Key Points
A novel FXI binding tripeptide motif has sequence Asp-Phe-Pro (DFP).
FXI complex crystal structures reveal DFP peptides bound to the apple 2 domain.
Abstract
Factor XI (FXI) is the zymogen of FXIa, which cleaves FIX in the intrinsic pathway of coagulation. FXI is known to exist as a dimer and interacts with multiple proteins via its 4 apple domains in the “saucer section” of the enzyme; however, to date, no complex crystal structure has been described. To investigate protein interactions of FXI, a large random peptide library consisting of 106 to 107 peptides was screened for FXI binding, which identified a series of FXI binding motifs containing the signature Asp-Phe-Pro (DFP) tripeptide. Motifs containing this core tripeptide were found in diverse proteins, including the known ligand high-molecular-weight kininogen (HK), as well as the extracellular matrix proteins laminin and collagen V. To define the binding site on FXI, we determined the crystal structure of FXI in complex with the HK-derived peptide NPISDFPDT. This revealed the location of the DFP peptide bound to the FXI apple 2 domain, and central to the interaction, the DFP phenylalanine side-chain inserts into a major hydrophobic pocket in the apple 2 domain and the isoleucine occupies a flanking minor pocket. Two further structures of FXI in complex with the laminin-derived peptide EFPDFP and a DFP peptide from the random screen demonstrated binding in the same pocket, although in a slightly different conformation, thus revealing some flexibility in the molecular interactions of the FXI apple 2 domain.
Introduction
The physiological function of coagulation factor XI (FXI) is poorly understood compared with the vitamin K–dependent coagulation proteases; people deficient in FXI do not show the strong bleeding phenotypes observed for FVIII and FIX deficiency.1 From a structural perspective, FXI is unique among the coagulation factors because it circulates as a disulfide-linked dimer.2-4 Each subunit has 607 amino acids, with four 90–amino acid apple domains (A1-A4) that, in the FXI monomer, assemble into a saucer-shaped disc, with the protease domain resting on top in an arrangement that we have described as a cup and saucer.5 Biochemical data have mapped protein interactions to the apple domains for diverse ligands, including substrate FIX,6 thrombin,7 platelet glycoprotein Ib,8 and high-molecular-weight kininogen (HK),9 but to date, no complex crystal structures have been determined to define the molecular basis of these interactions.10 FXI circulates in complex with cofactor HK.9 A close homolog of FXI is plasma coagulation factor prekallikrein (PK), which has the same domain structure, with 4 apple domains at the N-terminus. In common with FXI, PK also binds HK, but unlike FXI, PK is a monomeric protein.11
The 2 pathways contributing to blood coagulation in humans are the intrinsic pathway, which can be activated by FXIa cleaving FIX, and the extrinsic pathway, which is activated by tissue factor. In addition to the principal substrate FIX, FXIa has also been shown to cleave and inactivate tissue factor pathway inhibitor, promoting a pathway of intravascular thrombus formation.12 Several lines of evidence suggest that therapeutic intervention targeting FXI and HK has the potential for providing medicines with a safer anticoagulation profile than those currently available.1,13,14
Random combinatorial peptide libraries based on the one-bead-one-compound method have proved successful in identifying ligands for various macromolecular targets.15 By screening a random peptide library comprising 106 to 107 peptides, we have identified FXI binding peptides that all share the core motif Asp-Phe-Pro (DFP). Of importance, this motif can also be found in the cofactor HK, laminin, and collagen V. By determining 3 FXI-DFP peptide ligand complex structures, we show how synthetic DFP peptides interact with a pocket in the apple 2 domain and how the dimeric FXI orients 2 bound DFP peptides onto the same surface.
Methods
Synthesis and screening of peptide libraries
The one-bead-one-compound libraries were synthesized on TentaGel S-NH2 resin (Rapp Polymere, Tübingen, Germany) using the split-mix method.15,16 Two unbiased libraries (libraries 1 and 2) where screened against biotinylated FXI. Library 1 was fixed at 12 residues in length; library 2 had varying lengths. The format for library 1 was O12-resin, whereby O was D,N,2Q,A,S,T,G,P,H,K,R,Y,W,F,L,I,V and M and C were omitted. The format for library 2 was OO-Ψ-O0,1,2-Ψ-O0,1,2-Ψ-O0,1,2-Ψ-OO-resin, whereby O was D,N,E,Q,A,S,T,G,P,H,K,R,V and Ψ (only hydrophobic) was W,F,Y,L,V; the residues M and C were omitted. A total of 1 g of resin was used in a 5-mL syringe with filter frit. The resin was washed 5 times with 15 mM Tris pH 7.4, 150 mM sodium chloride (NaCl), 0.5% bovine serum albumin, and 0.05% Tween 20 (incubation buffer). For prescreening of libraries against streptavidin-alkaline phosphatase (Strep-AP; stock 1 mg/mL; Sigma), Strep-AP alone was added to the library to a final dilution of 1:10 000 and incubated for 30 minutes. This library was then washed 4 times with washing buffer (15 mM Tris pH 7.4, 150 mM NaCl, and 0.05% Tween 20) and washed twice with staining buffer (50 mM Tris pH 8.8, 0.15 M NaCl, 0.05% Tween 80, and 15 mM magnesium chloride). The substrate 6-chloro-3-indoxyl-phosphate (50 mg/mL in N-methyl-2-pyrrolidone; Biosynth AG) was added, and the reaction was allowed to proceed for 30 to 45 minutes. For screening of libraries against FXI, the resins were washed with 6 M guanidinium chloride pH 3.0 for 20 minutes, washed with washing buffer, and lastly washed twice with incubation buffer. Biotinylation of FXI was performed using 200 μg of FXI, which was gel filtrated using a NAP5 column equilibrated with 1% sodium bicarbonate pH 8. Added to this solution (500 μL) was 4 μL of a 7 mM solution of biotin N-hydroxysuccinimide ester (Sigma) in DMSO, and the reaction was allowed to proceed for 3 hours at 4°C. The FXI was then gel filtrated using a NAP5 column against 100 mM NaCl and Tris buffer pH 7.4 and stored in aliquots (30 μL) at −25°C. A total of 4 μL of biotinylated FXI (stock concentration 1.2 μM) plus 1 μL of Strep-AP was added to the library in 2 mL of incubation buffer and allowed to incubate for 2 hours. The resin was washed 3 times with washing buffer and then staining buffer, and 5-bromo-4-chloro-3-indolyl phosphate with nitro blue tetrazolium was added. After 1 hour, the reaction was stopped and the most intense blue beads were removed for sequencing. For the sequence determination, blue beads were removed under a microscope and sequenced by an Edman sequencer (Procise, Applied Biosystems). Peptide sequences were aligned using the program MEME17 to produce Figure 1B, and database searches for DFP motifs were performed using PROSITE.18 In Figure 3A, HK protein sequences were aligned with National Center for Biotechnology Information HOMOLOGENE server and shaded with the BOXSHADE server.
Protein crystallization and structure determination
Recombinant human untagged FXI was expressed and purified from Chinese hamster ovary cells as previously described.19 All synthetic peptides used for crystallization experiments were purchased from GenScript. FXI was concentrated to 8 mg/mL and added to an equal volume of peptide sequence YPRHIYPDFPTDTT (P39), FNPISDFPDTTS (HKP), or RLEFPDFPIDD (LP2) to achieve a 1:5 protein-to-peptide molar ratio for P39 and a 1:6 ratio for peptides HKP and LP2. The resulting solution was 4 mg/mL FXI in 50 mM Tris-HCl pH 7.6 and 75 mM NaCl. Crystals were grown in sitting drop from conditions of 0.1 M 2-(N-morpholino)ethanesulfonic acid pH 6.5 and 20% polyethylene glycol 4000 for the FXI-P39 complex and 0.1 M N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid pH 7.5 and 20% polyethylene glycol 1500 for the FXI-HKP and FXI-LP2 complexes. Data were collected from 2 crystals for FXI-P39 and on a single crystal for FXI-HKP and FXI-LP2 at the Diamond Light Source synchrotron using beamline I04. Data were processed with XDS20 and reduced in space group P43212 using the CCP4 software suite21 (Table 1). The structures were solved by molecular replacement using Phaser,22 with the crystal structure of FXI (Protein Data Bank code 2F83)5 as a search model. High-quality 2mFo − DFc and mFo − DFc electron density maps enabled direct location of the bound peptides (supplemental Figure 2, available on the Blood Web site). Manual rebuilding was performed using the program Coot,23 and refinement was performed using REFMAC, with statistics shown in Table 1. Protein structures in Figures 2A-B,D,F and 4 were generated using PyMOL, and those in Figure 2C,E used LIGPLOT.24
Crystallographic data collection and refinement statistics
| . | FXI-HKP . | FXI-P39 . | FXI-LP2 . |
|---|---|---|---|
| Data collection | |||
| Space group | P43212 | P43212 | P43212 |
| Cell dimensions | |||
| a, b, c, Å | 80.6, 80.6, 251.8 | 81.3, 81.3, 252.2 | 81.1, 81.1, 253.0 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution, Å | 39.8-2.85 (2.92-2.85) | 77.4-2.80 (2.90-2.80) | 77.3-3.0 (3.08-3.0) |
| Rsym* | 0.083 | 0.094 | 0.158 |
| I/sigI | 16.0 (3.0) | 17.1 (2.8) | 12.5 (3.1) |
| Completeness, % | 99.9 (100.0) | 95.1 (71.9) | 97.8 (97.2) |
| Redundancy | 6.0 (6.2) | 8.3 (3.5) | 9.8 (9.1) |
| Refinement | |||
| Number of reflections | 20 319 | 19 565 | 23 419 |
| Rwork /Rfree† | 0.226/0.285 | 0.201/0.264 | 0.210/0.279 |
| B-factors, Å2 | |||
| Protein | 81.5 | 74.6 | 80.30 |
| Ligands | 21.0 | 111.80 | 117.50 |
| Root mean square deviations | |||
| Bond lengths, Å | 0.011 | 0.015 | 0.022 |
| Bond angles, ° | 1.82 | 1.82 | 1.98 |
| Protein Data Bank code | 5I25 | 5EOK | 5EOD |
| . | FXI-HKP . | FXI-P39 . | FXI-LP2 . |
|---|---|---|---|
| Data collection | |||
| Space group | P43212 | P43212 | P43212 |
| Cell dimensions | |||
| a, b, c, Å | 80.6, 80.6, 251.8 | 81.3, 81.3, 252.2 | 81.1, 81.1, 253.0 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution, Å | 39.8-2.85 (2.92-2.85) | 77.4-2.80 (2.90-2.80) | 77.3-3.0 (3.08-3.0) |
| Rsym* | 0.083 | 0.094 | 0.158 |
| I/sigI | 16.0 (3.0) | 17.1 (2.8) | 12.5 (3.1) |
| Completeness, % | 99.9 (100.0) | 95.1 (71.9) | 97.8 (97.2) |
| Redundancy | 6.0 (6.2) | 8.3 (3.5) | 9.8 (9.1) |
| Refinement | |||
| Number of reflections | 20 319 | 19 565 | 23 419 |
| Rwork /Rfree† | 0.226/0.285 | 0.201/0.264 | 0.210/0.279 |
| B-factors, Å2 | |||
| Protein | 81.5 | 74.6 | 80.30 |
| Ligands | 21.0 | 111.80 | 117.50 |
| Root mean square deviations | |||
| Bond lengths, Å | 0.011 | 0.015 | 0.022 |
| Bond angles, ° | 1.82 | 1.82 | 1.98 |
| Protein Data Bank code | 5I25 | 5EOK | 5EOD |
Values in parentheses are for highest-resolution shell.
Rsym is Sum(h) [Sum(j) [I(hj) − <Ih>]/Sum(hj) < Ih>, where I is the observed intensity and <Ih> is the average intensity of multiple observations from symmetry-related reflections calculated with SCALA.
Rwork is Sum(h) ||Fo|h − |Fc|h|/Sum(h) |Fo|h, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree is computed as in Rwork, but only for (5%) randomly selected reflections, which were omitted in refinement, calculated using REFMAC.
SPR binding studies
Plasma-purified HK was purchased from Enzyme Research Laboratories. Laminin 111, 411, and 511 were purchased from BioLamina. HK, FXI, or laminin was immobilized onto a CM5 chip using an amine coupling kit (GE Healthcare), and experiments were performed on a Biacore 3000 instrument. The running buffer used for all experiments was 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid pH 7.5, 140 mM NaCl, 5 μM ETDA, and 0.005% polysorbate 20. To assess any nonspecific binding, the analyte (FXI or peptide) was also injected over an empty flow cell. Collagen V peptide of sequence KDLQLCHPDFPDGEB and HK peptide of sequence INPTTQMKESYYFDLTDGLS (HKC) were purchased from Biomatik. Binding curves were analyzed on the basis of the surface plasmon resonance (SPR) response units recorded at equilibrium for each analyte protein concentration, and a Hill plot was generated using Prism 6 (GraphPad Software). Competition assays were performed with increasing concentrations of peptide P39 (6-600 nM) mixed with a fixed concentration of FXI (6 nM).
Results
Identification of peptides binding FXI
To identify peptides and novel ligands for FXI, we screened a random peptide library containing greater than 3 million peptides and identified 52 FXI binding peptides. From the sequences identified (termed P1-P52), 37 were observed to contain the tripeptide motif Asp-Phe-Pro (DFP) and 4 contained a related DFD sequence, 2 contained DFS sequences, and 1 contained the DLP sequence. Figure 1A shows the DFP-containing peptides aligned, and supplemental Figure 1 contains all 52 peptide sequences. A hydrophobic side chain F/Y/I/W located 2 amino acids before DFP is preferred across all DFP peptides, with the exception of HxDFP, and there are no examples in which a charged K/R/D/E amino acid occupies this position. The most common motif is F/YxDFP, present in 29 peptides; 3 peptides contain IxDFP and 2 contain WxDFP or HxDFP. A preference for proline occurs between F/Y/I and the DFP tripeptide, with the second preference being an acidic residue. It is also common to observe 1 or 2 basic residues N-terminal to the hydrophobic F/Y/I/W residue. By contrast, the C-terminus does not show any conserved features, and several peptides have no residues present C-terminal to the DFP motif.
FXI binding peptides with the DFP motif. (A) Alignment of peptide sequences from the random screen, with 37 peptides shown containing the DFP tripeptide. (B) Peptide motif overview calculated using MEME software for the 37 FXI-binding DFP peptide sequences. (C) Location of DFP pentapeptide sequences in the proteins HK (KNG1_HUMAN), laminin (LAMA1_HUMAN), and collagen V (CO5A1_HUMAN) identified using PROSITE software.
FXI binding peptides with the DFP motif. (A) Alignment of peptide sequences from the random screen, with 37 peptides shown containing the DFP tripeptide. (B) Peptide motif overview calculated using MEME software for the 37 FXI-binding DFP peptide sequences. (C) Location of DFP pentapeptide sequences in the proteins HK (KNG1_HUMAN), laminin (LAMA1_HUMAN), and collagen V (CO5A1_HUMAN) identified using PROSITE software.
A search of the Swiss-Prot database for extracellular human proteins containing the DFP motif revealed that the FxDFP sequence occurs in 13 human proteins, YxDFP in 4, HxDFP in 3, WxDFP in 2, and IxDFP in 7 (supplemental Tables 1-3). The known binding partner for FXI (cofactor HK)25 contains the sequence ISDFP (residues 585-589; mature sequence numbering is used from the Swiss-Prot database entry name KNG1_HUMAN), which is located in the C-terminal domain 6. Other proteins that contained a close match to sequences from the screen include the extracellular matrix protein laminin α1 chain, with the sequence YRDFP (sequence LAMA1_HUMAN, residues 1081-1085, termed LP1) and FPDFP (residues 2191-2195, termed LP2). The laminin peptide LP2 most closely resembles the screen peptides, with FPDFP being the most common pentapeptide motif occurring in 8 peptides. Other proteins that closely match the pentapeptide preferences include matrix protein collagen V, with the sequence HPDFP (sequence CO5A1_HUMAN, residues 1646-1650) and collagen XI (sequence COBA1_HUMAN, residues 1614-1618) (Figure 1C).
Structures of F/YxDFP peptide FXI complexes
We next synthesized examples of peptides containing the DFP motif, including peptide P39 from the screen and DFP peptides derived from HK (HKP) and laminin (LP2), and performed cocrystallization experiments with FXI. The laminin-derived LP2 peptide with sequence RLEFPDFPIDD cocrystallized with FXI in a 1:6 molar mixture, and the structure was solved to 3.0 Å resolution, with clearly identifiable electron density for the sequence EFPDFP. The peptide forms contacts exclusively with the apple 2 domain through interactions with a major and minor pocket, burying a total surface area of 992 Å2 (Figure 2A). Central to the interaction is the DFP phenylalanine side chain that fits precisely into a major pocket, forming contacts with side chains from the apple domain β sheet (residues Leu148, Ile146, Thr132, and Ala134). The DFP proline ring packs against the His143 side chain from the β4-β5 loop. The DFP aspartate side chain forms a salt bridge to FXI Lys103 and a hydrogen bond to the FXI Asn106 side chain (Figure 2A). A second interaction site occurs from the peptide phenylalanine that is 2 residues N-terminal to the DFP sequence contacting a minor pocket formed by residues Tyr107, Asn108, and Ser109. For this peptide, the phenylalanine main-chain amide and carbonyl form hydrogen bonds to the FXI residue Asn106 main-chain carbonyl and side chain nitrogen, respectively. The peptide glutamate residue side chain forms a hydrogen bond to the side chain of Asn106.
FXI peptide complex structures. (A) Cartoon diagram showing only the apple 2 domain in white, with laminin LP2 peptide shown as sticks in purple. Electrostatic and hydrogen bonding interactions are shown as lavender dotted lines. Residues interacting with the peptide are shown as sticks in green. (B) Charged surface representation of the apple 2 domain (blue is positive, red is negative) bound to the P39 peptide (orange) shown as sticks. (C) Schematic diagram illustrating the P39 peptide (purple) bound to the apple 2 domain, calculated using LIGPLOT software, with major and minor pockets indicated. (D) Cartoon diagram of the apple 2 domain in white, with bound HK peptide in blue shown as sticks. (E) Schematic diagram illustrating HK peptide (purple) bound to the apple 2 domain, calculated using LIGPLOT. (F) Superposition of the FXI-P39 and FXI-HKP peptide apple 2 domain structures. The apple 2 domain is shown as a cartoon in white for the FXI-P39 structure, and the P39 and HKP peptides are shown as sticks in orange and purple, respectively.
FXI peptide complex structures. (A) Cartoon diagram showing only the apple 2 domain in white, with laminin LP2 peptide shown as sticks in purple. Electrostatic and hydrogen bonding interactions are shown as lavender dotted lines. Residues interacting with the peptide are shown as sticks in green. (B) Charged surface representation of the apple 2 domain (blue is positive, red is negative) bound to the P39 peptide (orange) shown as sticks. (C) Schematic diagram illustrating the P39 peptide (purple) bound to the apple 2 domain, calculated using LIGPLOT software, with major and minor pockets indicated. (D) Cartoon diagram of the apple 2 domain in white, with bound HK peptide in blue shown as sticks. (E) Schematic diagram illustrating HK peptide (purple) bound to the apple 2 domain, calculated using LIGPLOT. (F) Superposition of the FXI-P39 and FXI-HKP peptide apple 2 domain structures. The apple 2 domain is shown as a cartoon in white for the FXI-P39 structure, and the P39 and HKP peptides are shown as sticks in orange and purple, respectively.
In addition, we determined the 2.8-Å structure of FXI in complex with peptide P39 from the random screen using a similar protocol. Clearly identifiable electron density was observed for the 8-residue sequence IYPDFPTD (supplemental Figure 2). The peptide occupies the same elongated crevice as the laminin peptide, and the tyrosine aromatic side chain packs into the same minor pocket. At the C-terminus of the P39 structure, the Thr-Asp residues are observed to form minor van der Waals contacts with FXI, as shown in Figure 2B-C.
Structure of the HK DFP peptide FXI complex
The FXI complex structure with a 12-mer peptide containing HK amino acids 582 to 593 was solved to 2.85-Å resolution. The HK peptide HKP was also observed bound to the apple 2 domain, and the sequence NPISDFPD was successfully identified in the electron density (Figure 2D). The DFP motif occupies the same major pocket as P39 peptide, and the isoleucine side chain from HKP forms an equivalent packing interaction to the tyrosine side chain from P39, contacting the FXI apple 2 domain minor pocket (Figure 2E).
Figure 2F illustrates a superposition of the FXI-P39 and FXI-HKP structures, revealing differences between IxDFP and YxDFP, whereby the DFP Phe contact with the FXI apple 2 domain major pocket is identical; however, there is some flexibility in the positioning of the N-terminal amino acids contacting the minor pocket. The P39 peptide tyrosine main-chain backbone amide forms a hydrogen bond to the main-chain carbonyl of FXI Asn140, which does not occur in the HKP complex. In contrast, both the native FXI structure and the HKP complex have a water molecule bound to the side chain of Asn140. In the HKP complex, this water molecule is also coordinated by the DFP aspartate side chain, whereas in the laminin and P39 complex structure, there is no bound water present and the DFP aspartate side chain is slightly reoriented, forming an interaction with the Asn106 side chain.
Biological significance of the FXI-HKP complex structure
The FXI-HKP structure is supported by previous data: biochemical characterization of the interaction between FXI and HK has localized binding to the C-terminal portion of the HK domain 6, spanning at least 58 amino acids (HK residues 556-613) and containing the HKP sequence.25 An alignment of the HK C-terminal region of domain 6 sequences across several species shows that the ISDFP sequence in HKP is well conserved (Figure 3A). Amino acid substitutions in the region surrounding ISDFP show similarities to the conserved features identified in the random peptide screen. In common with the FXI binding peptide sequences from the random screen, (1) the aspartate from the HKP DFP is absolutely conserved, (2) a proline residue is present directly N-terminal to the DFP in elephant and mole rat sequences, (3) a lysine residue is present 4 amino acids N-terminal to the DFP in cow, mole rat, and mouse, and (4) the mole rat IPDLP sequence is similar to the screen peptide 7, which has sequence FPDLP (supplemental Figure 1). One significant difference with the random peptide screen is that the HK sequences have a conserved isoleucine residue occurring 2 amino acids N-terminal to the DFP that is not substituted to F/Y/W/H in any species.
HK amino acid sequences and interaction with FXI. (A) The HK C-terminal domain 6 amino acid sequence alignment derived from the National Center for Biotechnology Information HOMOLOGENE server for different species is indicated. Conserved residues across >50% of the sequences are shaded black (highly conserved) or gray (weak conservation) with the program BOXSHADE. Peptides P27 and P31 from the random screen that match sequences within HK domain 6 are shown aligned. Dashed lines represent the sequence of synthetic peptides HKP and HKC. ★ indicates residue Ile563, which is the location of a venous thrombosis risk factor–associated human missense mutation Ile563Thr, located directly N-terminal to the HK ISDFP sequence. Plots of SPR sensorgrams measured in response units (RU) illustrate peptide P39 (B) and HKP (C) binding to immobilized FXI, and FXI binding to immobilized HK (D). (E) Binding of the HKC peptide to immobilized FXI.
HK amino acid sequences and interaction with FXI. (A) The HK C-terminal domain 6 amino acid sequence alignment derived from the National Center for Biotechnology Information HOMOLOGENE server for different species is indicated. Conserved residues across >50% of the sequences are shaded black (highly conserved) or gray (weak conservation) with the program BOXSHADE. Peptides P27 and P31 from the random screen that match sequences within HK domain 6 are shown aligned. Dashed lines represent the sequence of synthetic peptides HKP and HKC. ★ indicates residue Ile563, which is the location of a venous thrombosis risk factor–associated human missense mutation Ile563Thr, located directly N-terminal to the HK ISDFP sequence. Plots of SPR sensorgrams measured in response units (RU) illustrate peptide P39 (B) and HKP (C) binding to immobilized FXI, and FXI binding to immobilized HK (D). (E) Binding of the HKC peptide to immobilized FXI.
Previously, it had been shown that monoclonal antibodies against the FXI apple 2 domain can inhibit the FXI-HK interaction, with binding constants in the low nanomolar range.26,27 To examine the interaction of FXI with HK and peptide P39, we developed an SPR assay (see “Methods”). Using this assay, the P39 peptide and the HKP peptide bound to immobilized FXI with a KD of 320 μM and 324 μM, respectively, which is similar to the 150 μM value previously reported for a 13-mer peptide25 from the HK sequence (Figure 3B-C). The binding of FXI to full-length HK resulted in a KD of ∼0.3 nM, which differs significantly from the published value of 69 nM that was measured for the isolated HK light-chain fragment in a solution-based assay.25 Furthermore, using the same SPR assay, the homologous protein PK binding to HK resulted in a KD of 18 nM (data not shown), which is in excellent agreement with the published value of 12 nM measuring the PK interaction with the HK light-chain fragment.25
Because previous data have described that at least 58 amino acids from the HK domain 6 contribute to FXI binding,25 we examined these amino acid stretches for further matches with the FXI binding peptides. This revealed a sequence similarity with the DFD FXI binding peptides in the far C-terminal region of HK (Figure 3A shows the peptide P31 sequence aligned). To test whether this sequence binds FXI, we synthesized a peptide spanning the C-terminal residues 607 to 626 of HK (HKC) and, in an SPR experiment, recorded specific binding with a measured KD of 90 μM.
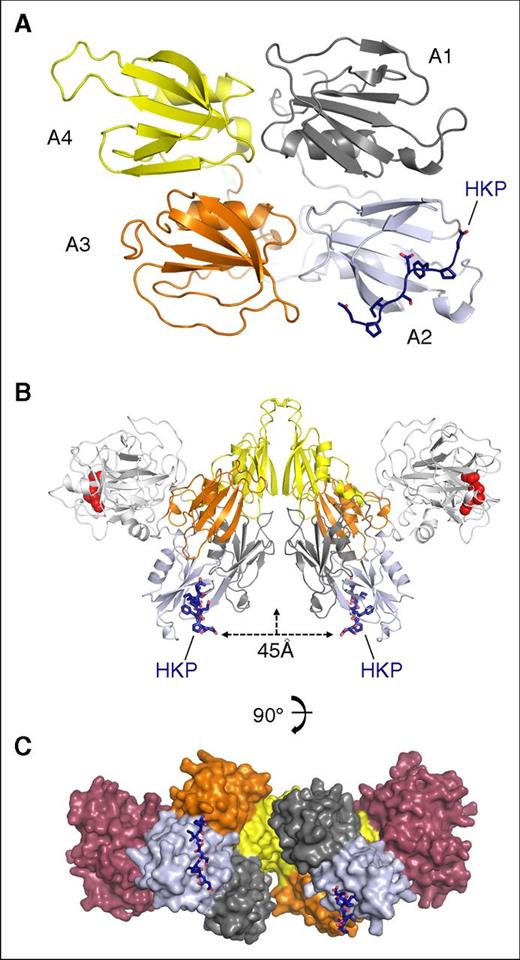
The FXI “saucer sections” organize DFP peptides onto the same surface
The structure of the HKP peptide bound to the apple 2 domain in the FXI saucer section is shown in Figure 4A. Two FXI monomers assemble into a disulfide-linked dimer through the apple 4 domain; Figure 4B shows that the 2 HKP peptides are organized onto an opposing surface in relation to the protease domain. The peptides are facing the dimer axis in an arrangement that effectively localizes the apple 2 domain binding pockets in the same plane 45 Å apart (Figure 4B). The apple 4 domain dimer interface is offset by an angle of 90° such that the orientation of the 2 bound HK peptides is approximately at right angles (Figure 4C; supplemental Video). This arrangement raises the question as to whether, in the biological complex, 2 HK polypeptides bind FXI using both apple 2 domains.
FXI dimer colocalization of bound HK peptides. (A) Cartoon diagram of the apple domain saucer section shown in gray, blue, orange, and yellow for the A1, A2, A3, and A4 domains, respectively. The HK peptide is colored in dark blue and shown as sticks. (B) The FXI dimer bound to the HK peptide (blue sticks) is shown with the protease domain (white) and active-site residues (red spheres) for His413, Asp462, and Ser557. The distance between the HK peptides is indicated as 45 Å, and the vertical arrow shows the dimer axis. (C) Surface representation of the FXI dimer, with each subunit colored the same as in panel A, except the protease domain is red.
FXI dimer colocalization of bound HK peptides. (A) Cartoon diagram of the apple domain saucer section shown in gray, blue, orange, and yellow for the A1, A2, A3, and A4 domains, respectively. The HK peptide is colored in dark blue and shown as sticks. (B) The FXI dimer bound to the HK peptide (blue sticks) is shown with the protease domain (white) and active-site residues (red spheres) for His413, Asp462, and Ser557. The distance between the HK peptides is indicated as 45 Å, and the vertical arrow shows the dimer axis. (C) Surface representation of the FXI dimer, with each subunit colored the same as in panel A, except the protease domain is red.
Early studies isolated the tightly bound FXI-HK complex from plasma, describing a complex of 380 kDa characterized by gel filtration.9 The disulfide-linked dimeric nature of FXI is well established biochemically and structurally,5 and on gel filtration, it elutes at ∼200 kDa, which is the same molecular weight described by nonreducing sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE). HK, by comparison, has been previously reported as both a disulfide-linked circular monomer28 and dimer.29,30 We repeated these gel filtration experiments to examine the stoichiometry of binding for the FXI-HK complex using a calibrated Superdex 200 column. This revealed that FXI elutes close to the expected molecular weight of the disulfide-linked dimer at ∼200 kDa (supplemental Figure 3). Gel filtration of FXI-HK mixtures with a varying ratio revealed a peak for a complex obtained with a molecular weight of ∼400 kDa, consistent with previously published results.9 SDS-PAGE of samples collected from the resulting gel filtration fractions of the FXI-HK complex revealed that both components were present, and nonreducing SDS-PAGE gels revealed FXI and HK migrating at the same molecular weight of ∼200 kDa (supplemental Figure 3). To quantitate FXI and HK in these experiments, a gel filtration experiment with a 1:1 molar ratio of FXI and HK was performed, and samples of the resulting single peak were analyzed by Edman degradation, confirming the equimolar presence of FXI and HK polypeptides in the 2:2 FXI-HK 400-kDa complex (supplemental Table 4). The lower plasma concentration of FXI dimer (0.031 μM) compared with HK (0.6363 μM) means that the 2:2 complex will be favored, and the structural data suggest that the 2 domains of HK domain 6 will be aligned in close proximity (Figure 4C).
FXI binding to laminin and a collagen V–derived DFP peptide
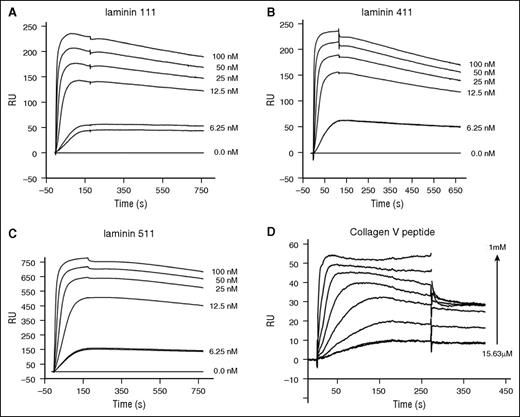
Laminins are large heterotrimeric proteins with α, β, and γ subunits and form a critical component of the basement membrane.31 Laminin 111 is important in development and occurs in the brain, whereas laminin 411 and 511 are abundant in the extracellular matrix of blood vessels and arteries.31 FXI has not been previously shown to bind proteins from the extracellular matrix; however, our observation that the FPDFP and YPDFP motifs are present in laminin prompted us to evaluate the binding of FXI to human laminin heterotrimers 111, 411, and 511 using SPR. These experiments demonstrated that FXI bound in a concentration-dependent manner to laminin 111, 411, and 511 with KD values of 11.1 nM, 9.2 nM, and 8.3 nM, respectively (plots shown in Figure 5; all KD values were determined using a Hill plot and averaged from triplicated experiments). We also found that laminin 111 purified from a native basement membrane (Engelbreth-Holm-Swarm murine sarcoma basement membrane; Sigma) bound FXI in a similar fashion to the recombinant laminins, with a KD of 18.8 nM (data not shown).
FXI binding to laminin and a collagen V peptide. Plots of SPR sensorgrams measured in response units (RU) illustrating FXI binding to laminin 111 (A), laminin 411 (B), or laminin 511 (C) immobilized on a CM5 sensor chip. (D) DFP peptide derived from the collagen V sequence binding to immobilized FXI.
FXI binding to laminin and a collagen V peptide. Plots of SPR sensorgrams measured in response units (RU) illustrating FXI binding to laminin 111 (A), laminin 411 (B), or laminin 511 (C) immobilized on a CM5 sensor chip. (D) DFP peptide derived from the collagen V sequence binding to immobilized FXI.
The HPDFP motif occurs in the C-terminal noncollagenous (NC) domains of collagen V and XI α1 chains (termed α1(V)) and does not occur in the fibrillar regions. To test whether a peptide derived from the α1(V) chain could bind FXI, we synthesized a peptide of sequence KDLQLCHPDFPDGE; SPR measurements revealed that this bound with a KD of 78 μM (Figure 5D).
Discussion
FXI contains 4 apple domains, which are members of the plasminogen, apple, and nematode family of protein folds found in the coagulation factors plasminogen, FXI, and PK, as well as in parasite cell binding proteins and hepatocyte growth factor.32 One of the fundamental and unanswered questions is how these apple domains recognize ligands.32 Here, we report that FXI binds peptides containing the DFP motif, with a preference for I/Y/F/W/HxDFP pentapeptides. Three complex crystal structures describe examples of I/Y/FxDFP peptides bound to the FXI apple 2 domain that utilize a major pocket in the center of the β sheet to bind the DFP; a minor pocket is occupied by the I/Y/F amino acid side chain. A second distinct group of FXI binding peptides were identified that contained the DFD motif, and these were found to have similarity to the C-terminal region of HK.
The data are biologically significant, in that an ISDFP pentapeptide occurs in the amino acid sequence of domain 6 from HK, which is a known cofactor of FXI. HK acts as a cofactor for FXI zymogen activation by FXIIa33,34 cleavage, and functional assays have shown that peptides derived from the HK domain 6 sequence inhibit kaolin-stimulated coagulation reactions.25 Monoclonal antibodies that bind to the FXI apple 2 domain have been shown to inhibit HK binding in vitro, as well as thrombus formation in vivo in animal models of cardiovascular disease.27 In addition to studies in animals, global genetic linkage analysis of mutations in humans has identified a common HK variant located in domain 6 that is linked to greater susceptibility to deep vein thrombosis and changes in coagulation.35 This missense mutation Ile563Thr is located directly N-terminal to the HK ISDFP sequence (highlighted in Figure 3A as a star). Directly C-terminal to the HK ISDFP sequence is the Cys596 residue, which is implicated in forming a disulfide bond with Cys10 from the HK N-terminus, although the complete disulfide bonding has been reported previously only for splice variant low-molecular-weight kininogen,36 and not for HK. The overall 3-dimensional structure of the HK domain 6 is unknown, and there are no homologous proteins in the database to allow molecular modeling.
We identified the HPDFP sequence in the C-terminal NC domain of collagen V. This sequence occurs in the α1(V) chain, and collagen V is normally present within the interiors of collagen fibers as α1(V)2α2(V) heterotrimers. Upon secretion, the collagen V C-terminal NC domains are cleaved off and secreted into the extracellular space.37 Excessive amounts of α1(V) chains have been shown to be deposited in atherosclerotic lesions, although it is unknown whether the C-terminal NC domains are also deposited.38,39
FPDFP and YRDFP pentapeptide sequences occur within the extracellular matrix protein laminin, and we demonstrated binding of FXI to native and recombinant laminin heterotrimers. There is increasing evidence of the importance of laminins in plasma coagulation and thrombus formation, and the interaction between platelet integrins and laminins has been well characterized.40,41 Efficient coagulation factor cleavage reactions typically take place only on a designated surface, and vitamin K–dependent coagulation factors have an N-terminal Gla domain that is capable of binding directly to negatively charged phospholipids present in activated platelets to fulfill this purpose. Laminin has been previously shown to provide a surface that can promote plasma coagulation and thrombus formation in the absence of platelets.42 HK and coagulation FXII (which activates FXI) have both previously been reported to bind laminin.43,44 It is notable that the substrate of FXIa is FIX which binds the basement membrane through a well-characterized interaction with collagen IV,45 and the interaction we observe for FXI with laminin may be important for colocalization of coagulation factors from the intrinsic pathway to sites in the extracellular matrix.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Diamond Light Source for access to beamline I04 (proposal number 10369). The authors thank Helen Phillipou (University of Leeds) for assistance with SPR data collection, Lars Thim (Novo Nordisk) for N-terminal sequencing, and Hanne Grøn (Novo Nordisk) for critical reading of the manuscript.
This work was supported by an industrial PhD studentship from the Danish Agency for Science Technology and Innovation (S.S.W.) and the British Heart Foundation Programme grant RG/12/9/29775 (J.E.).
Authorship
Contribution: S.S.W. and G.H. crystallized and determined the structures of FXI-P39, FXI-LP2, and FXI-HKP; S.Ø. performed the peptide screening experiments; S.S.W., C.L., and P.M.W. performed and analyzed the SPR experiments; and J.E. and H.S. initiated and designed the research, analyzed the results, and wrote the paper.
Conflict-of-interest disclosure: S.Ø. and H.S. are Novo Nordisk employees. The remaining authors declare no competing financial interests.
Correspondence: Jonas Emsley, Centre for Biomolecular Sciences, School of Pharmacy, University of Nottingham, Nottingham NG72RD, United Kingdom; e-mail: jonas.emsley@nottingham.ac.uk.