Key Points
Scaffolding adaptor protein GAB2 is required for BCR-ABL1–evoked myeloid and lymphoid leukemogenesis.
SHP2 and p85 binding to GAB2 activate distinct signaling pathways and are required differentially for myeloid and lymphoid leukemogenesis.
Abstract
Tyrosine kinase inhibitors (TKIs) directed against BCR-ABL1, the product of the Philadelphia (Ph) chromosome, have revolutionized treatment of patients with chronic myeloid leukemia (CML). However, acquired resistance to TKIs is a significant clinical problem in CML, and TKI therapy is much less effective against Ph+ B-cell acute lymphoblastic leukemia (B-ALL). BCR-ABL1, via phosphorylated Tyr177, recruits the adapter GRB2-associated binding protein 2 (GAB2) as part of a GRB2/GAB2 complex. We showed previously that GAB2 is essential for BCR-ABL1–evoked myeloid transformation in vitro. Using a genetic strategy and mouse models of CML and B-ALL, we show here that GAB2 is essential for myeloid and lymphoid leukemogenesis by BCR-ABL1. In the mouse model, recipients of BCR-ABL1–transduced Gab2−/− bone marrow failed to develop CML-like myeloproliferative neoplasia. Leukemogenesis was restored by expression of GAB2 but not by GAB2 mutants lacking binding sites for its effectors phosphatidylinositol 3-kinase (PI3K) or SRC homology 2–containing phosphotyrosine phosphatase 2 (SHP2). GAB2 deficiency also attenuated BCR-ABL1–induced B-ALL, but only the SHP2 binding site was required. The SHP2 and PI3K binding sites were differentially required for signaling downstream of GAB2. Hence, GAB2 transmits critical transforming signals from Tyr177 to PI3K and SHP2 for CML pathogenesis, whereas only the GAB2–SHP2 pathway is essential for lymphoid leukemogenesis. Given that GAB2 is dispensable for normal hematopoiesis, GAB2 and its effectors PI3K and SHP2 represent promising targets for therapy in Ph+ hematologic neoplasms.
Introduction
The product of the Philadelphia (Ph) chromosome translocation, BCR-ABL1, is a constitutively active protein-tyrosine kinase that is the direct molecular cause of chronic myeloid leukemia (CML) and Ph+ B-cell acute lymphoblastic leukemia (B-ALL). ABL1 tyrosine kinase inhibitors (TKIs), such as imatinib mesylate, induce hematologic and cytogenetic remissions in the majority of CML patients, but disease relapse occurs in most patients after imatinib is withdrawn,1 and a significant proportion of patients develop resistance to TKI therapy. Furthermore, patients with advanced stage CML or B-ALL respond poorly to TKIs.2,3 Hence, there is a need to identify and validate additional pharmacologic targets to prevent or overcome TKI resistance.
BCR-ABL1 activates multiple signaling networks in leukemic cells, including the RAS/extracellular signal-regulated kinase (ERK), signal transducer and activator of transcription (STAT), c-Jun N-terminal kinase (JNK)/stress-activated protein kinase, and phosphatidylinositol 3-kinase (PI3K) pathways.4 A major challenge has been determining which of these pathways are essential for leukemogenesis. Mouse models of BCR-ABL1–induced leukemia have proven useful in this effort, as both CML and Ph+ B-ALL can be reproduced faithfully by expressing BCR-ABL1 in bone marrow (BM) stem/progenitor cells through retroviral transduction, followed by bone marrow transplantation (BMT). Recipients of BCR-ABL1–transduced BM develop a fatal CML-like myeloproliferative neoplasm (MPN) that originates from hematopoietic stem cells (HSCs),5,6 can progress to blast crisis,7 and is responsive to TKI therapy.8 To model B-ALL in mice, BM is transduced in the absence of myeloid cytokines, resulting in B-lymphoblastic leukemia/lymphoma in transplant recipients.9,10 This disease originates from early lymphoid progenitors5 and is characterized by a block in B-cell differentiation at the pre-B stage.9
The contribution of the BCR region of BCR-ABL1 to leukemogenesis, specifically the autophosphorylation site at tyrosine 177 (Y177), has been the subject of intensive study. In CML cell lines, BCR-ABL1 is phosphorylated at Y177, forming a binding site for the SH2 domain of the adapter protein GRB2, which is critical for its recruitment to BCR-ABL1.11,12 We and others have shown that the BCR-ABL1-Y177F mutant is greatly attenuated in the ability to induce CML-like MPN in the murine BMT model.13-15 These findings demonstrate that Y177 and the signaling pathway(s) downstream of this site are critical for myeloid and perhaps also contribute to lymphoid leukemogenesis by BCR-ABL1.
Earlier, we reported a critical role for a GRB2/GAB2 complex in BCR-ABL1–induced transformation in vitro that is mediated by Y177.16 The GAB family (GAB1, GAB2, GAB3) consists of the pleckstrin homology (PH) domain containing scaffolding/adaptor proteins involved many signaling pathways.17,18 GAB2 undergoes phosphorylation on several tyrosine residues in response to multiple cell stimuli.19-23 Moreover, GAB2 is constitutively tyrosyl-phosphorylated in BCR-ABL1–transformed cells,19,20 forming binding sites for other SH2-containing signal relay proteins, including the p85 regulatory subunit of type IA PI3K and the protein-tyrosine phosphatase SHP2.20,24,25 Gab2−/− mice are viable and fertile, but have defects in their allergic response and in mast cell development.21,23 Although GAB2-deficient mice exhibit normal baseline hematopoiesis, long-term multilineage repopulation by HSCs is impaired, concomitant with defects in the PI3K and ERK signaling cascades.26 Roles for GAB2 in the pathogenesis of several other malignancies have also been reported.27-31
Here, we use a genetic strategy in mice to define the role of GAB2 in leukemogenesis by BCR-ABL1. Our results indicate an essential role for GAB2 in BCR-ABL1–induced MPN and B-ALL, but demonstrate that distinct GAB2-mediated signaling pathways are required for these 2 hematopoietic malignancies.
Methods
Mice
Gab2−/−mice were produced by homologous recombination in embryonic stem cells as described21 and backcrossed for >5 generations onto the Balb/c and C57Bl/6J backgrounds, respectively.
Retroviral constructs and stocks
The retroviral vectors p210MIGR1 and MINVneo were described previously.32,33 An HA-tagged Gab2WT cDNA was introduced into p210MIGR1 and p210MINV in place of the green fluorescent protein (GFP) or neomycin resistance genes, respectively, yielding p210-IRESwt-Gab2WT and p210-IRESmut-Gab2WT (Figure 2A). Site-specific mutagenesis was used to create the Gab2Δp85 (Y441/465/574F), Gab2ΔShp2 (Y604/633F), Gab2ΔPH (deletion of Gab2 amino acids 2-100), and Gab2ΔGrb2 (delP348-K354; P498/503A) constructs. Ecotropic retroviral stocks were generated by transient transfection of 293 cells using the kat packaging system,10 and stocks were matched for titer by transduction of NIH3T3 cells. Transduction frequency was assessed by flow cytometric detection of GFP34 or, for bicistronic viruses coexpressing BCR-ABL1 and GAB2, by intracellular staining for HA epitope-tagged GAB2 proteins (see below), as well as by Southern blot analysis of proviral DNA content in genomic DNA.34 Titers determined by either method were concordant.
Bone marrow transduction, transformation, and transplantation
For myeloid transformation and leukemogenesis assays, BM was collected from Gab2+/+ and Gab2−/− mice and transduced twice with retrovirus, as previously described.34 Transduced cells were plated in cytokine-free methylcellulose (M3234; Stem Cell Technologies) for myeloid colony assays or injected intravenously into lethally irradiated recipients for myeloid leukemogenesis, as previously described.9 For B-lymphoid transformation/leukemogenesis, BM from Gab2+/+ or Gab2−/− donors was transduced once with BCR-ABL1 retrovirus in the absence of cytokines. B-lymphoid transformation was assessed by pre-B-cell colony formation in agarose and stroma-dependent growth, as previously described.35,36 For lymphoid leukemogenesis assays, transduced cells were injected into sublethally irradiated recipients, as previously described.10 The clinical features and histopathology of BCR-ABL1–induced CML-like disease, B-ALL, T-cell lymphoblastic acute leukemia/lymphoma (T-ALL), and histiocytic sarcoma were described previously.10 All mouse experiments were approved by the Institutional Animal Use and Care Committees of Tufts Medical Center and University Health Network.
Immunoblotting
To characterize signaling downstream of GAB2 in BCR-ABL1 transformation, myeloid- or B lymphoid-enriched BM cells were transduced with viruses, as indicated. GFP+ or red fluorescent protein (RFP)+GFP+ cells were recovered by fluorescence-activated cell sorter, starved in Iscove's Modified Dulbecco's Medium 2% fetal bovine serum for 2 hours, and lysed in radioimmunoprecipitation assay buffer. Lysates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted, as previously described.10 For details on antibodies and sources, see supplemental Methods, available on the Blood Web site.
Southern blotting
Southern blot analysis of proviral integration and leukemia-initiating cell engraftment was performed as previously described,10 except that genomic DNA was digested with BamHI, and blots were hybridized with a radioactive probe from the 5′ end of the BCR gene.
Statistical analysis
Survival comparisons were performed by log-rank test. Band intensities in immunoblots from myeloid cells were compared by 2-sided Student t test; the null hypothesis was that the logarithm of the intensity ratio was 0. For lymphoid cell blots, band intensities were compared by 1-sided Student t test, owing to the results of the myeloid signaling experiments. Multiple comparison P value correction was performed with the Benjamini-Hochberg method.
Additional methods are found in the supplemental Methods.
Results
BCR-ABL1 cannot induce CML-like MPN in the absence of GAB2
When expressed in mouse myeloid progenitors, BCR-ABL1 can induce myeloid colony formation in the absence of exogenous cytokines.37 We showed earlier that progenitors from Gab2−/− BM donors (mixed 129Sv;C57Bl/6J background) are resistant to transformation by BCR-ABL1 in vitro.16 To assess the role of GAB2 in CML-like MPN, we bred the Gab2−/− mutation onto the C57Bl/6J and Balb/c backgrounds for >5 generations. For each strain, donor BM from congenic Gab2−/− mice or Gab2+/+ littermates was harvested, transduced with p210MIGFP retrovirus that coexpresses p210 BCR-ABL1 and GFP,32 and transplanted into lethally irradiated Gab2+/+ recipients. Hematopoietic stem/progenitor cells from Gab2−/− mice have deficient responses to cytokines26 and to cytotoxic drugs (G.M. and B.G.N., unpublished observations, June 2000), so donor mice were not treated with 5-fluorouracil (5-FU) prior to BM harvest.9
In C57Bl/6J mice, recipients of BCR-ABL1–transduced BM from Gab2+/+ donors efficiently developed fatal CML-like leukemia within 5 weeks of transplantation (supplemental Figure 1A), characterized by leukocytosis, splenomegaly, and infiltration of the spleen, liver, and lung parenchyma with maturing GFP+ myeloid cells (supplemental Figure 1B-E).9 By contrast, recipients of BCR-ABL1–transduced BM from Gab2−/− littermate donors did not develop clinical MPN. Instead, beginning about 3 months after transplantation, they succumbed to precursor T-ALL, characterized by thymic enlargement and mesenteric lymphadenopathy with infiltration of GFP+CD90+ lymphoblasts (supplemental Figure 1F).
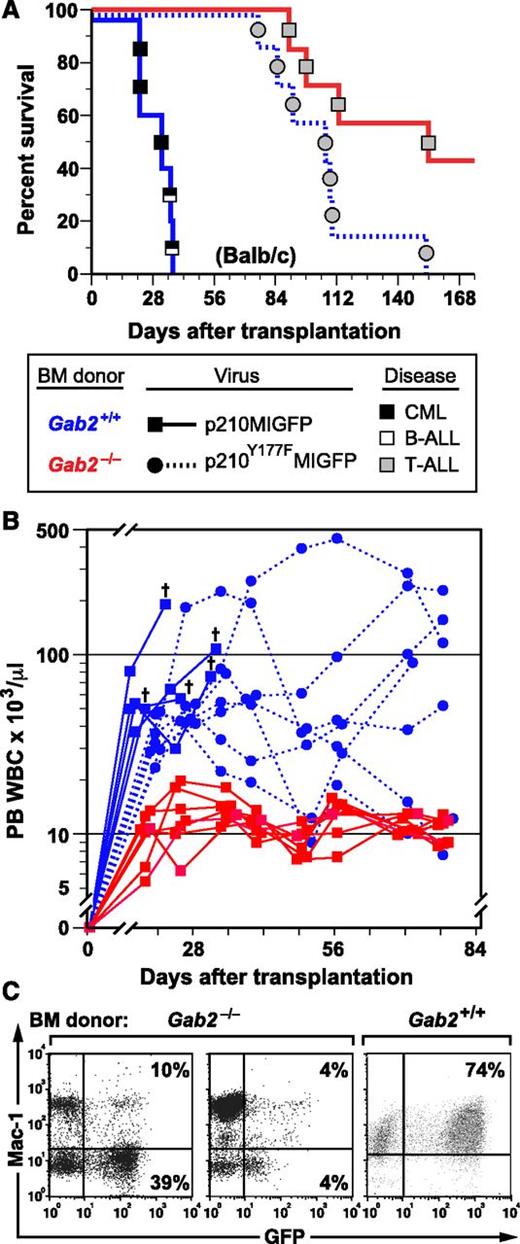
In parallel, we transplanted recipients with p210MIGFP-transduced BM from Gab2−/− and Gab2+/+ littermate donors on the Balb/c background and also transplanted a cohort of recipients with BCR-ABL1-Y177F–transduced Gab2+/+ BM. Balb/c recipients of BCR-ABL1–transduced Gab2+/+ donor BM also efficiently developed CML-like MPN, either alone or concomitant with B-ALL (Figure 1A). As reported previously,13-15 BCR-ABL1 Y177F, which lacks a GRB2 binding site, induced attenuated, nonfatal MPN in most recipients (Figure 1A-B). Similar to the results obtained using B6 mice, recipients of Balb/c BCR-ABL1–transduced Gab2−/− donor BM did not develop overt MPN, but tended to succumb to T-ALL after a prolonged latent period (Figure 1A). Although peripheral blood leukocyte (PBL) counts in recipients of BCR-ABL1–transduced Gab2−/− donor BM were predominantly within the normal range (Figure 1B), most had circulating GFP+ cells at 6 to 8 weeks after transplantation (Figure 1C), indicating engraftment of retrovirus-transduced donor cells. These results demonstrate that GAB2-deficient HSCs are completely resistant to induction of CML-like MPN by BCR-ABL1. The fact that donor Gab2 deficiency phenocopies the effects of BCR-ABL1-Y177F indicates that critical leukemogenic signals from Y177 of BCR-ABL1 are probably transmitted through a GRB2/GAB2 complex.
GAB2 is required for induction of CML-like MPN by BCR-ABL1. (A) Kaplan-Meier (K-M) survival curves for recipients of BM from Gab2−/− donors in the Balb/c background (>F9 generation backcross) transduced with p210MIGFP (red line, n = 7) or p210Y177FMIGFP (blue dotted line, n = 7) retrovirus and from wild-type Balb/c donors (blue solid line, n = 5) transduced with p210MIGFP retrovirus. Symbols represent individual mice (squares for recipients of Gab2+/+ BM, circles for recipients of Gab2−/− BM); disease phenotype is indicated by shading (black, CML-like MPN; white, B-ALL; gray, T-ALL). For p210MIGFP-transduced BM, the difference in survival between the Gab2−/− and Gab2+/+ recipient cohorts was significant (P = .0003, Wilcoxon test). There was no significant survival difference between recipients of p210MIGFP-transduced Gab2−/− BM and p210Y177FMIGFP-transduced Gab2+/+ BM (P = .058). (B) Scatter plot of PBL counts in the 3 cohorts from the transplants in A at various times after transplantation. The cross indicates death of a recipient of p210MIGFP-transduced Gab2+/+ BM. Note the attenuated and nonfatal leukocytosis induced in recipients of p210Y177FMIGFP-transduced Gab2+/+ BM. (C) Flow cytometric plot of expression of GFP (x-axis) and the myeloid marker Mac-1 (y-axis) in PBLs from 2 representative recipients of p210MIGFP-transduced BM from Gab2−/− donors (left) and from a recipient of p210MIGFP-transduced Gab2+/+ BM with CML-like MPN (right), obtained 1 month after transplantation.
GAB2 is required for induction of CML-like MPN by BCR-ABL1. (A) Kaplan-Meier (K-M) survival curves for recipients of BM from Gab2−/− donors in the Balb/c background (>F9 generation backcross) transduced with p210MIGFP (red line, n = 7) or p210Y177FMIGFP (blue dotted line, n = 7) retrovirus and from wild-type Balb/c donors (blue solid line, n = 5) transduced with p210MIGFP retrovirus. Symbols represent individual mice (squares for recipients of Gab2+/+ BM, circles for recipients of Gab2−/− BM); disease phenotype is indicated by shading (black, CML-like MPN; white, B-ALL; gray, T-ALL). For p210MIGFP-transduced BM, the difference in survival between the Gab2−/− and Gab2+/+ recipient cohorts was significant (P = .0003, Wilcoxon test). There was no significant survival difference between recipients of p210MIGFP-transduced Gab2−/− BM and p210Y177FMIGFP-transduced Gab2+/+ BM (P = .058). (B) Scatter plot of PBL counts in the 3 cohorts from the transplants in A at various times after transplantation. The cross indicates death of a recipient of p210MIGFP-transduced Gab2+/+ BM. Note the attenuated and nonfatal leukocytosis induced in recipients of p210Y177FMIGFP-transduced Gab2+/+ BM. (C) Flow cytometric plot of expression of GFP (x-axis) and the myeloid marker Mac-1 (y-axis) in PBLs from 2 representative recipients of p210MIGFP-transduced BM from Gab2−/− donors (left) and from a recipient of p210MIGFP-transduced Gab2+/+ BM with CML-like MPN (right), obtained 1 month after transplantation.
Complementation of the Gab2−/− mutation by retroviral GAB2 expression
The failure of recipients of BCR-ABL1–transduced BM from Gab2−/− donors to develop MPN could indicate that signal(s) emanating from GAB2 is/are required for the massive expansion of BCR-ABL1–expressing hematopoietic stem/progenitor cells (HSPCs). Alternatively, GAB2 deficiency could decrease the number of target cells for BCR-ABL1–evoked MPN, resulting in smaller initial numbers of leukemia-initiating cells.5,38
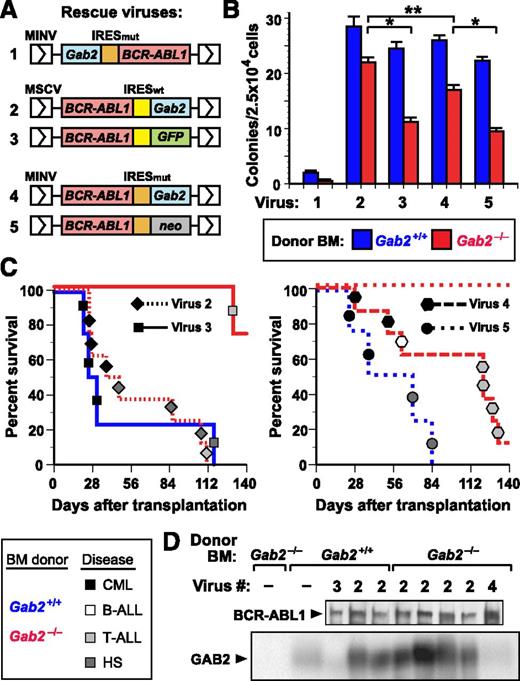
To distinguish between these possibilities, we tested whether retroviral coexpression of wild-type (WT) GAB2 and BCR-ABL1 could restore leukemogenic potential to Gab2−/− cells. We designed a series of retroviral vectors wherein BCR-ABL1 and Gab2 are coexpressed via an internal ribosome entry site (IRES) (Figure 2A), and screened them for their ability to restore efficient transformation of Gab2−/− BM (Figure 2B). We first tested an MINV-based vector33 with Gab2 placed 5′ of a mutant form of the encephalomyocarditis virus IRES (IRESmut) and BCR-ABL1 in the 3′ position, a configuration permitting 5- to 10-fold higher levels of expression of the 5′ gene. Not only did this virus fail to restore BCR-ABL1–evoked transformation of Gab2−/− BM, it significantly compromised transformation of Gab2+/+ BM (Figure 2B). By contrast, when Gab2 was positioned 3′ of IRESmut or IRESwt in the MINV- or MSCV-based vector, we observed rescue of myeloid transformation in vitro (Figure 2B), although the p210-IRESwt-Gab2 vector was more efficient. Finally, we compared the vectors for their ability to rescue CML-like MPN in vivo (Figure 2C). Both Gab2-expressing vectors restored CML-like MPN to recipients of transduced Gab2−/− donor BM, although p210-IRESwt-Gab2 was more efficient, inducing CML-like MPN in half of recipients. Therefore, we chose this vector for all subsequent experiments. Immunoblots of primary myeloid leukemic cell extracts demonstrated that p210-IRESwt-Gab2 virus led to GAB2 levels approximately threefold higher than endogenous GAB2 levels in normal BM (Figure 2D). Southern blot analysis of leukemic cell genomic DNA revealed that the MPN in recipients of BCR-ABL1– and BCR-ABL1-Y177F–transduced Gab2+/+ BM, as well as p210-Gab2WT–transduced Gab2−/− BM, derived from 1 to 2 distinct proviral clones (supplemental Figure 2, lanes 1-12), demonstrating repopulation of these cohorts with similar numbers of leukemia-initiating cells. These results show that comparable numbers of target cells for BCR-ABL1–induced MPN are present in Gab2−/− BM and WT BM and establish that GAB2 is required for BCR-ABL1 to induce MPN.
Complementation of Gab2 deficiency by retroviral GAB2 expression. (A) Schematic of retroviral constructs coexpressing BCR-ABL1 and GAB2 or a reporter gene via IRES sequences. A mouse Gab2 cDNA (blue box) was cloned either 5′ or 3′ of a mutant form of the encephalomyocarditis virus IRES (IRESmut, orange box) in the retroviral vector Minnal virus (MINV)33 or 3′ of the wild-type IRES (IRESwt, yellow box) in murine stem cell virus (MSCV)-IRES-GFP (MIGR1)32 ; the BCR-ABL1 cDNA was placed in the other position. As a control vector, either GFP (for IRESwt) or the neomycin resistance gene (for IRESmut) was cloned in place of Gab2. (B) Coexpression of GAB2 and BCR-ABL1 restores efficient myeloid cell transformation in vitro. BM from the indicated Balb/c donor mice (blue, Gab2+/+; red, Gab2−/−) was transduced with the indicated viral stock from A and plated in methylcellulose without cytokines. The number of cytokine-independent mixed myeloid colonies (CFU-G, -M, -GM, and -GEMM) from Gab2−/− BM was increased by transduction with viruses 2 or 4, relative to viruses 3 or 5 (P < .0001 and P = .0005, respectively, Student t tests), whereas there was no significant difference in the efficiency of transformation of Gab2+/+ BM with viruses 2 to 5. Virus 2 (p210-IRESwt-Gab2) induced significantly more colonies from Gab2−/− BM than virus 4 (p210-IRESmut-Gab2; P = .0082, Student t test), whereas the difference in transformation of Gab2+/+ and Gab2−/− BM by virus 2 was not significant (P = .167, Student t test). In these experiments, the number of cytokine-independent colonies induced in Gab2-deficient BM was somewhat higher than previously reported,16 possibly due to lower retroviral titers or the different genetic background (129;B6) in our earlier study. (C) Rescue of CML-like MPN by coexpression of GAB2 and BCR-ABL1 in BM. K-M curves for recipients of BM from Gab2+/+ (blue lines) or Gab2−/− (red lines) donors, transduced with p210-IRESwt-Gab2 or p210-IRESwt-GFP retroviruses (viruses 2 and 3 from A, respectively; left) or with p210-IRESmut-Gab2 or p210-IRESmut-neo retroviruses (viruses 4 and 5 from A, respectively; right). Symbol nomenclature is as in Figure 1A. Histiocytic sarcoma is a BCR-ABL1–induced disease of monocyte/macrophage proliferation that can be seen in recipients of BCR-ABL1–transduced BM from WT donors that have not been treated with 5-FU.9 (D) Immunoblot of protein extracts from primary MPN cells from the transplants in C probed with anti-ABL1 (upper) and anti-GAB2 (lower) antibodies.
Complementation of Gab2 deficiency by retroviral GAB2 expression. (A) Schematic of retroviral constructs coexpressing BCR-ABL1 and GAB2 or a reporter gene via IRES sequences. A mouse Gab2 cDNA (blue box) was cloned either 5′ or 3′ of a mutant form of the encephalomyocarditis virus IRES (IRESmut, orange box) in the retroviral vector Minnal virus (MINV)33 or 3′ of the wild-type IRES (IRESwt, yellow box) in murine stem cell virus (MSCV)-IRES-GFP (MIGR1)32 ; the BCR-ABL1 cDNA was placed in the other position. As a control vector, either GFP (for IRESwt) or the neomycin resistance gene (for IRESmut) was cloned in place of Gab2. (B) Coexpression of GAB2 and BCR-ABL1 restores efficient myeloid cell transformation in vitro. BM from the indicated Balb/c donor mice (blue, Gab2+/+; red, Gab2−/−) was transduced with the indicated viral stock from A and plated in methylcellulose without cytokines. The number of cytokine-independent mixed myeloid colonies (CFU-G, -M, -GM, and -GEMM) from Gab2−/− BM was increased by transduction with viruses 2 or 4, relative to viruses 3 or 5 (P < .0001 and P = .0005, respectively, Student t tests), whereas there was no significant difference in the efficiency of transformation of Gab2+/+ BM with viruses 2 to 5. Virus 2 (p210-IRESwt-Gab2) induced significantly more colonies from Gab2−/− BM than virus 4 (p210-IRESmut-Gab2; P = .0082, Student t test), whereas the difference in transformation of Gab2+/+ and Gab2−/− BM by virus 2 was not significant (P = .167, Student t test). In these experiments, the number of cytokine-independent colonies induced in Gab2-deficient BM was somewhat higher than previously reported,16 possibly due to lower retroviral titers or the different genetic background (129;B6) in our earlier study. (C) Rescue of CML-like MPN by coexpression of GAB2 and BCR-ABL1 in BM. K-M curves for recipients of BM from Gab2+/+ (blue lines) or Gab2−/− (red lines) donors, transduced with p210-IRESwt-Gab2 or p210-IRESwt-GFP retroviruses (viruses 2 and 3 from A, respectively; left) or with p210-IRESmut-Gab2 or p210-IRESmut-neo retroviruses (viruses 4 and 5 from A, respectively; right). Symbol nomenclature is as in Figure 1A. Histiocytic sarcoma is a BCR-ABL1–induced disease of monocyte/macrophage proliferation that can be seen in recipients of BCR-ABL1–transduced BM from WT donors that have not been treated with 5-FU.9 (D) Immunoblot of protein extracts from primary MPN cells from the transplants in C probed with anti-ABL1 (upper) and anti-GAB2 (lower) antibodies.
Multiple GAB2 domains are required for BCR-ABL1–evoked MPN
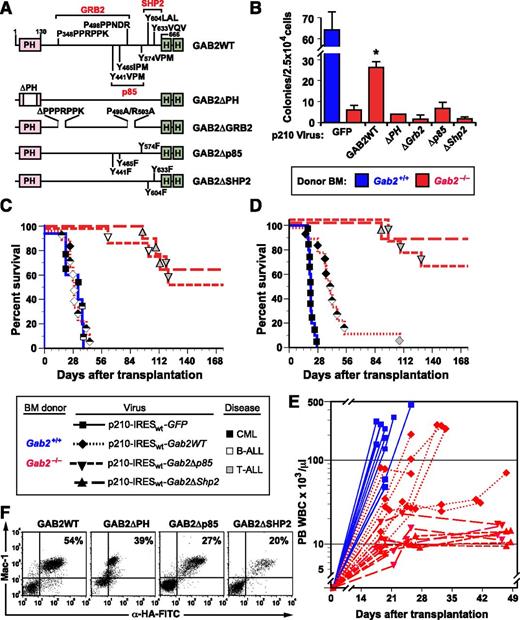
The ability to “rescue” the CML phenotype in Gab2-deficient BM provided an opportunity to identify the structural features of GAB2 required for leukemogenesis. We cloned cDNAs encoding GAB2 with deletion of its Pleckstrin homology domain (ΔPH), mutations in the bipartite proline-rich binding sites for the GRB2 SH3 domains (ΔGrb2),39 or Tyr-to-Phe substitutions in its SH2 binding sites40 for the p85 subunit (Δp85) of PI3K or for SHP2 (ΔShp2) into the 3′ position of p210-IRESwt-Gab2 (vector 2 in Figure 2A). All GAB2 constructs also encoded tandem influenza hemagglutinin (HA) epitopes at their COOH termini (Figure 3A). Transduction of Gab2−/− BM with the Gab2WT virus efficiently restored cytokine-independent myeloid colony formation; by contrast, none of the Gab2-mutant viruses transformed Gab2−/− BM in vitro (Figure 3B).
GAB2 structural domains required for induction of CML-like MPN by BCR-ABL1. (A) Scheme showing GAB2 mutants used in this experiment. PH, pleckstrin homology domain; HA, hemagglutinin epitope tag. Binding sites for the GRB2 SH3 domains and SH2 domains of p85 PI3K and SHP2 are indicated. (B) GAB2 mutants fail to rescue myeloid cell transformation in vitro. BM from the indicated donor mice (blue, Gab2+/+; red, Gab2−/−) was transduced with retrovirus coexpressing BCR-ABL1 and the different GAB2 variants depicted in A and plated in methylcellulose without exogenous cytokines. The difference in colony numbers between Gab2−/− BM transduced with p210-IRESwt-Gab2WT virus, compared with all Gab2 mutant viruses, was significant (P ≤ .004, Student t tests). (C) K-M curves of recipients of BM from Gab2+/+ littermates (blue solid line, n = 5) transduced with p210MIGFP retrovirus and of Gab2−/− donors (red lines), transduced with p210-IRESwt-Gab2WT retrovirus (dotted line, n = 9) or the indicated Gab2 mutant retrovirus (dashed lines, n = 8 for each cohort). There was no difference in survival between recipients of p210MIGFP-transduced Gab2+/+ BM and p210-IRESwt-Gab2WT–transduced Gab2−/− BM. All recipients of Gab2 mutant-transduced BM showed prolonged survival (P < .0001, Mantel-Cox tests). (D) Survival curve as in C, except that the transduced BM was lineage depleted prior to transplantation, as described in Methods. (E) Scatter plot of peripheral blood (PB) leukocyte counts in the 4 cohorts from the transplant in D vs time after transplantation. (F) Flow cytometric detection of the retrovirally expressed HA-tagged GAB2 proteins in PBLs from the indicated recipients of transduced Gab2−/− BM. Note the discrete populations of Mac-1+HA+ myeloid cells. Leukocytes from nontransduced Gab2−/− donors had no detectable fluorescein isothiocyanate fluorescence (data not shown). Detection of the HA epitope tag on the GAB2ΔPH protein by intracellular antibody staining was consistently less efficient than for the other GAB2 proteins, despite equivalent protein expression (Figure 5C).
GAB2 structural domains required for induction of CML-like MPN by BCR-ABL1. (A) Scheme showing GAB2 mutants used in this experiment. PH, pleckstrin homology domain; HA, hemagglutinin epitope tag. Binding sites for the GRB2 SH3 domains and SH2 domains of p85 PI3K and SHP2 are indicated. (B) GAB2 mutants fail to rescue myeloid cell transformation in vitro. BM from the indicated donor mice (blue, Gab2+/+; red, Gab2−/−) was transduced with retrovirus coexpressing BCR-ABL1 and the different GAB2 variants depicted in A and plated in methylcellulose without exogenous cytokines. The difference in colony numbers between Gab2−/− BM transduced with p210-IRESwt-Gab2WT virus, compared with all Gab2 mutant viruses, was significant (P ≤ .004, Student t tests). (C) K-M curves of recipients of BM from Gab2+/+ littermates (blue solid line, n = 5) transduced with p210MIGFP retrovirus and of Gab2−/− donors (red lines), transduced with p210-IRESwt-Gab2WT retrovirus (dotted line, n = 9) or the indicated Gab2 mutant retrovirus (dashed lines, n = 8 for each cohort). There was no difference in survival between recipients of p210MIGFP-transduced Gab2+/+ BM and p210-IRESwt-Gab2WT–transduced Gab2−/− BM. All recipients of Gab2 mutant-transduced BM showed prolonged survival (P < .0001, Mantel-Cox tests). (D) Survival curve as in C, except that the transduced BM was lineage depleted prior to transplantation, as described in Methods. (E) Scatter plot of peripheral blood (PB) leukocyte counts in the 4 cohorts from the transplant in D vs time after transplantation. (F) Flow cytometric detection of the retrovirally expressed HA-tagged GAB2 proteins in PBLs from the indicated recipients of transduced Gab2−/− BM. Note the discrete populations of Mac-1+HA+ myeloid cells. Leukocytes from nontransduced Gab2−/− donors had no detectable fluorescein isothiocyanate fluorescence (data not shown). Detection of the HA epitope tag on the GAB2ΔPH protein by intracellular antibody staining was consistently less efficient than for the other GAB2 proteins, despite equivalent protein expression (Figure 5C).
Recipients of Gab2−/− BM (Balb/c background) transduced with the Gab2WT virus also efficiently developed leukemia on transplantation, with most recipients developing mixed MPN and B-ALL. By contrast, MPN was not observed in recipients of Gab2−/− BM transduced with any of the Gab2-mutant viruses (Figure 3C; supplemental Figure 3). These recipients tended to succumb principally to T-ALL, beginning ∼3 months after transplantation.
To reduce the incidence of recipients with mixed disease, we repeated these experiments but lineage depleted the donor BM prior to transplantation41 to remove early lymphoid progenitors, which are the target cells for BCR-ABL1–induced B-ALL.5 As expected, we observed more efficient rescue of CML-like MPN on coexpression of GAB2 and BCR-ABL1 in Gab2−/− BM, and about half the recipients succumbed to CML-like disease without evidence of B-ALL (Figure 3D-E). As before, coexpression of BCR-ABL1 and GAB2 lacking its p85 or SHP2 binding sites failed to cause MPN, as reflected by modestly elevated PBL counts that were consistently ∼10 to 15 × 103/μL (Figure 3E). Southern blot analysis of PBL DNA showed that, similar to recipients of p210-Gab2WT–transduced Gab2−/− BM, evaluable recipients of p210-Gab2Δp85– or p210-Gab2ΔShp2–transduced Gab2−/− BM were repopulated with provirus-containing cells (supplemental Figure 2). Also, flow cytometry and intracellular staining for the HA epitope tag revealed similar levels of expression of virus-derived GAB2 proteins in circulating myeloid cells from all cohorts (Figure 3F). These results are consistent with comparable levels of engraftment by Gab2-deficient BM coexpressing BCR-ABL1 and the different GAB2 mutants and demonstrate that the PH domain, the GRB2 SH3 domain-binding site, and the binding sites for PI3K and SHP2 SH2 domains are each necessary for GAB2 to mediate signals required for BCR-ABL1–evoked MPN.
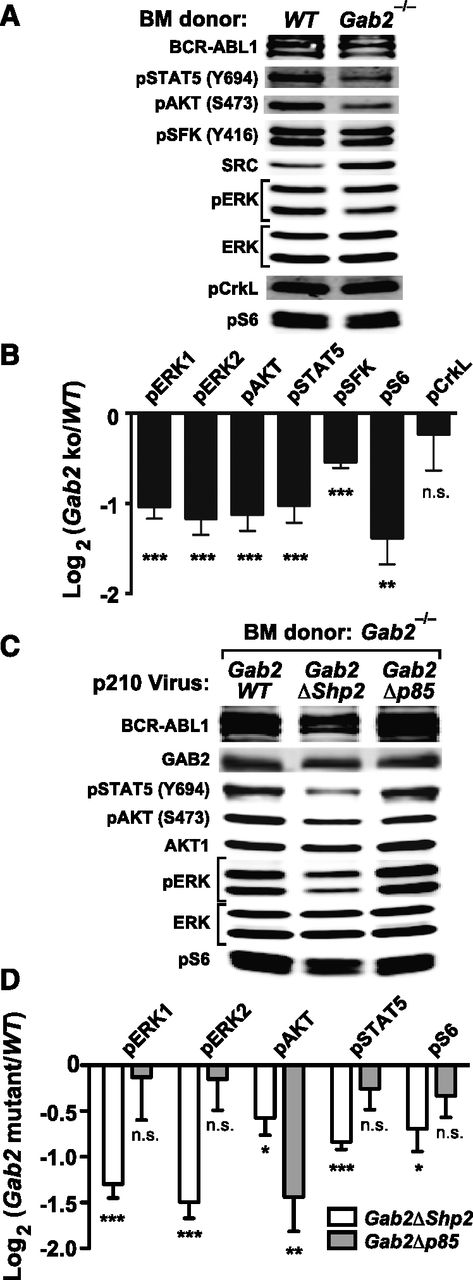
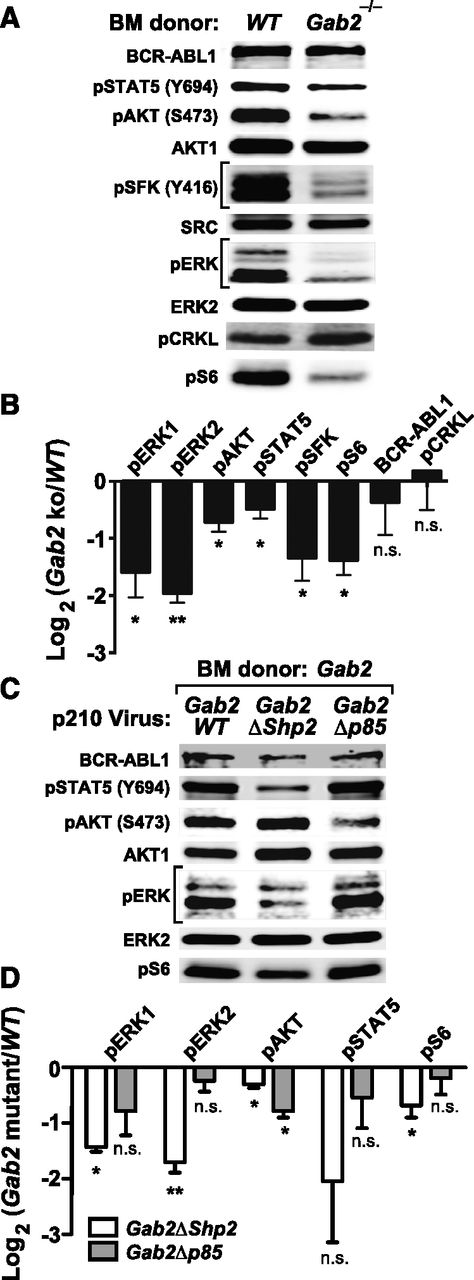
To analyze signaling downstream of GAB2 in the CML-like MPN induced by BCR-ABL1, we infected lineage-depleted WT or Gab2−/− BM cells with p210MIGFP virus and immunoblotted extracts from transduced (GFP+) cells. Compared with WT cells, Gab2−/− cells had lower levels of phospho-ERK1/ERK2, phospho–Ak strain transforming (AKT) (S473), phospho-STAT5 (Y694), phospho-SFK (Y416), and phospho-S6 (S235/236) (Figure 4A-B; supplemental Figure 4A). By contrast, phosphorylation of CRKL, a direct substrate of BCR-ABL1, was unaffected by GAB2 deficiency (Figure 4A-B). We then assessed signaling events affected by mutation of the GAB2 binding sites for SHP2 and p85. We cotransduced lineage-depleted Gab2−/− BM cells with pMSCV-BCR/ABL1-IRES-RFP and pMSCV-GAB2-IRES-GFP (Gab2WT, Gab2ΔShp2, or Gab2Δp85) viruses and immunoblotted extracts from doubly infected cells. Compared with Gab2WT-reconstituted cells, Gab2ΔShp2-reconstituted cells had lower levels of phospho-ERK1/ERK2, phospho-AKT, phospho-STAT5, and phospho-S6; by contrast, only phospho-AKT levels were reduced in Gab2Δp85-reconstituted cells. Notably, phospho-AKT levels in the latter cells were even lower than those found in Gab2ΔShp2-reconstituted cells (Figure 4C-D; supplemental Figure 4B). Thus, GAB2 is a critical mediator of multiple pathways downstream of BCR-ABL1, and the SHP2 and p85 binding sites on GAB2 play distinct roles: SHP2 binding is required for full activation of multiple downstream effectors (especially ERK and STAT5), whereas p85 binding is more specifically and critically required for full activation of the AKT pathway.
Analysis of signaling events in primary BCR-ABL1–expressing myeloid cells. (A) Representative immunoblot of primary Lin–GFP+ cell extracts from WT or Gab2−/− BM cells transduced with BCR-ABL1 p210MIGFP retrovirus; similar results were obtained in ≥6 independent biological replicates. (B) Statistical analysis of signaling events in A, pooled from ≥6 biological replicates (***P < .001, **P < .01, *P < .05, n.s., not significant; 2-sided Student t tests). (C) Representative immunoblot of primary Lin–GFP+RFP+ cell extracts from Gab2−/− BM cells cotransduced with p210-IRES-RFP and Gab2-IRES-GFP viruses, as indicated. Similar results were obtained in ≥5 biological replicates. (D) Statistical analysis of signaling events pooled from ≥5 biological replicates of the experiment shown in C (***P < .001, **P < .01, *P < .05, n.s., not significant; all vs Gab2WT cotransduced samples; 2-sided Student t tests).
Analysis of signaling events in primary BCR-ABL1–expressing myeloid cells. (A) Representative immunoblot of primary Lin–GFP+ cell extracts from WT or Gab2−/− BM cells transduced with BCR-ABL1 p210MIGFP retrovirus; similar results were obtained in ≥6 independent biological replicates. (B) Statistical analysis of signaling events in A, pooled from ≥6 biological replicates (***P < .001, **P < .01, *P < .05, n.s., not significant; 2-sided Student t tests). (C) Representative immunoblot of primary Lin–GFP+RFP+ cell extracts from Gab2−/− BM cells cotransduced with p210-IRES-RFP and Gab2-IRES-GFP viruses, as indicated. Similar results were obtained in ≥5 biological replicates. (D) Statistical analysis of signaling events pooled from ≥5 biological replicates of the experiment shown in C (***P < .001, **P < .01, *P < .05, n.s., not significant; all vs Gab2WT cotransduced samples; 2-sided Student t tests).
Different requirements for GAB2 binding proteins in BCR-ABL1–induced B-lymphoid transformation
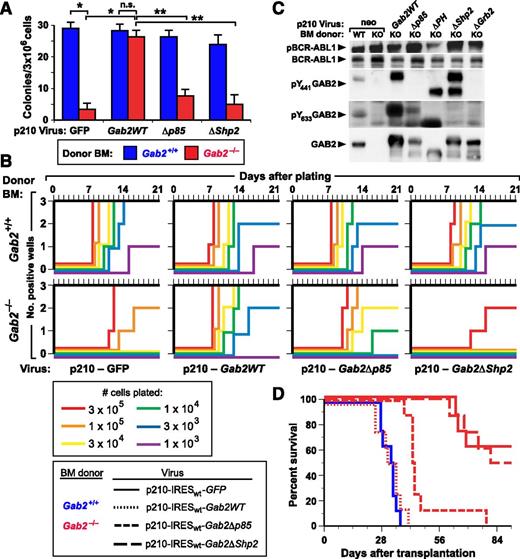
We then analyzed the impact of GAB2 deficiency on the ability of BCR-ABL1 to transform immature lymphoid progenitors in vitro using 2 quantitative assays: colony formation in agarose42 and stroma-dependent growth.43 In the colony assay, GAB2-deficient BM was profoundly refractory to BCR-ABL1 transformation, giving rise to only ∼10% of the colonies as WT BM (Figure 5A). Transformation of GAB2-deficient BM was restored by transduction with p210-Gab2WT retrovirus, but not by Gab2 mutants lacking either their p85 or SHP2 binding sites. In the second assay, BCR-ABL1 stimulates the outgrowth of B-lymphoid progenitors that are stroma dependent and not highly leukemogenic in mice.36,43 As shown earlier,16 BCR-ABL1–transduced Gab2−/− BM showed much lower outgrowth than transduced Gab2+/+ BM (Figure 5B). Coexpression of BCR-ABL1 and Gab2WT restored efficient outgrowth of Gab2−/− BM, whereas coexpression of the Gab2ΔShp2 mutant did not. Interestingly, the Gab2Δp85 mutant also rescued B-lymphoid transformation in vitro, albeit to a lesser extent than Gab2WT (Figure 5B). Immunoblot analysis of extracts from the transformed cells demonstrated that the various retrovirally encoded GAB2 proteins were expressed and phosphorylated on the appropriate sites (Figure 5C).
In vitro lymphoid transformation and in vivo leukemogenesis evoked by BCR-ABL1 are dependent on GAB2. (A) Agarose colony assays. BM from the indicated Balb/c donor (blue, Gab2+/+; red, Gab2−/−) mice was transduced with retrovirus coexpressing p210 BCR-ABL1 and either GFP or the indicated GAB2 variant. Transduced cells were plated directly in agarose as described in Methods, and transformed pre-B-lymphoid colonies were counted at day 10. The differences in colony numbers between p210-GFP–transduced Gab2+/+ and Gab2−/− BM and between p210-GFP–transduced and p210-Gab2WT–transduced Gab2−/− BM, were significant (P < .0001 and P = .001, respectively, unpaired Student t tests), whereas there was no difference between p210-GFP–transduced Gab2+/+ and p210-Gab2WT–transduced Gab2−/− BM (P = .224). The difference in colony numbers between Gab2−/− BM transduced with p210-Gab2WT retrovirus and either p210-Gab2Δp85 or p210-Gab2ΔShp2 retrovirus was significant (P = .0004 and P = .0002, respectively, unpaired Student t tests), whereas the difference between Gab2−/− BM transduced with p210-Gab2Δp85 and p210-Gab2ΔShp2 retrovirus did not reach significance (P = .0647, unpaired Student t test). (B) Whitlock-Witte assays. Freshly harvested Balb/c BM from Gab2+/+ (top) and Gab2−/− (bottom) donors was transduced with retrovirus coexpressing p210 BCR-ABL1 and either GFP or the indicated GAB2 variant, and plated in triplicate at the indicated cell numbers per well. Nontransduced cells were added to provide 106 total cells for stromal support. Wells were scored as positive when cell number reached 106 viable nonadherent cells per well, as described in Methods. (C) Extracts from immortalized BCR-ABL1–transformed B-lymphoid cell lines (from Figure 5B) from Gab2+/+ (WT) or Gab2−/− (knockout) donors were analyzed for GAB2 protein expression and phosphorylation, as described in Methods. (D) K-M curves of recipients of BM from Gab2+/+ littermates transduced with p210MIGFP retrovirus (blue solid line, n = 8), and of Gab2−/− donors (red lines) transduced with p210-IRESwt-Gab2WT retrovirus (dotted line, n = 8) or the indicated Gab2 mutant retrovirus (dashed lines, n = 8 for each cohort). All deaths were due to B-ALL. There was no significant difference in survival between recipients of p210MIGFP-transduced Gab2+/+ BM and p210-IRESwt-Gab2WT–transduced Gab2−/− BM (P = .493) or between recipients of p210MIGFP-transduced Gab2−/− BM and p210-IRESwt-Gab2ΔShp2–transduced Gab2−/− BM (P = .734). By contrast, the survival of recipients of p210MIGFP-transduced Gab2+/+ BM and p210MIGFP-transduced Gab2−/− BM, and of recipients p210-IRESwt-Gab2WT–transduced and p210-IRESwt-Gab2Δp85–transduced Gab2−/− BM, was different (P < .0001 and P = .001, respectively, Mantel-Cox tests).
In vitro lymphoid transformation and in vivo leukemogenesis evoked by BCR-ABL1 are dependent on GAB2. (A) Agarose colony assays. BM from the indicated Balb/c donor (blue, Gab2+/+; red, Gab2−/−) mice was transduced with retrovirus coexpressing p210 BCR-ABL1 and either GFP or the indicated GAB2 variant. Transduced cells were plated directly in agarose as described in Methods, and transformed pre-B-lymphoid colonies were counted at day 10. The differences in colony numbers between p210-GFP–transduced Gab2+/+ and Gab2−/− BM and between p210-GFP–transduced and p210-Gab2WT–transduced Gab2−/− BM, were significant (P < .0001 and P = .001, respectively, unpaired Student t tests), whereas there was no difference between p210-GFP–transduced Gab2+/+ and p210-Gab2WT–transduced Gab2−/− BM (P = .224). The difference in colony numbers between Gab2−/− BM transduced with p210-Gab2WT retrovirus and either p210-Gab2Δp85 or p210-Gab2ΔShp2 retrovirus was significant (P = .0004 and P = .0002, respectively, unpaired Student t tests), whereas the difference between Gab2−/− BM transduced with p210-Gab2Δp85 and p210-Gab2ΔShp2 retrovirus did not reach significance (P = .0647, unpaired Student t test). (B) Whitlock-Witte assays. Freshly harvested Balb/c BM from Gab2+/+ (top) and Gab2−/− (bottom) donors was transduced with retrovirus coexpressing p210 BCR-ABL1 and either GFP or the indicated GAB2 variant, and plated in triplicate at the indicated cell numbers per well. Nontransduced cells were added to provide 106 total cells for stromal support. Wells were scored as positive when cell number reached 106 viable nonadherent cells per well, as described in Methods. (C) Extracts from immortalized BCR-ABL1–transformed B-lymphoid cell lines (from Figure 5B) from Gab2+/+ (WT) or Gab2−/− (knockout) donors were analyzed for GAB2 protein expression and phosphorylation, as described in Methods. (D) K-M curves of recipients of BM from Gab2+/+ littermates transduced with p210MIGFP retrovirus (blue solid line, n = 8), and of Gab2−/− donors (red lines) transduced with p210-IRESwt-Gab2WT retrovirus (dotted line, n = 8) or the indicated Gab2 mutant retrovirus (dashed lines, n = 8 for each cohort). All deaths were due to B-ALL. There was no significant difference in survival between recipients of p210MIGFP-transduced Gab2+/+ BM and p210-IRESwt-Gab2WT–transduced Gab2−/− BM (P = .493) or between recipients of p210MIGFP-transduced Gab2−/− BM and p210-IRESwt-Gab2ΔShp2–transduced Gab2−/− BM (P = .734). By contrast, the survival of recipients of p210MIGFP-transduced Gab2+/+ BM and p210MIGFP-transduced Gab2−/− BM, and of recipients p210-IRESwt-Gab2WT–transduced and p210-IRESwt-Gab2Δp85–transduced Gab2−/− BM, was different (P < .0001 and P = .001, respectively, Mantel-Cox tests).
BCR-ABL1 induces B-ALL through the GAB2-SHP2 signaling pathway
We also tested the role of GAB2 in B-lymphoid leukemogenesis by transducing BM from Balb/c Gab2+/+ and Gab2−/− donors without cytokine stimulation and transplanting transduced cells into lethally irradiated recipients. In this model, recipients of BCR-ABL1–transduced WT BM develop B-ALL, characterized by involvement of BM, spleen, and lymph nodes, along with malignant pleural effusions containing B220+ lymphoblasts.10 In contrast to the effect of GAB2 deficiency on myeloid leukemogenesis, wherein no recipient of BCR-ABL1–transduced Gab2−/− BM developed CML-like MPN (Figure 1), about half of the recipients of Gab2−/− BM developed B-ALL, but only after a significantly prolonged latent period (Figure 5D). Coexpression of WT Gab2WT or the Gab2Δp85 mutant with BCR-ABL1 restored induction of B-ALL in all recipients of Gab2−/− BM, although the survival of the Gab2Δp85 cohort was significantly longer. By contrast, the Gab2ΔShp2 mutant failed to rescue the lymphoid leukemogenesis defect. Together with the results of the in vitro growth assays (Figure 5B), these data show that GAB2 is not absolutely required for BCR-ABL1–induced B-lymphoid transformation or leukemogenesis, but GAB2-SHP2 signaling contributes significantly to the pathogenesis of BCR-ABL1–induced B-ALL.
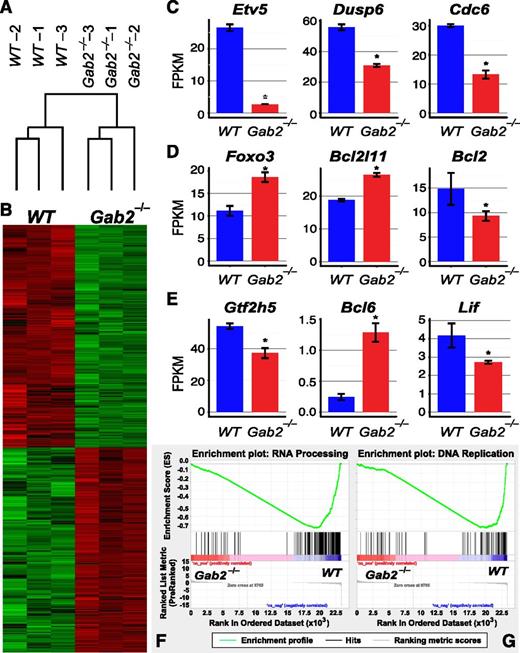
To analyze GAB2-dependent signaling pathways in BCR-ABL1–transformed B-lymphoid cells, we performed immunoblot analysis. Similar to its effects in myeloid transformation, GAB2 was required for full activation of ERK, AKT, STAT5, SFK, and S6 (Figure 6A-B; supplemental Figure 5A). We also performed RNA-seq analysis to assess whether these changes in signaling pathways altered downstream transcriptional events. Unsupervised hierarchical clustering revealed good separation of WT and Gab2−/− samples (Figure 7A), consistent with significant effects of Gab2 deficiency on the transcriptome. Supervised comparison revealed ∼6000 differentially expressed genes in WT and Gab2−/− pre-B cells (Figure 7B). Consistent with the observed alterations in proximal signaling events, multiple downstream transcriptional targets of the ERK, AKT, and STAT5 pathways were significantly altered (Figure 7C-E), including Etv5, Dusp6, and Cdc6 (ERK), Foxo3, Bcl2l11, and Bcl2 (AKT), and Gtf2h, Bcl6, and Lif (STAT5). Gene Ontology enrichment analysis showed that genes involved in RNA processing and DNA replication also were significantly downregulated in Gab2−/− cells (Figure 7F-G). Finally, we used Enrichr, coupled with the Encyclopedia of DNA Elements (ENCODE) ChIP-seq database, to identify downregulation of potential targets of transcription factors (TFs) including MYC, ELK, FOS, FOXM1, and STAT5 (supplemental Tables 1 and 2), reflecting the observed changes in BCR-ABL1 signaling (Figure 6A-B).
Analysis of signaling events in primary BCR-ABL1–expressing B-lymphoid progenitors. (A) Representative immunoblot of primary CD127+GFP+ cell extracts from Gab2+/+ (WT) or Gab2−/− BM cells transduced with BCR-ABL1 p210MIGFP retrovirus; similar results were obtained in 3 independent biological replicates. (B) Statistical analysis of signaling events pooled from 3 biological replicates of the experiment shown in A (**P < .01, *P < .05; 1-sided Student t tests). (C) Representative immunoblot of primary CD127+GFP+RFP+ cell extracts from Gab2−/− BM cells cotransduced with p210-IRES-RFP and Gab2-IRES-GFP viruses, as indicated. Similar results were obtained in 3 biological replicates. (D) Statistical analysis of signaling events pooled from 3 biological replicates of the experiment shown in C (**P < .01, *P < .05; P = .1 for p-STAT5 between Gab2ΔShp2 and Gab2WT; 1-sided Student t tests).
Analysis of signaling events in primary BCR-ABL1–expressing B-lymphoid progenitors. (A) Representative immunoblot of primary CD127+GFP+ cell extracts from Gab2+/+ (WT) or Gab2−/− BM cells transduced with BCR-ABL1 p210MIGFP retrovirus; similar results were obtained in 3 independent biological replicates. (B) Statistical analysis of signaling events pooled from 3 biological replicates of the experiment shown in A (**P < .01, *P < .05; 1-sided Student t tests). (C) Representative immunoblot of primary CD127+GFP+RFP+ cell extracts from Gab2−/− BM cells cotransduced with p210-IRES-RFP and Gab2-IRES-GFP viruses, as indicated. Similar results were obtained in 3 biological replicates. (D) Statistical analysis of signaling events pooled from 3 biological replicates of the experiment shown in C (**P < .01, *P < .05; P = .1 for p-STAT5 between Gab2ΔShp2 and Gab2WT; 1-sided Student t tests).
GAB2 deficiency affects transcription of downstream effectors. RNA-seq was performed on BCR-ABL1–transformed WT or Gab2−/− pre-B cells. (A) Unsupervised clustering of samples based on expression of all genes. (B) Heat map showing differential gene expression (supervised analysis) in WT and Gab2−/− cells. (C-E) mRNA levels of typical transcriptional targets downstream of the (C) ERK, (D) AKT, or (E) STAT5 pathway were altered (*FDR < 0.05, CuffDiff). (F-G) gene set enrichment analysis of Gene Ontology term enrichment in Gab2−/− compared with WT cells. Gene sets in biological processes of (F) RNA processing and (G) DNA replication were significantly enriched in downregulated genes in Gab2−/− cells. FDR < 0.01 for both (F) and (G).
GAB2 deficiency affects transcription of downstream effectors. RNA-seq was performed on BCR-ABL1–transformed WT or Gab2−/− pre-B cells. (A) Unsupervised clustering of samples based on expression of all genes. (B) Heat map showing differential gene expression (supervised analysis) in WT and Gab2−/− cells. (C-E) mRNA levels of typical transcriptional targets downstream of the (C) ERK, (D) AKT, or (E) STAT5 pathway were altered (*FDR < 0.05, CuffDiff). (F-G) gene set enrichment analysis of Gene Ontology term enrichment in Gab2−/− compared with WT cells. Gene sets in biological processes of (F) RNA processing and (G) DNA replication were significantly enriched in downregulated genes in Gab2−/− cells. FDR < 0.01 for both (F) and (G).
Reconstitution studies using Gab2 mutants indicated that SHP2 binding was required for ERK, AKT, S6, and possibly STAT5 activation, whereas p85 binding was required for activation of AKT (Figure 6C-D; supplemental Figure 5B). Hence, GAB2 activates similar downstream signaling events in BCR-ABL1–transformed myeloid and lymphoid cells, but the requirement of these pathways for transformation differs in the 2 hematopoietic neoplasms.
Discussion
Substantial effort has been made to identify key signaling pathways in Ph+ leukemias whose blockade can prevent or overcome TKI resistance and perhaps lead to cures.44 Mouse models provide invaluable tools for such studies, in part because they allow genetic manipulation of leukemic cells that is difficult to achieve using human cell lines or leukemia cells from patients. Here, we studied the effects of GAB2 deficiency in mouse models of CML and B-ALL and found that by acting through distinct signaling pathways, this scaffolding adaptor is required for myeloid and B-lymphoid leukemogenesis by BCR-ABL1.
We found that Gab2−/− BM was resistant to induction of CML-like MPN by BCR-ABL1, suggesting that signals from GAB2 are essential for the characteristic marked expansion of myeloid cells in this disease. GAB2 deficiency might have other effects that abrogate leukemogenesis in this model, including altered stem cell abundance, transduction efficiency, or engraftment ability. Several lines of evidence argue against these possibilities. We observed equivalent engraftment of provirus-positive cells (supplemental Figure 2), and circulating GFP+ myeloid cells (Figure 1C) were present in recipients of transduced Gab2−/− BM, indicating that they had been repopulated with retrovirus-transduced cells. Most importantly, the mouse Gab2-null allele was complemented by coexpression of GAB2 and BCR-ABL1 in transduced stem cells, which resulted in efficient restoration of MPN (Figure 3C-D).
The GRB2 binding site at Y177 of BCR-ABL1 is required for efficient induction of CML-like MPN in mice, and subsequent analyses have confirmed that pY177 has an analogous role in BCR-ABL1–expressing primary human CD34+ cells.45 Our previous studies demonstrated that GAB2 phosphorylation was diminished in leukemic cells expressing BCR-ABL1 Y177F, and in vitro transformation of GAB2-deficient myeloid and lymphoid progenitors by BCR-ABL1 also was decreased. The results here demonstrate that GAB2 is required for both myeloid and lymphoid leukemogenesis by BCR-ABL1 and argue that the critical signals flow through a pY177-GRB2-GAB2 complex. Although our data clearly show that GRB2/GAB2 complexes are essential, we do not exclude a role for SOS, another GRB2 effector, which mediates RAS activation, in CML pathogenesis. A direct role for GAB2 in human CML is also suggested by the demonstration that siRNA knockdown of GAB2 selectively impairs colony formation by CML progenitors in vitro.46 As hematopoiesis is essentially normal in Gab2−/− mice, these results suggest that inhibiting GAB2 function might be an effective treatment of Ph+ leukemia with little adverse effects on normal blood cells. Other work suggests that GAB2 might also be a therapeutic target in AML and some solid tumors.27-31
In CML patients, leukemic stem cells are relatively resistant to TKI therapy.47-49 Several observations suggest that GAB2 might be required for maintenance of BCR-ABL1+ leukemic stem cells. Although we were able to detect provirus-containing, Gab2-deficient myeloid cells in peripheral blood at 4 to 12 weeks after transplantation, these cells generally disappeared from the circulation at longer time points, and we were unable to serially transplant BCR-ABL1–expressing Gab2-deficient BM (data not shown). Although further studies are needed, these observations suggest that targeting GAB2 or GAB2-dependent pathways might be an effective strategy for eradication of CML, in addition to reversing the myeloproliferative phenotype.
Previous studies of BCR-ABL1 leukemogenesis have suggested that different signaling pathways are involved in CML and B-ALL pathogenesis.8,10 The ability to rescue BCR-ABL1 leukemogenesis in Gab2-deficient BM by coexpression of GAB2 allowed us to ask which functional domains of GAB2 are required. In the CML model, rescue of leukemogenesis by GAB2 required GRB2 binding, the PH domain, and SH2 domain binding sites for both p85 and SHP2. By contrast, B-lymphoid transformation and leukemogenesis were largely restored by the Gab2Δp85 mutant but not by Gab2ΔShp2. Together, these results emphasize that distinct signaling pathways are essential for the pathogenesis of BCR-ABL1+ myeloid and lymphoid leukemias. In particular, our results validate SHP2, which is required for the pathogenesis of both leukemia types, as an important new target for therapy of Ph+ leukemia. In parallel, another study investigated the role of GAB2 overexpression in conferring resistance of CML cells to ABL1 TKIs and found that GAB2-mediated TKI resistance is also dependent on its interaction with SHP2 and p85.50
Analysis of GAB2-dependent signals in BCR-ABL1–expressing primary myeloid and B-lymphoid cells revealed distinct roles for the GAB2 SHP2 and p85 binding sites in BCR-ABL1–induced transformation. In agreement with our previous observations,16 AKT and ERK activation were decreased in Gab2−/− myeloid cells expressing BCR-ABL1. We also found that GAB2 is required for full activation of SFKs and STAT5 by BCR-ABL1. As a direct target of SFKs, decreased STAT5 activation in Gab2-deficient cells could be explained by lower SFK activation. Alternatively, SHP2 might promote STAT5 activation by evoking cytokine production and consequent activation of JAK kinases; indeed, we have found that expression of a leukemogenic PTPN11 mutant in TF-1 myeloid cells promotes STAT activation via such an autocrine pathway (S.G. and B.G.N., unpublished data, August 2014). Conversely, activation of pathways downstream of GAB2 by activated STAT5 in myeloid leukemia cells has also been reported.51
RNA-seq analysis of WT and Gab2−/− BCR-ABL1+ pre-B cells revealed that these signaling changes led to corresponding alterations in downstream transcription. ENCODE TF-target enrichment analysis indicated that the activities of transcription factors such as MYC, FOS, ELK, FOXM1, and STAT5 are downregulated in Gab2−/− lymphoblasts, consistent with the lower phospho-ERK, phospho-AKT, and phospho-STAT5 in these cells. Gene Ontology term enrichment analysis indicated that biological processes such as RNA processing and DNA replication are downregulated in Gab2−/− cells. Changes in RNA processing might be attributable to lower MYC activity in Gab2−/− cells,52 whereas DNA replication can additionally be regulated by other transcription factors, such as FOS, FOXM1, and STAT5.53-55
We also discriminated the roles of SHP2 and PI3K binding to GAB2 in BCR-ABL1 signaling; whereas SHP2 binding is required for activation of ERK, S6, and STAT5, PI3K binding is required only for AKT activation. The failure of Gab2Δp85 to rescue myeloid leukemogenesis clearly implicates type IA PI3K as a potential therapeutic target in this disease, but whether AKT is also a valid therapeutic target in CML is unclear. In this regard, we have found that CML-like MPN can be induced efficiently in BM from mice with deficiency in any 1 of the 3 AKT isoforms (W.W.C. and R.A.V.E., unpublished observations, January 2013). Notably, our data indicate that p85 binding to GAB2 is less critical for BCR-ABL1–evoked lymphoid leukemogenesis, although type IA PI3Ks are required for this process.56 Conceivably, myeloid progenitors might have a higher dependence on GAB2 for activation of PI3K than do B-lymphoid progenitors, due to parallel mechanisms for type 1A PI3K activation in lymphoid, but not myeloid, progenitors. Alternatively, myeloid cells might require higher levels of PI3K activity than lymphoid cells to be transformed by BCR-ABL1, such that residual levels of activation in GAB2-deficient lymphoid cells are sufficient.
In conclusion, we identified a critical role for GAB2 in the pathogenesis of CML and Ph+ B-ALL and showed that leukemogenic signals from Y177 of BCR-ABL1 flow through distinct pathways in these hematologic malignancies. Our results validate GAB2 and downstream signaling molecules, particularly SHP2, as novel targets for therapy, whose inhibition might complement TKIs in the effort to prevent relapse of leukemia and permanently cure patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Katherine Lazarides and Monica Betancur for excellent technical assistance and Princess Margaret Cancer Center for RNA-seq analyses.
This work was supported by grants from the National Institutes of Health, National Cancer Institute (R01 CA090576) and the Leukemia & Lymphoma Society to R.A.V.E., from the Leukemia & Lymphoma Society and Worldwide Cancer Research to G.M., and by National Institutes of Health, National Cancer Institute grants R01 CA11494 and R37 CA049152 to B.G.N. Work in the B.G.N. laboratory was partially supported by the Ontario Ministry of Health and Long Term Care and the Princess Margaret Hospital Foundation. B.G.N. also was a Tier I Canada Research Chair.
Authorship
Contribution: S.G., W.W.C., G.M., J.R., B.G.N., and R.A.V.E. designed and performed the experiments; A.S., Z.L., and C.V. analyzed RNA-seq data; S.L. provided essential advice; and S.G., W.W.C., G.M., B.G.N., and R.A.V.E. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for G.M. is State University of New York System (SUNY) Upstate Medical University, Syracuse, NY.
The current affiliation for J.R. is University of Pittsburgh, Pittsburgh, PA.
The current affiliation for B.G.N. is Laura and Isaac Perlmutter Cancer Center, NYU Langone Medical Center, New York, NY.
The current affiliation for R.A.V.E. is Chao Family Comprehensive Cancer Center, University of California, Irvine, CA.
Correspondence: Richard A. Van Etten, Chao Family Comprehensive Cancer Center, University of California, Irvine, 839 Medical Sciences Court, Sprague Hall 124, Irvine, CA 92697; e-mail: vanetten@uci.edu; and Benjamin G. Neel, Laura & Isaac Perlmutter Cancer Center, 522 First Ave, Smilow Building 12th Floor, Suite 1201, New York, NY 10016; e-mail: benjamin.neel@nyumc.org.
References
Author notes
S.G., W.W.C., and G.M. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal