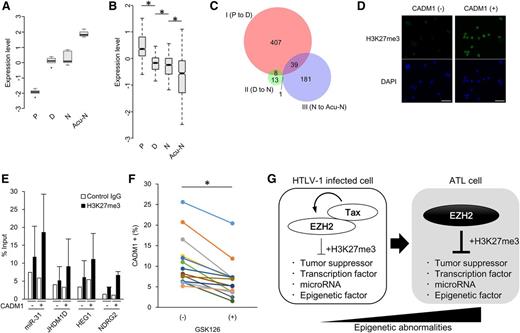
The epigenetic landscape is sensitive to an abundance of the associated factors.31 We previously determined the whole transcriptome of ATL cells.11 Detailed analysis of the genes encoding epigenetic factors defined a “gene signature” of ATL. Core components of PRC2, such as EZH2, SUZ12, EED, and RBBP4, were significantly highly expressed in acute-type ATL cells (n = 26) compared with normal CD4+ T cells (n = 21; Figure 1A), as identified previously.11,20 PRC2 was a unique complex, all core components of which showed increased expression in ATL. Additional genes also exhibited altered expression, including components of PRC1, TrxG, and SWI/SNF. Other genes also varied widely in expression. Remarkably, EZH2 was the most significantly upregulated among genes encoding histone methyltransferase (supplemental Figure 1A). Copy number variation data revealed chromosomal amplification at 7q in 21.4% (36/168) of ATL cases, but there is no focal amplification within EZH2 (supplemental Figure 1B). We performed Sanger sequencing of EZH2 in ATL samples and nevertheless found no ATL clones with the Y641, A677, or A687 mutations that play a critical role in lymphomagenesis (0/50, 0%; Figure 1B).32-34 However, depletion of EZH2 or SUZ12 reduced the growth activity of ATL cell models carrying EZH2 wild-type (WT), suggesting that the overexpression of PRC2 components is epigenetically involved in ATL cell survival (Figure 1C). Treatment for 14 days with GSK126, a specific inhibitor EZH2, reduced the viability of ATL cells. The half-maximal inhibitory concentration (IC50) of GSK126 in TL-Om1 cells was 60.9 nM, which is comparable with that previously observed in diffuse large B-cell lymphoma (DLBCL) cells bearing a gain-of-function EZH2 mutation (Figure 1D). A similar result was obtained in another ATL cell line ATN-1 (supplemental Figure 1C). Interestingly, only 2 to 4 days of treatment significantly reduced viability after drug withdrawal (supplemental Figure 1D). The EZH2 inhibition induced dramatic apoptosis in an ATL cell line (Figure 1E). To test its efficacy in a more relevant model, we treated patient-derived primary ATL cells with an EZH2 inhibitor. Widespread apoptosis was detected among ATL cells but not among normal CD4+ T cells (Figure 1F and supplemental Figure 1E). Furthermore, H3K27me3 was diminished by GSK126 treatment (Figure 1G-H).
ATL-specific transcriptome of epigenetic regulators. (A) Expression pattern of epigenetic factors shown by heat map (left) and mean expression levels (right graph) based on gene-expression profiles of acute-type ATL cells (n = 26) and normal CD4+ T cells (n = 21). (B) Summary of EZH2 mutation status at Y641, A677, and A687 in patient samples from ATL (n = 50) and cell lines. (C) ATL cell lines were transduced with lentivirus vectors carrying short hairpin RNA (shRNA) targeting EZH2 or SUZ12, followed by long-term culture in complete medium. Results as percentage of each Venus-positive population transduced with shRNA series are plotted (n = 3, mean ± SD). (D) Dose-dependent effect of an EZH2 inhibitor GSK126 on cell proliferation in TL-Om1 (ATL-derived) and WSU-DLCL2 (GCB-DLBCL–derived) cells. (E) Apoptosis induction by GSK126 treatment. TL-Om1 cells were treated with 5 μM of GSK126 for 6 days. The apoptotic cells were detected by Annexin V staining. (F) GSK126-dependent apoptosis induction in primary ATL cells. PBMCs from ATL patients (n = 8) were treated with 5 µM of GSK126 for 4 days. The CD4+/Annexin V+ cells were calculated by flow cytometry (P < .05). (G-H) Western blots showing H3K27me3 level in TL-Om1 cells (G) and primary ATL cells (H) treated with GSK126 at indicated concentrations for 6 days. Mean intensity of H3K27me3 blot from 3 ATL samples is shown.
ATL-specific transcriptome of epigenetic regulators. (A) Expression pattern of epigenetic factors shown by heat map (left) and mean expression levels (right graph) based on gene-expression profiles of acute-type ATL cells (n = 26) and normal CD4+ T cells (n = 21). (B) Summary of EZH2 mutation status at Y641, A677, and A687 in patient samples from ATL (n = 50) and cell lines. (C) ATL cell lines were transduced with lentivirus vectors carrying short hairpin RNA (shRNA) targeting EZH2 or SUZ12, followed by long-term culture in complete medium. Results as percentage of each Venus-positive population transduced with shRNA series are plotted (n = 3, mean ± SD). (D) Dose-dependent effect of an EZH2 inhibitor GSK126 on cell proliferation in TL-Om1 (ATL-derived) and WSU-DLCL2 (GCB-DLBCL–derived) cells. (E) Apoptosis induction by GSK126 treatment. TL-Om1 cells were treated with 5 μM of GSK126 for 6 days. The apoptotic cells were detected by Annexin V staining. (F) GSK126-dependent apoptosis induction in primary ATL cells. PBMCs from ATL patients (n = 8) were treated with 5 µM of GSK126 for 4 days. The CD4+/Annexin V+ cells were calculated by flow cytometry (P < .05). (G-H) Western blots showing H3K27me3 level in TL-Om1 cells (G) and primary ATL cells (H) treated with GSK126 at indicated concentrations for 6 days. Mean intensity of H3K27me3 blot from 3 ATL samples is shown.
We identified genes upregulated by EZH2 inhibition (supplemental Figure 2A). The drug-sensitive genes showed high H3K27me3 levels, suggesting that EZH2 inhibition can target genes with accumulated histone methylation (supplemental Figure 2B). EZH2 inhibition diminished H3K27me3 level on the target loci, which in turn reactivated expression of functionally important genes (supplemental Figure 2C-D). The drug effect was promoted by cotreatment with HDAC inhibitor and/or DNA demethylation agent.
These data collectively suggest that ATL cells have abnormal epigenetic profiles depending on the PRC2 deregulation necessary for leukemic cell survival.
Epigenetic characteristics of ATL
We comprehensively analyzed the histone methylation pattern using a Coc assay that covered ∼21 000 gene promoters and internal regions. To ensure the accuracy and biological/clinical significance of the analysis and to extract disease-associated epigenetic variations, we used primary ATL cells with a proviral load of >100% (n = 3) and >94% pure resting CD4+ T cells isolated from healthy volunteers (n = 2). We confirmed the validity of the Coc results relative to those derived from normal T cells by comparing them with archival ChIP-seq data (supplemental Figure 3A).35
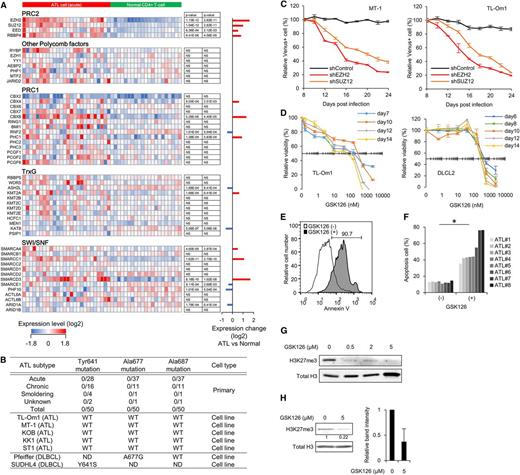
A direct comparison revealed significant changes of H3K27me3 at half of the genes in ATL cells, among which genes associated with H3K27me3 gain were frequent (Figure 2A and supplemental Figure 3B-C). Methylation intensity profiling clearly showed epigenetic convergence in ATL cells. H3K27-hypomethylated regions in resting T cells were hypermethylated in ATL cells (Figure 2B and supplemental Figure 3D).36 Hierarchical clustering based on the histone methylation pattern identified common regions with altered H3K27me3/H3K4me3 and revealed the phenotypic threshold (Figure 2C). ATL frequently showed abnormal accumulation of H3K27me3, which was associated with H3K4me3 loss (Figure 2D and supplemental Figure 3E-F). A gene subset specifically associated with H3K27me3 in ATL cells overlapped poorly with those of embryonic stem cells or DLBCL cells (Figure 2E). Thus, ATL cells exhibit a genome-wide alteration of H3K27me3, the pattern of which differs from previously identified EZH2-dependent cell types.
Epigenetic alterations in primary ATL cells. (A) Overall percentage of genes with H3K27me3 states in ATL cells compared with normal CD4+ T cells. The averaged methylation values at each gene (fold change [FC] > ±1.5 vs normal CD4+ T cells) were calculated from normalized probes. (B) Methylation intensity profiling of normalized each probe in ATL cells (n = 3) and normal CD4+ T cells (n = 2). Relative H3K27me3 levels of each probe (P < .05) are shown in histogram. (C) Heat maps and hierarchical clustering of H3K27me3 (left) and H3K4me3 (right) levels at regions (LogFC > 3) in primary ATL cells, normal T cells, and TL-Om1 cells. (D) The number of probes showing gain (LogFC > 3) or loss (LogFC < −3) of H3K27me3 and H3K4me3 in ATL cells compared with normal CD4+ T cells. (E) Venn diagram showing the overlap between genes associated with H3K27me3 in ATL cells (FC > 2 vs normal CD4+ T cells), embryonic stem cells,36 and DLBCL cells.32 (F-G) Composite enrichment profile of H3K27me3 relative to the closest transcriptional start site (TSS). Graphs show relative H3K27me3 levels calculated from all normalized probes (F) and frequency of H3K27me3 gain in ATL compared with normal CD4+ T cells (G; closed circles indicate gain in ATL [P < .05]; open circles indicate loss in ATL [P < .05]). (H) Box plots showing relative H3K27me3 levels at silenced promoter region (expression FC < −2 in acute-type ATL vs normal CD4+ T cells, P < .001) and gene-enhancer region (defined by H3K4me1+H3K27ac marks in CD4+ T cells14 ) in ATL and normal T cells.
Epigenetic alterations in primary ATL cells. (A) Overall percentage of genes with H3K27me3 states in ATL cells compared with normal CD4+ T cells. The averaged methylation values at each gene (fold change [FC] > ±1.5 vs normal CD4+ T cells) were calculated from normalized probes. (B) Methylation intensity profiling of normalized each probe in ATL cells (n = 3) and normal CD4+ T cells (n = 2). Relative H3K27me3 levels of each probe (P < .05) are shown in histogram. (C) Heat maps and hierarchical clustering of H3K27me3 (left) and H3K4me3 (right) levels at regions (LogFC > 3) in primary ATL cells, normal T cells, and TL-Om1 cells. (D) The number of probes showing gain (LogFC > 3) or loss (LogFC < −3) of H3K27me3 and H3K4me3 in ATL cells compared with normal CD4+ T cells. (E) Venn diagram showing the overlap between genes associated with H3K27me3 in ATL cells (FC > 2 vs normal CD4+ T cells), embryonic stem cells,36 and DLBCL cells.32 (F-G) Composite enrichment profile of H3K27me3 relative to the closest transcriptional start site (TSS). Graphs show relative H3K27me3 levels calculated from all normalized probes (F) and frequency of H3K27me3 gain in ATL compared with normal CD4+ T cells (G; closed circles indicate gain in ATL [P < .05]; open circles indicate loss in ATL [P < .05]). (H) Box plots showing relative H3K27me3 levels at silenced promoter region (expression FC < −2 in acute-type ATL vs normal CD4+ T cells, P < .001) and gene-enhancer region (defined by H3K4me1+H3K27ac marks in CD4+ T cells14 ) in ATL and normal T cells.
H3K27me3 peaks were enriched around the transcriptional start site (TSS) (Figure 2F). ATL-specific methylation gains were enriched at around −3000 bp region from the TSS, a region that carries multiple sequence elements related to general transcriptional regulation (Figure 2G). The methylation gains were detected at silenced promoters, gene enhancers (Figure 2H), and genes with CpG islands (supplemental Figure 3G). The basal methylation level was high at genes without CpG islands in both cases.
Epigenetic reprograming of ATL-specific gene expression
We combined the Coc results with the gene expression data. More than half (54.6%) of the genes downregulated in ATL cells were associated with H3K27me3 gain (Figure 3A). The H3K27me3 level showed a moderate inverse correlation with their expression (r = −0.36, P < 1E−15; Figure 3B-C, supplemental Figure 4, and supplemental Table 2). H3K27me3 gain was associated with downregulation of gene expression. This trend highlighted the gene signature of ATL; ATL-specific H3K27me3 gains were observed at frequently downregulated genes, which were involved in the characteristics of ATL such as NDRG2, CDKN1A, ZEB1, BCL2L11 (BIM), CD7, and other tumor suppressors.12,13,37-40 Conversely, H3K27me3 loss was associated with the upregulation of genes such as the ATL-specific surface marker CADM123,41 and Hedgehog signaling-linked EVC1/242 (supplemental Table 3). These H3K27me3-gain–associated genes showed a stepwise decrease during disease progression, suggesting the involvement of EZH2-dependent epigenetic alteration in the development and pathogenesis of ATL (Figure 3D). The majority of the methylated gene set was commonly suppressed in both indolent and aggressive ATL (Figure 3E and supplemental Table 2). Suppression intensity was associated with disease progression. The clinical subtypes of ATL could be stratified based on the epigenetically silenced gene set.
Integrative analysis of gene expression and H3K27me3 profiles. (A) Overall percentage of H3K27me3 alterations (Coc analysis FC > ±1.5 in ATL samples with >100% proviral load vs normal CD4+ T cells) in downregulated genes (expression profile FC < −1.5 in acute-type ATL vs normal CD4+ T cells, P < .001) in ATL cells. (B) Scatter plot showing gene expression changes (x-axis, P < .001) and their relative H3K27me3 changes (y-axis, averaged values from normalized probes) in ATL cells compared with normal CD4+ T cells. (C) Box plots showing relative expression levels of genes (ATL vs normal CD4+ T cells, P < .001) with H3K27me3 gain (Coc FC > 1.5 in ATL) and loss (Coc FC < −1.5 in ATL) in acute-type ATL and normal CD4+ T cells (*P < .001). (D) Box plot showing relative expression levels of H3K27me3-associated silenced gene set in ATL cells with each clinical subtype compared with normal CD4+ T cells (*P < 1E−10). (E) Venn diagram showing H3K27me3-associated (Coc FC > 1) suppressed gene set (expression FC < −1.5, P < .05) during clinical progression of ATL. (F) Gene ontology analysis of H3K27me3-associated suppressed 856 genes (defined in supplemental Figure 4). P values (left) and gene counts (right, clinical subtypes are indicated by color) of the top 10 categories are shown. (G) Epigenetic reprogramming of KDM family genes in ATL. The heat map and middle graph show expression patterns of KDM family in acute-type ATL cells and normal CD4+ T cells. H3K27me3 alterations are shown in the right graph. (H) Venn diagram showing miRNAs downregulated in ATL (white circle, miRNA array FC < −1.5, P < .05) and those associated with H3K27me3 gain in ATL cells compared with normal CD4+ T cells (gray circle, Coc FC > 1). (I) Sequence motif analysis at miRNA gene regions epigenetically downregulated in ATL. Top 5 transcription factors and Fisher scores are provided. (J) Relative expression levels of H3K27me3-associated suppressed 75 miRNAs defined in (H) (*P < .0001). (K) ATL-specific epigenetic reprogramming of HOXA cluster. Top, expression levels of HOXA genes in ATL cells relative to normal CD4+ T cells (solid bars indicate P < .05). Bottom, relative H3K27me3 levels at HOXA loci in primary ATL cells, TL-Om1 cells, and normal CD4+ T cells.
Integrative analysis of gene expression and H3K27me3 profiles. (A) Overall percentage of H3K27me3 alterations (Coc analysis FC > ±1.5 in ATL samples with >100% proviral load vs normal CD4+ T cells) in downregulated genes (expression profile FC < −1.5 in acute-type ATL vs normal CD4+ T cells, P < .001) in ATL cells. (B) Scatter plot showing gene expression changes (x-axis, P < .001) and their relative H3K27me3 changes (y-axis, averaged values from normalized probes) in ATL cells compared with normal CD4+ T cells. (C) Box plots showing relative expression levels of genes (ATL vs normal CD4+ T cells, P < .001) with H3K27me3 gain (Coc FC > 1.5 in ATL) and loss (Coc FC < −1.5 in ATL) in acute-type ATL and normal CD4+ T cells (*P < .001). (D) Box plot showing relative expression levels of H3K27me3-associated silenced gene set in ATL cells with each clinical subtype compared with normal CD4+ T cells (*P < 1E−10). (E) Venn diagram showing H3K27me3-associated (Coc FC > 1) suppressed gene set (expression FC < −1.5, P < .05) during clinical progression of ATL. (F) Gene ontology analysis of H3K27me3-associated suppressed 856 genes (defined in supplemental Figure 4). P values (left) and gene counts (right, clinical subtypes are indicated by color) of the top 10 categories are shown. (G) Epigenetic reprogramming of KDM family genes in ATL. The heat map and middle graph show expression patterns of KDM family in acute-type ATL cells and normal CD4+ T cells. H3K27me3 alterations are shown in the right graph. (H) Venn diagram showing miRNAs downregulated in ATL (white circle, miRNA array FC < −1.5, P < .05) and those associated with H3K27me3 gain in ATL cells compared with normal CD4+ T cells (gray circle, Coc FC > 1). (I) Sequence motif analysis at miRNA gene regions epigenetically downregulated in ATL. Top 5 transcription factors and Fisher scores are provided. (J) Relative expression levels of H3K27me3-associated suppressed 75 miRNAs defined in (H) (*P < .0001). (K) ATL-specific epigenetic reprogramming of HOXA cluster. Top, expression levels of HOXA genes in ATL cells relative to normal CD4+ T cells (solid bars indicate P < .05). Bottom, relative H3K27me3 levels at HOXA loci in primary ATL cells, TL-Om1 cells, and normal CD4+ T cells.
GO analysis revealed that genes epigenetically suppressed by H3K27me3 were enriched in some biological processes, including transcriptional regulation and lymphocyte activation (Figure 3F). The gene set comprises various transcription factors and zinc finger proteins, the functions of a large part of which remain unknown (supplemental Figure 5A). The suppression was specific to ATL and not enriched in embryonic stem and DLBCL cells (supplemental Figure 5B). In addition, genes encoding histone demethylases were newly identified as suppressed by H3K27me3 (Figure 3G and supplemental Figure 5C). Expression of KDM6B, which encodes a JMJD3 demethylase of H3K27me3,43 was significantly downregulated with H3K27me3 gain in all cases (supplemental Figure 5D-F). KDM6B knockdown resulted in H3K27me3 accumulation, suggesting that the generation and maintenance of global H3K27me3 mark is further supported by H3K27me3-dependent KDM6B silencing (supplemental Figure 5G). The reciprocal relationship between KDM6B and EZH2 was not observed in other cancers that depend on H3K27me3 (Oncomine database). ATL cells appear to acquire the coherent pattern that produces and maintains the systematic abnormality of H3K27me3.
We previously observed a global loss of miRNAs in ATL.11 An integrative analysis of H3K27me3 mapping and miRNA pattern showed that miRNAs downregulated in ATL clearly overlapped with those associated with high H3K27me3 (P < 1E−30; Figure 3H and supplemental Figure 6A). Promoter regions of the suppressed miRNAs harbored specific motifs recognized by transcription factors such as ZEB1, MZF1, and YY1 (Figure 3I). Genes harboring YY1 binding sites showed slightly higher H3K27me3 level in both ATL and normal T cells (supplemental Figure 6B) but not in DLBCL cells.44 The epigenetic suppression of miRNAs may be associated with early ATL development through potential interactions with several signaling pathways (Figure 3J and supplemental Figure 6C).
The epigenetic reprogramming also included specific H3K27me3 loss (Figure 2A,C). Interestingly, the loss of H3K27me3 was associated with HOXA cluster transcription in ATL (Figure 3K). HOXA genes are known to be involved in lymphocyte differentiation and survival in normal and malignant cells.45,46 Acute-type ATL cells showed higher levels of HOXA (supplemental Figure 6D). Collectively, global changes of H3K27me3 are involved in the determination of an ATL-specific gene-expression program that includes transcription factors, epigenetic modifiers, miRNAs, and developmental genes.
Constitutively active NF-κB pathway induces EZH2 overexpression
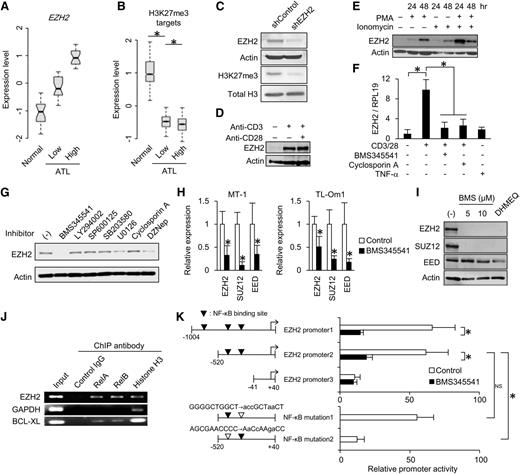
Given that EZH2 upregulation is critical for H3K27me3-mediated epigenetic repression (Figure 4A-B)31,47 and maintaining overall H3K27me3 level (Figure 4C), increased EZH2 expression may be indispensable for the maintenance of H3K27me3 in ATL. We identified a close relationship between lymphocyte activation and EZH2 transcription. Although the level of EZH2 expression was low in resting T cells (Figure 1A),48 lymphocyte activation by anti-CD3/CD28 stimulation or polymethacrylate/ionomycin dramatically induced EZH2 expression (Figure 4D-E). Pre-inhibition of NF-κB or Ca2+, which are involved in important pathways downstream of the T-cell receptor, abolished EZH2 induction (Figure 4F). Solo stimulation with tumor necrosis factor-α did not increase EZH2 expression, suggesting that sufficient EZH2 induction requires the complex T-cell signaling network in physiologic condition. In parallel, we found that NF-κB activation plays a critical role in the chronic expression of EZH2 in ATL cells. The IKKβ inhibitor BMS345541 significantly repressed EZH2 expression (Figure 4G-H).49 Furthermore, 2 independent NF-κB inhibitors effectively reduced EZH2 in leukemic cells derived from 3 patients with ATL (Figure 4I and supplemental Figure 7). Interestingly, BMS345541 inhibited the expression of SUZ12 and EED that were overexpressed in ATL cells (Figure 1A). ChIP assay demonstrated the direct binding of both RelA and RelB to the promoter region of EZH2 (Figure 4J). An EZH2 promoter reporter assay revealed that a NF-κB binding site within −500 bp upstream of EZH2 TSS significantly contributed to its transcription, which was effectively prevented by IKKβ inhibition (Figure 4K). These results demonstrate that EZH2 overexpression is induced by sustained NF-κB activation.
Regulatory mechanism of EZH2 expression. (A-B) Relationship between the levels of EZH2 and epigenetically suppressed gene set. ATL patient samples were divided into 2 groups according to EZH2 level. Box plots show expression levels of EZH2 (A) and H3K27me3 target genes (B) in normal CD4+ T cells (n = 21) and ATL cells (EZH2 low, n = 25; EZH2 high, n = 25) (*P < .001). (C) EZH2 and H3K27me3 levels in TL-Om1 with shcontrol or shEZH2 analyzed by western blotting. (D) EZH2 expression level in CD4+ T cells that were unstimulated, stimulated with anti-CD3 antibody, or stimulated with anti-CD3/CD28 antibodies for 48 hours. (E) EZH2 expression level in CD4+ T cells that were unstimulated or stimulated with polymethacrylate and/or ionomycin for indicated times. (F) EZH2 mRNA level in CD3+ T cells that were unstimulated or stimulated with anti-CD3/CD28 antibodies or tumor necrosis factor-α in the presence or absence of pretreatment (2 hours before stimulation) with NF-κB or NFAT inhibitor (n = 3, mean ± SD, P < .05). (G) EZH2 level in MT-1 cells that were treated with intracellular signaling inhibitor series (10 μM BMS345541, 20 μM LY294002, 20 μM SP600125, 20 μM SB203580, 20 μM U0126, 1 μg/mL cyclosporin A) for 48 hours. DZNep (5 μM) was used as a positive control.49 (H) mRNA levels of PRC2 core components in MT-1 and TL-Om1 cells in the presence or absence of IKKβ inhibitor BMS345541 for 12 hours (n = 3, mean ± SD, P < .05). (I) Western blots showing PRC2 core component levels in primary ATL samples that were treated with BMS345541 (5 or 10 μM) and DHMEQ (10 μg/mL) for 48 hours. (J) PCR-ChIP assay detected RelA and RelB bindings on EZH2 promoter region in TL-Om1 cells. Promoter regions of GAPDH and BCL-XL genes were used as a negative or a positive control, respectively. (K) EZH2-promoter activity in the presence or absence of IKKβ inhibitor BMS345541 for 12 hours analyzed by dual-luciferase reporter assay (n = 4, mean ± SD, P < .05). The role of putative NF-κB binding sites was evaluated by mutant reporters (indicated by open triangles).
Regulatory mechanism of EZH2 expression. (A-B) Relationship between the levels of EZH2 and epigenetically suppressed gene set. ATL patient samples were divided into 2 groups according to EZH2 level. Box plots show expression levels of EZH2 (A) and H3K27me3 target genes (B) in normal CD4+ T cells (n = 21) and ATL cells (EZH2 low, n = 25; EZH2 high, n = 25) (*P < .001). (C) EZH2 and H3K27me3 levels in TL-Om1 with shcontrol or shEZH2 analyzed by western blotting. (D) EZH2 expression level in CD4+ T cells that were unstimulated, stimulated with anti-CD3 antibody, or stimulated with anti-CD3/CD28 antibodies for 48 hours. (E) EZH2 expression level in CD4+ T cells that were unstimulated or stimulated with polymethacrylate and/or ionomycin for indicated times. (F) EZH2 mRNA level in CD3+ T cells that were unstimulated or stimulated with anti-CD3/CD28 antibodies or tumor necrosis factor-α in the presence or absence of pretreatment (2 hours before stimulation) with NF-κB or NFAT inhibitor (n = 3, mean ± SD, P < .05). (G) EZH2 level in MT-1 cells that were treated with intracellular signaling inhibitor series (10 μM BMS345541, 20 μM LY294002, 20 μM SP600125, 20 μM SB203580, 20 μM U0126, 1 μg/mL cyclosporin A) for 48 hours. DZNep (5 μM) was used as a positive control.49 (H) mRNA levels of PRC2 core components in MT-1 and TL-Om1 cells in the presence or absence of IKKβ inhibitor BMS345541 for 12 hours (n = 3, mean ± SD, P < .05). (I) Western blots showing PRC2 core component levels in primary ATL samples that were treated with BMS345541 (5 or 10 μM) and DHMEQ (10 μg/mL) for 48 hours. (J) PCR-ChIP assay detected RelA and RelB bindings on EZH2 promoter region in TL-Om1 cells. Promoter regions of GAPDH and BCL-XL genes were used as a negative or a positive control, respectively. (K) EZH2-promoter activity in the presence or absence of IKKβ inhibitor BMS345541 for 12 hours analyzed by dual-luciferase reporter assay (n = 4, mean ± SD, P < .05). The role of putative NF-κB binding sites was evaluated by mutant reporters (indicated by open triangles).
HTLV-1 Tax disrupts the host epigenome
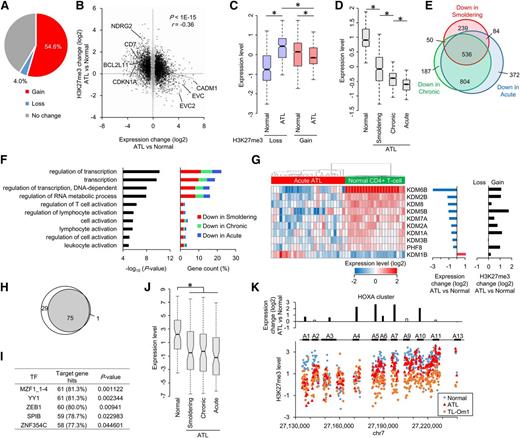
Inspired by the mechanism of EZH2 deregulation, we next addressed the role of Tax in active EZH2 transcription. Tax, but not other HTLV-1 proteins such as Rex, p30, or HBZ, enhanced EZH2 promoter activity (Figure 5A). This induction was blocked by treatment with DHMEQ or the MEK inhibitor U0126, as well as by NF-κB–deficient mutation (M22) (Figure 5B).
Biological interaction between EZH2 and HTLV-1 Tax. (A) EZH2 promoter activity in the presence or absence of HTLV-1 genes. 293FT cells were transfected with plasmid vectors carrying EZH2 promoter reporter and indicated HTLV-1 cDNAs. The luciferase activities were quantified at 48 hours post-transfection (n = 3, mean ± SD, P < .05). (B) Tax-dependent EZH2 gene activation analyzed by luciferase reporter assay. Signaling inhibitors (DHEMQ for NF-κB inhibition; cyclosporin A [CsA] for NAFT inhibition; U0126 for ERK1/2 inhibition) were cotreated. A Tax M22 was used as a NF-κB–deficient mutant (n = 3, mean ± SD, P < .01). (C-D) Interaction between Tax and PRC2. 293T cells were transfected with plasmids for expression of FLAG-Tax (C), or FLAG-EZH2 and untagged Tax (D), respectively. Nuclear cell extracts from each sample were immunoprecipitated with an anti-FLAG antibody. Coprecipitated proteins were prepared and analyzed by western blotting with indicated antibodies. H.C., IgG heavy chain. (E) Pull-down assay with GST-EZH2. Expression vectors for deletion mutant series of GST-tagged EZH2 were established (top). 293T cells were transfected with the plasmids encoding GST-EZH2 series and untagged Tax, and then subjected in GST pull-down assay. Purified GST-EZH2 series and copurified PRC2 components were detected by western blotting with indicated antibodies (bottom). (F) Co-immunoprecipitation (Co-IP) between endogenous Tax and EZH2 in HTLV-1–infected HUT102 cells. The same amounts of anti-EZH2 antibody and non-specific IgG were used in the immunoprecipitation, followed by detection with indicated antibodies. (G) Confocal microscopy data showing subcellular localizations of endogenous EZH2 and Tax in HUT102. The scale bar indicates 10 μm. Arrowheads indicate colocalized spots. (H) Confocal microscopy data showing subcellular localizations of endogenous EZH2 and ectopically expressed viral proteins that were detected by anti-EZH2 and anti-FLAG antibodies, respectively, in HeLa cells. The scale bars indicate 10 μm. Arrowheads indicate colocalized spots. (I) Venn diagram showing the overlap between genes (Coc analysis FC > 0 vs control IgG) associated with EZH2 occupancy, H3K27me3 accumulation, and Tax binding in HTLV-1–infected MT-2 cells. (J) Composite enrichment profiles of Tax, EZH2, and H3K27me3 relative to the closest TSS at Tax-bound genes (solid line, 5377 genes, Coc FC > 0 vs control IgG) and Tax-unbound genes (dashed line, 14 937 genes, Coc FC < 0 vs control IgG) in MT-2 cells.
Biological interaction between EZH2 and HTLV-1 Tax. (A) EZH2 promoter activity in the presence or absence of HTLV-1 genes. 293FT cells were transfected with plasmid vectors carrying EZH2 promoter reporter and indicated HTLV-1 cDNAs. The luciferase activities were quantified at 48 hours post-transfection (n = 3, mean ± SD, P < .05). (B) Tax-dependent EZH2 gene activation analyzed by luciferase reporter assay. Signaling inhibitors (DHEMQ for NF-κB inhibition; cyclosporin A [CsA] for NAFT inhibition; U0126 for ERK1/2 inhibition) were cotreated. A Tax M22 was used as a NF-κB–deficient mutant (n = 3, mean ± SD, P < .01). (C-D) Interaction between Tax and PRC2. 293T cells were transfected with plasmids for expression of FLAG-Tax (C), or FLAG-EZH2 and untagged Tax (D), respectively. Nuclear cell extracts from each sample were immunoprecipitated with an anti-FLAG antibody. Coprecipitated proteins were prepared and analyzed by western blotting with indicated antibodies. H.C., IgG heavy chain. (E) Pull-down assay with GST-EZH2. Expression vectors for deletion mutant series of GST-tagged EZH2 were established (top). 293T cells were transfected with the plasmids encoding GST-EZH2 series and untagged Tax, and then subjected in GST pull-down assay. Purified GST-EZH2 series and copurified PRC2 components were detected by western blotting with indicated antibodies (bottom). (F) Co-immunoprecipitation (Co-IP) between endogenous Tax and EZH2 in HTLV-1–infected HUT102 cells. The same amounts of anti-EZH2 antibody and non-specific IgG were used in the immunoprecipitation, followed by detection with indicated antibodies. (G) Confocal microscopy data showing subcellular localizations of endogenous EZH2 and Tax in HUT102. The scale bar indicates 10 μm. Arrowheads indicate colocalized spots. (H) Confocal microscopy data showing subcellular localizations of endogenous EZH2 and ectopically expressed viral proteins that were detected by anti-EZH2 and anti-FLAG antibodies, respectively, in HeLa cells. The scale bars indicate 10 μm. Arrowheads indicate colocalized spots. (I) Venn diagram showing the overlap between genes (Coc analysis FC > 0 vs control IgG) associated with EZH2 occupancy, H3K27me3 accumulation, and Tax binding in HTLV-1–infected MT-2 cells. (J) Composite enrichment profiles of Tax, EZH2, and H3K27me3 relative to the closest TSS at Tax-bound genes (solid line, 5377 genes, Coc FC > 0 vs control IgG) and Tax-unbound genes (dashed line, 14 937 genes, Coc FC < 0 vs control IgG) in MT-2 cells.
During pathologic processes, Tax can associate with numerous host factors, including histone modifiers.12,50 We performed coimmunoprecipitation assays and found that all of the core components of PRC2 could associate with Tax (Figure 5C-D). We generated a series of GST-EZH2 mutants and applied them in a pull-down assay. Tax physically interacted with the N- and C-termini of EZH2 and did not affect PRC2 composition (Figure 5E). This association between Tax and endogenous PRC2 components was detected in HTLV-1–infected cells (Figure 5F-G). Only Tax colocalized with endogenous EZH2 (Figure 5H and supplemental Figure 8).
To address the Tax-dependent epigenome, we performed Coc analysis in Tax-expressing MT-2 cells by using anti-H3K27me3, anti-EZH2, and anti-Tax antibodies (supplemental Figure 9A). More than half of the Tax-bound genes overlapped with EZH2- or H3K27me3-associated genes (Figure 5I). Remarkably, Tax binding was positively correlated with EZH2 occupancy and H3K27me3 accumulation (Figure 5J).
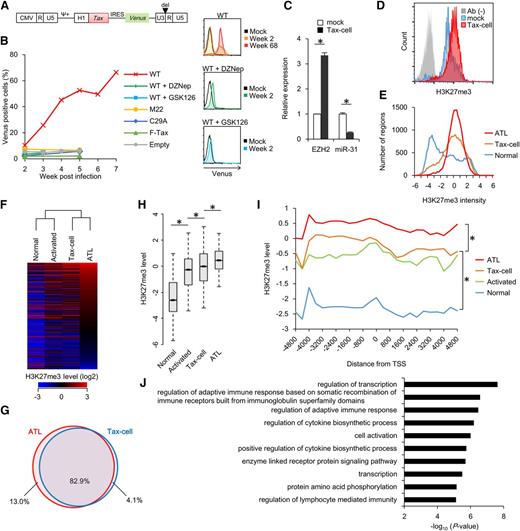
To further investigate the role of Tax on the host epigenome, we established Tax-expressing lentiviral vector series and introduced them into primary PBMCs (Figure 6A). The Venus-positive transduced cells rapidly expanded and were then immortalized by WT Tax expression (Figure 6B and supplemental Figure 9B).51 The functional importance of NF-κB and cAMP response element-binding protein activities in this process was supported by results from Tax mutants.52,53 EZH2 inhibition completely prevented cell growth, indicating that the enzymatic activity of EZH2 is necessary for Tax-dependent cell growth and subsequent immortalization, which is a relevant surrogate for studying the epigenome of preleukemic cells. In the immortalized cells (Tax-cells), EZH2 expression was increased and miR-31 expression was reciprocally decreased (Figure 6C). In addition, nuclear localization of NF-κB was detected (supplemental Figure 9C). Intracellular staining revealed higher H3K27me3 level in Tax-cells (Figure 6D).
Epigenetic alterations caused by HTLV-1 Tax. (A) Construction of Tax-expressing lentiviral vector. Tax-IRES-Venus expression is controlled by EF1α promoter. (B) Left: graph showing proportion of Venus+ cells in PBMC cultures that were transduced with Tax WT, Tax M22 (NF-κB–deficient), Tax C29A (nuclear localization–deficient), F-Tax (CREB-deficient), or empty lentivirus. DZNep (500 nM) or GSK126 (1 μM) were cotreated for 21 days to inhibit endogenous EZH2. Right: representative flow cytometry showing expansion of Venus+ (Tax-expressing) population by Tax expression, which was inhibited by GSK126 or DZNep treatment. PBMCs from 1 of 4 healthy donors was reproducibly immortalized by Tax expression, which is consistent with a previous report.51 (C) Expression levels of EZH2 and miR-31 in Tax-dependent immortalized cells (Tax-cell, 5 weeks post lentivirus transduction) quantified by quantitative reverse-transcription PCR (n = 3, mean ± SD, P < .05). (D) Representative flow cytometry showing H3K27me3 level in mock (activated PBMCs) and Tax-cell (30 weeks post-transduction). Staining without primary antibody was used as a negative control. (E) Methylation intensity profiling of each probe in ATL cells, normal CD4+ T cells, and Tax-cell. Relative H3K27me3 levels of normalized each probe (P < .05) are shown in histogram. (F) Heat map and hierarchical clustering of H3K27me3 level at regions (P < .05) in ATL cells, Tax-cell, activated CD4+ T cells, and control resting CD4+ T cells. (G) Venn diagram showing overlap between regions with H3K27me3 gain (P < .05) in ATL cells and Tax-cell compared with normal CD4+ T cells. (H) Box pot showing relative H3K27me3 levels at the overlapped 82.9% regions defined in (G) in resting and activated CD4+ T cells, Tax-cell, and ATL cells (*P < 1E−10). (I) Composite H3K27 methylation profiles of 12 050 probes that showed H3K27me3 gain in ATL cells compared with normal CD4+ T cells. (J) P value showing results of gene ontology analysis of genes that were associated with H3K27me3 gain in ATL and Tax-cells defined in (G) and downregulated in acute-type ATL (expression profile FC < −1.5, P < .05) compared with normal CD4+ T cells.
Epigenetic alterations caused by HTLV-1 Tax. (A) Construction of Tax-expressing lentiviral vector. Tax-IRES-Venus expression is controlled by EF1α promoter. (B) Left: graph showing proportion of Venus+ cells in PBMC cultures that were transduced with Tax WT, Tax M22 (NF-κB–deficient), Tax C29A (nuclear localization–deficient), F-Tax (CREB-deficient), or empty lentivirus. DZNep (500 nM) or GSK126 (1 μM) were cotreated for 21 days to inhibit endogenous EZH2. Right: representative flow cytometry showing expansion of Venus+ (Tax-expressing) population by Tax expression, which was inhibited by GSK126 or DZNep treatment. PBMCs from 1 of 4 healthy donors was reproducibly immortalized by Tax expression, which is consistent with a previous report.51 (C) Expression levels of EZH2 and miR-31 in Tax-dependent immortalized cells (Tax-cell, 5 weeks post lentivirus transduction) quantified by quantitative reverse-transcription PCR (n = 3, mean ± SD, P < .05). (D) Representative flow cytometry showing H3K27me3 level in mock (activated PBMCs) and Tax-cell (30 weeks post-transduction). Staining without primary antibody was used as a negative control. (E) Methylation intensity profiling of each probe in ATL cells, normal CD4+ T cells, and Tax-cell. Relative H3K27me3 levels of normalized each probe (P < .05) are shown in histogram. (F) Heat map and hierarchical clustering of H3K27me3 level at regions (P < .05) in ATL cells, Tax-cell, activated CD4+ T cells, and control resting CD4+ T cells. (G) Venn diagram showing overlap between regions with H3K27me3 gain (P < .05) in ATL cells and Tax-cell compared with normal CD4+ T cells. (H) Box pot showing relative H3K27me3 levels at the overlapped 82.9% regions defined in (G) in resting and activated CD4+ T cells, Tax-cell, and ATL cells (*P < 1E−10). (I) Composite H3K27 methylation profiles of 12 050 probes that showed H3K27me3 gain in ATL cells compared with normal CD4+ T cells. (J) P value showing results of gene ontology analysis of genes that were associated with H3K27me3 gain in ATL and Tax-cells defined in (G) and downregulated in acute-type ATL (expression profile FC < −1.5, P < .05) compared with normal CD4+ T cells.
Coc profiling identified aberrant H3K27me3 gains in Tax-cells (Figure 6E). The genome-wide H3K27me3 pattern in Tax-cells was significantly similar to that of ATL cells (Figure 6F). The regions of H3K27me3 accumulation significantly overlapped in ATL cells and Tax-cells (Figure 6G). In the common region, H3K27me3 increased stepwise during immortalization and disease progression (Figure 6H). H3K27me3 accumulation was observed at around −3000 bp from the TSS in Tax-cells, although the level of this accumulation was lower than that observed in ATL cells (Figure 6I), implying a Tax-independent mechanism during ATL development.
The genes with high H3K27me3 level in both ATL cells and Tax-cells were significantly associated with transcription regulation and other biological processes, including NDRG2, BCL2L11, and CDKN1A (Figure 6J). The CDKN1A promoter region exhibited a high level of H3K27me3, and EZH2 inhibition restored the CDKN1A expression in Tax-cells (Figure 3B and supplemental Figure 9D-G). Thus, it is conceivable that Tax contributes to ATL development by disrupting the host epigenetic machinery.
Epigenetic alterations already occur in HTLV-1–infected cells
Regarding the epigenetic characteristics of ATL and Tax-expressing cells, we hypothesized that epigenetic abnormalities may accumulate in leukemic cells from indolent-type ATL and in HTLV-1–infected cells from asymptomatic HTLV-1 carriers. To directly assess the “epigenetic hit” in the subpopulation, we exploited experimentally validated specific surface markers.23 CD4+ T cells from HTLV-1–infected carriers were sorted into 3 subpopulations according to their cell surface CD7 and CADM1 expression patterns (P, CD7+/CADM1−; D, CD7+/CADM1+; N, CD7−/CADM1+). The HTLV-1–infected cells are enriched in the D and N fractions, in which miR-31 is significantly silenced.23 EZH2 expression was fourfold higher in the D and N fractions than in the P fraction of the cells from indolent ATL and HTLV-1 carriers (Figure 7A). The N fraction from aggressive acute-type ATL showed the highest EZH2 expression. The gene set comprising the epigenetically silenced genes in aggressive ATL was selectively decreased in HTLV-1–infected D and N subpopulations from indolent ATL and HTLV-1 carriers (Figure 7B). We classified the gene set into 3 subgroups according to disease stages. The majority of downregulated genes with high H3K27me3 in ATL belonged to stage I, providing further evidence that HTLV-1 infection initiates H3K27me3 accumulation in HTLV-1–infected individuals (Figure 7C). The gene set downregulated at stage I showed higher methylation level and corresponding lower expression at the acute phase (supplemental Figure 10A).
Epigenetic alteration in HTLV-1–infected cells within asymptomatic carriers. (A) Box plot showing EZH2 expression in subpopulations from HTLV-1–infected individuals. P, D, and N represent subpopulations from indolent ATL (n = 3) and asymptomatic HTLV-1 carriers (n = 2). P indicates CD7+/CADM1− cells; D indicates CD7+/CADM1+ cells; N indicates CD7−/CADM1+ cells. Acu-N indicates CD7−/CADM1+ cells from acute-type ATL. (B) Box plot showing relative expression levels of the H3K27me3-dependent silenced 856 genes (defined in supplemental Figure 4) in the subpopulations from HTLV-1–infected individuals and acute-type ATL (*P < 1E−4). (C) Venn diagram showing the relationship between disease progression and H3K27me3 gain. The 3 subgroups were classified according to the disease stages (stage I indicates P to D progression; stage II indicates D to N progression; stage III indicates indolent N to acute-type N progression). Each gene set was defined as (1) H3K27me3 gain (LogFC > 1) in acute-type ATL compared with normal CD4+ T cells and (2) decreased expression (FC < −1.5, P < .05) at the stages. (D) Microscopy data showing representative H3K27me3 level in CD4+/CADM1+ and CD4+/CADM1− subpopulations within asymptomatic carriers (n = 3). The scale bars indicate 20 μm. (E) Graph showing the results of μChIP assay with anti-H3K27me3 antibody and control IgG in CD4+/CADM1+ and CD4+/CADM1− subpopulations derived from asymptomatic carriers (n = 3, mean ± SD). Promoter region of the H3K27me3-regulated genes were tested. (F) Graph showing the population size of CD4+/CADM1+ cells in the presence or absence of 5 μM GSK126 treatment of 9 days (n = 14; *P < .05).
Epigenetic alteration in HTLV-1–infected cells within asymptomatic carriers. (A) Box plot showing EZH2 expression in subpopulations from HTLV-1–infected individuals. P, D, and N represent subpopulations from indolent ATL (n = 3) and asymptomatic HTLV-1 carriers (n = 2). P indicates CD7+/CADM1− cells; D indicates CD7+/CADM1+ cells; N indicates CD7−/CADM1+ cells. Acu-N indicates CD7−/CADM1+ cells from acute-type ATL. (B) Box plot showing relative expression levels of the H3K27me3-dependent silenced 856 genes (defined in supplemental Figure 4) in the subpopulations from HTLV-1–infected individuals and acute-type ATL (*P < 1E−4). (C) Venn diagram showing the relationship between disease progression and H3K27me3 gain. The 3 subgroups were classified according to the disease stages (stage I indicates P to D progression; stage II indicates D to N progression; stage III indicates indolent N to acute-type N progression). Each gene set was defined as (1) H3K27me3 gain (LogFC > 1) in acute-type ATL compared with normal CD4+ T cells and (2) decreased expression (FC < −1.5, P < .05) at the stages. (D) Microscopy data showing representative H3K27me3 level in CD4+/CADM1+ and CD4+/CADM1− subpopulations within asymptomatic carriers (n = 3). The scale bars indicate 20 μm. (E) Graph showing the results of μChIP assay with anti-H3K27me3 antibody and control IgG in CD4+/CADM1+ and CD4+/CADM1− subpopulations derived from asymptomatic carriers (n = 3, mean ± SD). Promoter region of the H3K27me3-regulated genes were tested. (F) Graph showing the population size of CD4+/CADM1+ cells in the presence or absence of 5 μM GSK126 treatment of 9 days (n = 14; *P < .05).
We detected epigenetic accumulation in the infected cells from asymptomatic carriers. Immunofluorescence microscopy analysis after cell sorting showed H3K27me3 accumulation in the CD4+/CADM1+ subpopulation (Figure 7D). PCR-ChIP assay suggested that the epigenetic change specifically occurred in the CADM1+–infected cells (Figure 7E). Finally, we observed differential sensitivity to the EZH2 inhibitor depending on HTLV-1 infection status. Long-term treatment with GSK126 selectively reduced the CD4+/CADM1+ proportion of HTLV-1 carriers (Figure 7F and supplemental Figure 10B). Taken together, it was assumed that EZH2-dependent epigenetic abnormalities contribute to the survival of HTLV-1–infected cells.
Discussion
Epigenetic program is one of the molecular bases of cellular identity and functional processes. Here we showed the histone methylation pattern of ATL, which appears distinct from other cancer types and PRC2-dependent cell lineages. The global methylation gain at H3K27 causes silencing of a lot of genes, supporting the leukemic cell characteristics. Tracing the epigenetic marks and expression patterns in samples obtained from patients of various diagnostic categories as well as in other biologically relevant models has unveiled that epigenetic reprogramming arises at an early stage of ATL development, probably just after HTLV-1 infection (Figure 7G). Furthermore, we showed that aggressive progression is associated with a unique methylation pattern. Thus, PRC2-mediated epigenetic reprogramming is a fundamental molecular hallmark of ATL to achieve its specific identity.
From the integration of patient-derived datasets, several unprecedented aspects of ATL have emerged. First, we observed the abnormal transcriptome of epigenetic regulators. We suggested that the global alteration of the H3K27me3 mark depended on the abundance of EZH2 and other core components of PRC2; the epigenetic deregulation was not based on EZH2 somatic mutation.54 In addition, we identified frequent silencing of histone demethylases accompanied with H3K27me3 gain. Other histone modifications (eg, H3K4me3 and lysine acetylation) and DNA methylation (5-mC) also appear to be involved in ATL pathogenesis.6,12,20,54-56 The unique epigenomic architecture is attributed to an orchestrated balance of key regulators.
Second, we clarified the role of the ATL epigenome. H3K27me3 profiling of typical ATL samples demonstrated that more than half of the genomic regions neighboring the TSS were associated with the reprogrammed histone code. H3K27me3 gain was dominant and was broadly associated with the basal expression pattern. Importantly, the PRC2 target signature was commonly silenced in ATL of both the indolent and aggressive subtypes. Thus, PRC2-mediated epigenetic reprogramming appears to be involved in the early development of ATL. Disease progression was linked to the suppressive magnitude and additional gain of specific methylation. From the perspective of the epigenome, we suggest that the rough orientation of leukemic cell fate was decided at the very early stage of the clinical course. This concept is lent further support by our results in Tax-dependent immortalized cells and in the infected cells within asymptomatic carriers.
Third, in-depth investigations revealed the PRC2 target signature of ATL, which selectively includes genes involved in transcription regulation and functional miRNAs. In embryonic stem cells, many bivalent (H3K27me3 and H3K4me3) genes encode transcription factors involved in lineage commitment57 and do not overlap with those suppressed in ATL cells. Conventional studies have shown the epigenetic silencing of specific miRNA and its biological significance in several cancers. This study demonstrated the PcG-mediated hierarchical regulation of dozens of miRNA loci. Our findings strongly suggest that in addition to the direct regulation of focal chromatin structure, PRC2 generates diverse outcomes by the remote regulation of a broad spectrum of targets, including the on/off switches, buffers, and fine tuners of gene expression, which may increase the propensity for malignant transformation and clonal evolution of ATL. We also identified KDM family genes as PRC2 targets. Particularly, KDM6B, which encodes the H3K27me3 demethylase JMJD3, was significantly silenced in ATL. The ATL-specific coherent relationship comprising the epigenetic “writer” and “eraser” may contribute to locking or stabilizing the epigenetic states.58
We also approached the regulatory mechanisms of EZH2 abundance and genomic occupancy. A series of experiments in normal T cells and in ATL models demonstrated the necessity of a complicated signaling network for sufficient EZH2 induction. In addition to the marked contribution of NF-κB, sustained activity of other signaling cascades downstream of T-cell receptors is deductively suggested in ATL cells. Interestingly, EZH2 contributes to NF-κB activation through miR-31 silencing and NIK induction. Considering our results, we propose a positive feedback loop comprising miRNA, NF-κB, and an EZH2-dependent mechanism. The robustness of the pathologic system may be established during ATL development.
Using 2 biological models, we showed that HTLV-1 Tax affects the EZH2-dependent epigenome. The significant overlap of methylated genes in Tax-cells and ATL cells strongly suggests that epigenetic reprogramming was caused by HTLV-1 infection. This is supported by the results dealing with the epigenetic hit in the infected cells from HTLV-1 carriers. These findings imply a new function of HTLV-1, the global modification of host gene expression by regulating host epigenetic machinery. By affecting EZH2, Tax can indirectly suppress diverse target genes. This emerging concept is probably critical for phenotypic consequences because EZH2 inhibition completely prevents Tax-dependent immortalization and can diminish the size of the infected cells.
Understanding of the ATL epigenome has supported the efficacy and relevance of targeting PRC2. Relief of the cumulative methylation of genome-wide chromatin may restore the aberrant transcriptome to an ideal expression signature, permitting favorable treatments. According to the expression profiling, EZH2 may represent the first choice as a druggable target. In addition, peripheral T cells highly express EZH1 that compensates for the EZH2 function as an H3K27me3 writer and a chromatin regulator.48,59,60 In vivo study based on ATL development will be helpful for the design of proof-of-concept for epigenetic treatments. Future drug discovery and innovative progress of molecular biology from multidisciplinary approaches will achieve potent and selective synthetic lethality by targeting the regulators of H3K27me3 in aggressive and indolent ATL cells, as well as in clonally expanded infected cells, thus improving medical care and the prevention of disease onset.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Hiroyuki Miyoshi and Atsushi Miyawaki for providing the Venus-encoding lentivirus vectors, Dr Kazuo Umezawa for providing an NF-κB inhibitor DHMEQ, and Ms Takako Akashi for support and maintenance of JSPFAD.
This work was supported by AMED grant numbers 15ck0106133h002 (T.W., A.U., K.U.) and 15im0210101 (T.W.); JSPS KAKENHI grant numbers 24790436 (M.Y.), 15K06907 (M.Y.), 26293226 (M.Y., T.W., K.U., N.K., T.Y.), and 221S0001 (T.W., Y.T.); JSPS Fellows grant number 14J08236 (D.F.), and a grant from the Uehara Memorial Foundation (M.Y.).
Authorship
Contribution: D.F. designed and performed Coc analysis, analyzed data, and wrote the paper; S.N. analyzed EZH2 regulation and performed Sanger sequencing; M.H. performed Coc analysis and analyzed data; N.K. and A.S. analyzed EZH2-Tax interaction; K.N. performed expression array and contributed to data analysis; M.N. prepared clinical samples; T.Y. measured HTLV-1 proviral load; S.K. and K.U. developed CADM1/CD7-based flow cytometry method, provided clinical samples, and gave advice; Y.T. provided anti-Tax antibody; M.I. maintained JSPFAD and analyzed epidemiologic data; A.U. provided clinical samples and gave advice; M.Y. conceived and supervised the project, designed and performed experiments, analyzed data, and wrote the paper; and T.W. established the ATL cohort study and supervised all research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Makoto Yamagishi, Laboratory of Tumor Cell Biology, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, 4-6-1, Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail: myamagishi@mgs.k.u-tokyo.ac.jp; and Toshiki Watanabe, Laboratory of Tumor Cell Biology, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, 4-6-1, Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail: tnabe@ims.u-tokyo.ac.jp.

![Figure 2. Epigenetic alterations in primary ATL cells. (A) Overall percentage of genes with H3K27me3 states in ATL cells compared with normal CD4+ T cells. The averaged methylation values at each gene (fold change [FC] > ±1.5 vs normal CD4+ T cells) were calculated from normalized probes. (B) Methylation intensity profiling of normalized each probe in ATL cells (n = 3) and normal CD4+ T cells (n = 2). Relative H3K27me3 levels of each probe (P < .05) are shown in histogram. (C) Heat maps and hierarchical clustering of H3K27me3 (left) and H3K4me3 (right) levels at regions (LogFC > 3) in primary ATL cells, normal T cells, and TL-Om1 cells. (D) The number of probes showing gain (LogFC > 3) or loss (LogFC < −3) of H3K27me3 and H3K4me3 in ATL cells compared with normal CD4+ T cells. (E) Venn diagram showing the overlap between genes associated with H3K27me3 in ATL cells (FC > 2 vs normal CD4+ T cells), embryonic stem cells,36 and DLBCL cells.32 (F-G) Composite enrichment profile of H3K27me3 relative to the closest transcriptional start site (TSS). Graphs show relative H3K27me3 levels calculated from all normalized probes (F) and frequency of H3K27me3 gain in ATL compared with normal CD4+ T cells (G; closed circles indicate gain in ATL [P < .05]; open circles indicate loss in ATL [P < .05]). (H) Box plots showing relative H3K27me3 levels at silenced promoter region (expression FC < −2 in acute-type ATL vs normal CD4+ T cells, P < .001) and gene-enhancer region (defined by H3K4me1+H3K27ac marks in CD4+ T cells14) in ATL and normal T cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/14/10.1182_blood-2015-08-662593/4/m_1790f2.jpeg?Expires=1770914429&Signature=wdoEwlhTsk1Aoj6kqcnCqpgoVIwJF1wllLcHHFOOTKczn-1fT5CaTXtypM0mUweLZrBPJ9vc1WJWKlSrowxA6JF1NMwA2sf3JOhvl~MrItd55BvMZb64rSWI3cVz652qzLX9yOykOINTBxonCvBi-twOspco5265e9TuuHakk-2ycZkRMNWskhT5XpDUjDJTGojjA0FMiaXVeXWlX0p088gYByy4EDYBkokCYEbP0vgfJoCaXZb3bGMjk82paQyJJ-vZe-yI8jt-sQwKoTWb5fkJi4ols584NMqkn3RT0aFPKJCNf5luFhEvFInFiz7hygmDnLehUmYF0LjfkBDOpg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Biological interaction between EZH2 and HTLV-1 Tax. (A) EZH2 promoter activity in the presence or absence of HTLV-1 genes. 293FT cells were transfected with plasmid vectors carrying EZH2 promoter reporter and indicated HTLV-1 cDNAs. The luciferase activities were quantified at 48 hours post-transfection (n = 3, mean ± SD, P < .05). (B) Tax-dependent EZH2 gene activation analyzed by luciferase reporter assay. Signaling inhibitors (DHEMQ for NF-κB inhibition; cyclosporin A [CsA] for NAFT inhibition; U0126 for ERK1/2 inhibition) were cotreated. A Tax M22 was used as a NF-κB–deficient mutant (n = 3, mean ± SD, P < .01). (C-D) Interaction between Tax and PRC2. 293T cells were transfected with plasmids for expression of FLAG-Tax (C), or FLAG-EZH2 and untagged Tax (D), respectively. Nuclear cell extracts from each sample were immunoprecipitated with an anti-FLAG antibody. Coprecipitated proteins were prepared and analyzed by western blotting with indicated antibodies. H.C., IgG heavy chain. (E) Pull-down assay with GST-EZH2. Expression vectors for deletion mutant series of GST-tagged EZH2 were established (top). 293T cells were transfected with the plasmids encoding GST-EZH2 series and untagged Tax, and then subjected in GST pull-down assay. Purified GST-EZH2 series and copurified PRC2 components were detected by western blotting with indicated antibodies (bottom). (F) Co-immunoprecipitation (Co-IP) between endogenous Tax and EZH2 in HTLV-1–infected HUT102 cells. The same amounts of anti-EZH2 antibody and non-specific IgG were used in the immunoprecipitation, followed by detection with indicated antibodies. (G) Confocal microscopy data showing subcellular localizations of endogenous EZH2 and Tax in HUT102. The scale bar indicates 10 μm. Arrowheads indicate colocalized spots. (H) Confocal microscopy data showing subcellular localizations of endogenous EZH2 and ectopically expressed viral proteins that were detected by anti-EZH2 and anti-FLAG antibodies, respectively, in HeLa cells. The scale bars indicate 10 μm. Arrowheads indicate colocalized spots. (I) Venn diagram showing the overlap between genes (Coc analysis FC > 0 vs control IgG) associated with EZH2 occupancy, H3K27me3 accumulation, and Tax binding in HTLV-1–infected MT-2 cells. (J) Composite enrichment profiles of Tax, EZH2, and H3K27me3 relative to the closest TSS at Tax-bound genes (solid line, 5377 genes, Coc FC > 0 vs control IgG) and Tax-unbound genes (dashed line, 14 937 genes, Coc FC < 0 vs control IgG) in MT-2 cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/14/10.1182_blood-2015-08-662593/4/m_1790f5.jpeg?Expires=1770914429&Signature=uJeSUQuAPhROpB5ay4V79W7ZMUeOvxP3sGeMkOlk53CjKTZHzX92W5DldjjfhfiH-3bUti8zl97aDlc2pE9F9lkf7JoPKNnYKayxvYeRTopqmYbeNvFatvF1L~gtggKIzRViaDAJ1Ex6LW-YNno~LSLgXeCmY-ztlGIcjXf3RqxqvrZlUaWwQuytrw~KGKSlAQpTUlvzs9DuqBKbMk~9TvgMl77D8jXW~C3PX5Zv6wT4Hnh9O-ODTm4nbriss5RZclSGJ6fSIQDO3Qf8LMDPqe--kYenz40asGgsLpjB-cAJTjqwxYKs-cA~jNeH-QVZrUPvhZP4Hv6ESdYsTyVIsQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)