Key Points
PECAM-1 is enriched at endothelial cell intercellular junctions, where it regulates leukocyte trafficking and vascular permeability.
An atomic-level model of junctional PECAM-1 has been built based on a 2.8-Å resolution structure of its homophilic-binding domain.
Abstract
Platelet endothelial cell adhesion molecule-1 (PECAM-1) is a 130-kDa member of the immunoglobulin gene superfamily (IgSF) that is present on the surface of circulating platelets and leukocytes, and highly expressed at the junctions of confluent endothelial cell monolayers. PECAM-1–mediated homophilic interactions, known to be mediated by its 2 amino-terminal immunoglobulin homology domains, are essential for concentrating PECAM-1 at endothelial cell intercellular junctions, where it functions to facilitate diapedesis, maintain vascular integrity, and transmit survival signals into the cell. Given the importance of PECAM-1–mediated homophilic interactions in mediating each of these cell physiological events, and to reveal the nature and orientation of the PECAM-1–PECAM-1 homophilic-binding interface, we undertook studies aimed at determining the crystal structure of the PECAM-1 homophilic-binding domain, which is composed of amino-terminal immunoglobulin homology domains 1 and 2 (IgD1 and IgD2). The crystal structure revealed that both IgD1 and IgD2 exhibit a classical IgSF fold, having a β-sandwich topology formed by 2 sheets of antiparallel β strands stabilized by the hallmark disulfide bond between the B and F strands. Interestingly, despite previous assignment to the C2 class of immunoglobulin-like domains, the structure of IgD1 reveals that it actually belongs to the I2 set of IgSF folds. Both IgD1 and IgD2 participate importantly in the formation of the trans homophilic-binding interface, with a total buried interface area of >2300 Å2. These and other unique structural features of PECAM-1 allow for the development of an atomic-level model of the interactions that PECAM-1 forms during assembly of endothelial cell intercellular junctions.
Introduction
Platelet endothelial cell adhesion molecule-1 (PECAM-1; CD31) is a 130-kDa member of the immunoglobulin gene superfamily (IgSF) present on the surface of circulating platelets and leukocytes, and highly expressed at the junctions of confluent endothelial cell monolayers.1-3 The extracellular domain of PECAM-1 is composed of 6 immunoglobulin homology domains, followed by a 19-residue single-pass transmembrane domain and a 118-aa cytoplasmic tail. Like intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1), the extracellular domain of PECAM-1 functions to mediate cell-cell interactions; though ICAM-1, -2, and -3 and VCAM-1 each bind cell surface integrins via a signature motif present in a turn between immunoglobulin domain β sheets,4,5 PECAM-1 has no such integrin-binding sequence, and thus does not interact with integrins.6 Instead, PECAM-1 primarily binds other PECAM-1 molecules via homophilic-binding interactions known to require amino-terminal immunoglobulin homology domains 1 and 2 (IgD1 and IgD2).6-8
PECAM-1–mediated homophilic interactions are known to be essential for concentrating PECAM-1 at endothelial cell intercellular junctions,9,10 where it functions to maintain vascular integrity under conditions of inflammatory or thrombotic stress,11-15 in survival signaling in endothelial cells subjected to apoptotic stimuli,16,17 and in mediating leukocyte/endothelial cell interactions during the process of diapedesis.18-20 Mutagenesis studies have implicated amino acids on either side of the face of IgD1 in homophilic binding,8 however, the spatial orientation of this domain, the role that IgD2 plays in positioning IgD1 to participate in PECAM-1 homophilic interactions, and the contribution of its sialylated glycans to homophilic PECAM-1/PECAM-1 interactions21-23 remain to be critically explored. Complicating things further, PECAM-1 forms dimers and oligomers within the plane of the plasma membrane.24,25 Finally, studies using PECAM-1–containing nanodiscs and intact endothelial cells suggest that the affinity of PECAM-1/PECAM-1 homophilic interactions may be additionally regulated by heterophilic ligands that bind membrane-proximal PECAM-1 IgD6.26
Given the importance of PECAM-1–mediated homophilic interactions in mediating each of these cell physiological events, and to reveal the nature and orientation of the PECAM-1–PECAM-1 homophilic-binding interface, we undertook studies aimed at determining the crystal structure of the homophilic-binding domain of PECAM-1. Our results yield novel insights and suggest a model for the molecular nature of the interactions that PECAM-1 forms during assembly of endothelial cell intercellular junctions.
Materials and methods
Generation of PECAM-1 IgD1+D2
A complementary DNA sequence encoding PECAM-1 IgD1 and IgD2 (amino acids 1-202) was inserted into the vector pMT/BiP/V5-His A (Life Technologies, Grand Island, NY), and transfected into Drosophila S2 cells together with 0.2 μg of pCoBlast (Life Technologies). Protein expression of stably transfected cells was induced by CuSO4, and the recombinant protein purified on a HisTalon (Clonetech, Mountain View, CA) column. After removing the V5-His tag with tobacco etch virus protease, the protein was further purified by Sephacryl S-100 size-exclusion chromatography (supplemental Figure 1A, available on the Blood Web site), and concentrated to 8 to 10 mg/mL for crystallization. Selenomethionine (SeMet)-labeled protein was prepared by culturing the cells in ESF921 protein-free, methionine-deficient medium (Expression Systems, Davis, CA) for 6 hours before addition of 50 μg/mL SeMet (Thermo Fisher Scientific, Waltham, MA) and 700 μM CuSO4 for induction of protein expression. Additional SeMet was added at 24, 48, and 72 hours, and recombinant PECAM-1 IgD1-D2 purified from the cell supernatant as described in this paragraph. The degree of selenium incorporation was quantified by mass spectrometry (supplemental Figure 2).
Dimerization analysis
Gel-filtered PECAM-1 IgD1-D2 was dialyzed into 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES] pH 7.4, 150 mM NaCl), diluted to a concentration of either 2 or 6 mg/mL, and cross-linked by addition of ethylene glycol bis(succinimidyl succinate) (Sigma-Aldrich, St. Louis, MO) at concentrations of 0.05, 0.1, 0.5, 1, 2, and 5 mM. After 1 hour on ice, the reaction was stopped by addition of 20 mM Tris, pH 7.4, 150 mM NaCl. Crosslinked products were analyzed under reducing conditions by sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Crystallization, data collection, and structure determination
Initial crystallization conditions were determined by high-throughput Microbatch-Under-Oil screening at the Hauptman-Woodward Medical Research Institute (Buffalo, NY) using cocktails that differ in salt, pH, and polyethylene glycol (PEG) concentrations.27 Data were analyzed with Auto Sherlock28 to optimize the crystallization conditions. Diffraction-quality crystals were generated using the hanging drop method, with final conditions of 1 to 1.5 μL of PECAM-1 IgD1+D2 protein (8 mg/mL) and 0.66 to 1 μL of a Reservoir solution composed of 0.1 M Tris, pH 8.5, 0.1 M Mg(NO3)2, 18% to 20% PEG 3350, 0% to 1% glycerol at 19°C. Single crystals were harvested by soaking in 25% glycerol before flash-freezing in liquid nitrogen.
Diffraction data collected at the Life Sciences Collaborative Access Team (LS-CAT) beamline 21-ID-D at Argonne National Laboratory were processed with XDS.29 The resolution cutoff was based on the consideration of both CC1/2 and I/σI. Using a model of PECAM-1 IgD1-D2 generated by the PHYRE2 server,30 molecular replacement (MR) performed with the MR-Rosetta31,32 module of the Phenix package33 yielded a partial model with an Rwork of 0.378 and Rfree of 0.484 that was used for phasing. Multi-wavelength anomalous diffraction or single-wavelength anomalous diffraction (SAD) data from single SeMet-labeled crystals gave weak anomalous signals (supplemental Figure 3) owing to incomplete incorporation of SeMet (supplemental Figure 2), thus multicrystal anomalous diffraction was used for phasing.34 Selenium SAD data sets collected from 16 single crystals at the same wavelength were processed separately by XDS (supplemental Table 1) and then combined at a resolution cutoff of 3.0 Å using POINTLESS and SCALA35 within the CCP4 package36 (supplemental Figure 3; Table 1).
Statistics of x-ray diffraction data and structure refinement
| . | PECAM-1 D1-D2 . | ||
|---|---|---|---|
| Native . | Se SAD (single) . | Se SAD (combined) . | |
| Data collection | |||
| Space group | I4122 | I4122 | I4122 |
| Unit cell | |||
| a, b, c, Å | 103.6, 103.6, 286.1 | 104.0, 104.0, 281.8 | 103.6, 103.6, 280.3 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength, Å | 0.97872 | 0.97624 | 0.97624 |
| Resolution, Å* | 20-3.0 (3.5-3.0) | 20.0-2.8 (3.2-2.8) | 20-3.0 (3.2-3.0) |
| Rmerge,† (%)* | 19.5 (50.5) | 12.5 (49.3) | 48.3 (79.6) |
| No. of reflections, measured/unique | 177 694/29 711 | 223 739/36 261 | 4 827 647/16 005 |
| <I/σI>* | 8.7 (2.3) | 11.8 (2.4) | 36.3 (8.5) |
| Completeness (%)* | 99.6 (99.8) | 99.6 (99.8) | 98.9 (94.8) |
| Redundancy* | 6.0 (4.7) | 6.2 (4.8) | 301.6 (232.7) |
| Refinement | |||
| Resolution, Å | 20.0-2.80 | ||
| Unique reflections, work/free | 36 246/1 014 | ||
| Rwork/Rfree‡ | 0.238/0.279 | ||
| No. of atoms: protein/carbohydrates/water | 3 232/109/27 | ||
| Molecules/asymmetric unit | 2 | ||
| RMSD from ideal | |||
| Bond lengths, Å | 0.015 | ||
| Bond angles, ° | 1.736 | ||
| Ramachandran plot: favored/allowed/outliers, % | 94.2/5.5/0.3 | ||
| PDB accession no. | 5C14 | ||
| . | PECAM-1 D1-D2 . | ||
|---|---|---|---|
| Native . | Se SAD (single) . | Se SAD (combined) . | |
| Data collection | |||
| Space group | I4122 | I4122 | I4122 |
| Unit cell | |||
| a, b, c, Å | 103.6, 103.6, 286.1 | 104.0, 104.0, 281.8 | 103.6, 103.6, 280.3 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength, Å | 0.97872 | 0.97624 | 0.97624 |
| Resolution, Å* | 20-3.0 (3.5-3.0) | 20.0-2.8 (3.2-2.8) | 20-3.0 (3.2-3.0) |
| Rmerge,† (%)* | 19.5 (50.5) | 12.5 (49.3) | 48.3 (79.6) |
| No. of reflections, measured/unique | 177 694/29 711 | 223 739/36 261 | 4 827 647/16 005 |
| <I/σI>* | 8.7 (2.3) | 11.8 (2.4) | 36.3 (8.5) |
| Completeness (%)* | 99.6 (99.8) | 99.6 (99.8) | 98.9 (94.8) |
| Redundancy* | 6.0 (4.7) | 6.2 (4.8) | 301.6 (232.7) |
| Refinement | |||
| Resolution, Å | 20.0-2.80 | ||
| Unique reflections, work/free | 36 246/1 014 | ||
| Rwork/Rfree‡ | 0.238/0.279 | ||
| No. of atoms: protein/carbohydrates/water | 3 232/109/27 | ||
| Molecules/asymmetric unit | 2 | ||
| RMSD from ideal | |||
| Bond lengths, Å | 0.015 | ||
| Bond angles, ° | 1.736 | ||
| Ramachandran plot: favored/allowed/outliers, % | 94.2/5.5/0.3 | ||
| PDB accession no. | 5C14 | ||
PDB, Protein Data Bank; RMSD, root-mean-square deviation.
The highest resolution shell is shown in parentheses.
Rmerge = ΣhΣi|Ii−〈I〉|/ΣhΣiIi, where Ii is the observed intensity of the i-th measurement of reflection h, and〈I〉is the average intensity of that reflection obtained from multiple observations.
R = Σ||Fo|−|Fc||/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively, calculated for all data. Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 2.8% subset of data excluded from refinement.
The combined multicrystal data set analyzed using Phenix.Xtriage showed anomalous measurability up to 0.6 at low resolution, extending to ∼5-Å resolution (supplemental Figure 3). The selenium-containing substructure was determined using Phaser-EP37 within the Phenix package. Phaser-EP used in the MR-SAD mode, together with the partial MR model mentioned in the previous paragraph, facilitated substructure determination. The resulting heavy atom sites were used in AutoSol38 for phase calculation. The initial phase was calculated using Phaser, and an initial model was built with both Resolve and Buccaneer within AutoSol. Autobuild39 was then used to build a nearly complete model containing 418 aa with an Rwork of 0.30 and Rfree of 0.33. The complete model was manually built using COOT.40 An SAD data set processed to 2.8-Å resolution was used for final structure refinement with both Refmac5 and Phenix.refine.41 The final structural model was validated with Molprobity.42
Epitope mapping of the murine anti-PECAM-1 mAb PECAM-1.3
Ten-residue overlapping peptides corresponding to PECAM-1 IgD1 were synthesized directly onto a membrane with the SPOTS Epitope Mapping Kit (Sigma-Aldrich) and probed with monoclonal antibody (mAb) PECAM-1.3 to identify sequences that defined its epitope.
Results
PECAM-1 IgD1-D2 crystalizes as a strand-swapped cis dimer
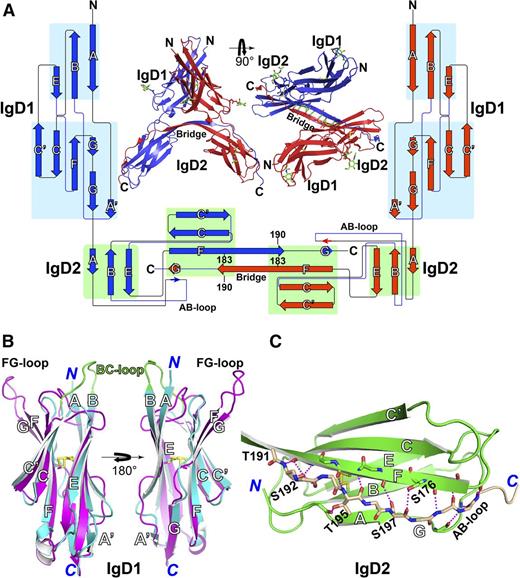
The structure of IgD1-D2 was determined by multicrystal anomalous diffraction and refined at 2.8-Å resolution with Rwork and Rfree of 23.8% and 27.9%, respectively (Table 1). Each asymmetric unit contains 2 copies of PECAM-1 IgD1-D2, each of which adopts a classical 2-layer β-sandwich topology for both immunoglobulin domains (Figure 1A43 ). Interestingly, IgD1-D2 exists as a cis homodimer formed by exchange of IgD2 C-terminal strand G (Figure 1A). The 2 IgD1-D2 molecules are highly superimposable except for a slight conformational diversity in strand G (supplemental Figure 4A). Strand swapping is facilitated by extension of the C termini of β-strand F toward its counterpart to form an antiparallel β-sheet that extends over 8 residues at the bridge region where the 2 strands meet (Figure 1A). Besides the strand-exchange region, there are no other close contacts between the 2 IgD1-D2 molecules in the strand-swapped dimer (Figure 1A).
Structure of the PECAM-1 homophilic-binding domain. (A) Side and top views of the structure of IgD1-D2 in the asymmetric unit. N-linked glycans are shown as sticks with green carbons and red oxygens. The backbone hydrogen bonds at the hinge region are dashed in green. The protein topology diagram was generated with Pro-origami43 with manual modifications. The lengths of the β-strand symbols, but not their linkers, are proportional to the lengths of amino acid sequences. Two IgD1-D2 molecules in blue and red form a dimer through swapping of the G β strand at the C terminus of IgD2 domain. (B) Superimposition of PECAM-1 IgD1 (in cyan) and human ICAM-1 IgD2 (in magenta, PDB code, 1IC1), with disulfide bonds represented by yellow sticks. (C) Structure of PECAM-1 IgD2. The swapped G strand is shown in wheat carbons. Oxygens and nitrogens are red and blue, respectively. The hydrogen bonds are dashed in magenta.
Structure of the PECAM-1 homophilic-binding domain. (A) Side and top views of the structure of IgD1-D2 in the asymmetric unit. N-linked glycans are shown as sticks with green carbons and red oxygens. The backbone hydrogen bonds at the hinge region are dashed in green. The protein topology diagram was generated with Pro-origami43 with manual modifications. The lengths of the β-strand symbols, but not their linkers, are proportional to the lengths of amino acid sequences. Two IgD1-D2 molecules in blue and red form a dimer through swapping of the G β strand at the C terminus of IgD2 domain. (B) Superimposition of PECAM-1 IgD1 (in cyan) and human ICAM-1 IgD2 (in magenta, PDB code, 1IC1), with disulfide bonds represented by yellow sticks. (C) Structure of PECAM-1 IgD2. The swapped G strand is shown in wheat carbons. Oxygens and nitrogens are red and blue, respectively. The hydrogen bonds are dashed in magenta.
Both IgD1 and IgD2 exhibit a classical IgSF fold, having a β-sandwich topology formed by 2 sheets of antiparallel β strands stabilized by the hallmark disulfide bond between the B and F strands (Figure 1B-C; supplemental Figure 4B). Despite previous assignment to the C2 class of immunoglobulin-like domains, the structure of IgD1 reveals that it actually belongs to the I2 set of IgSF folds, which was first defined in the structure of ICAM-1 IgD244 and has been found in the IgSF domains of other cell adhesion molecules, such as Dscam.45 The I2 set differs from the C2 fold by the presence of an A′ β strand (Figure 1B).46 Thus, 1 β-sheet of PECAM-1 IgD1 is composed of strands A, B, and E, whereas the other contains strands A,′ G, F, C, and C′. A short 310 helix is present within the EF loop, whereas strand G contains 2 small β-strand segments separated by a flexible loop (Figure 1B; supplemental Figure 4B). PECAM-1 IgD1 has high structural similarity with human ICAM-1 IgD2, with a root mean square deviation of only 2.2 Å over 81 Cα atoms (Figure 1B). In contrast to ICAM-1 IgD2, which uses its FG loop to mediate its homophilic interactions,44 PECAM-1 IgD1 has a markedly longer protrusion of the BC loop, which contains the homophilic-binding blocking epitope for the mAb PECAM-1.3.47
IgD2 is intermediate between the C2 and I2 sets of IgSF domains because although it does not contain a typical A′ strand, its AB loop interacts with the G stand through 2 backbone hydrogen bonds (Figure 1C). The majority of strand G in IgD2 adopts a loop conformation that forms backbone hydrogen bonds with strand F (Figure 1C). The side chains of Ser176, Ser192, and Ser197 also contribute to the hydrogen-bond network between the F and G strands (Figure 1C). The side chain of Thr195 in G-strand hydrogen bonds to a backbone oxygen of A strand (Figure 1C). The FG linker region in IgD2 exhibits a β-strand conformation stabilized by an antiparallel interaction with its counterpart FG linker (Figure 1A). In the absence of strand swapping, this FG linker might have formed a loop as seen in a typical immunoglobulin fold. The secondary structure of PECAM-1 IgD1 and IgD2 in relation to the amino acid sequence of these 2 domains is depicted schematically in Figure 2.48-50
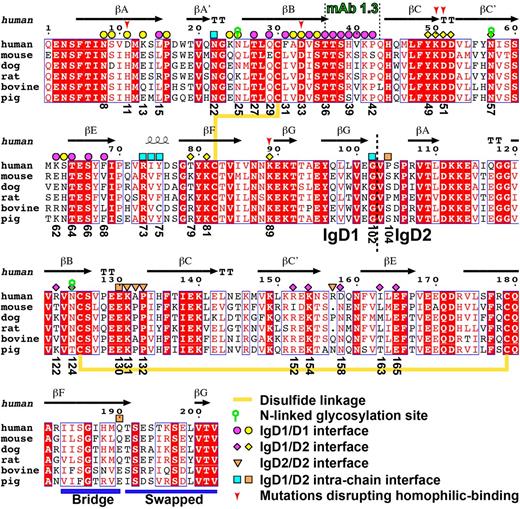
Sequence alignment of PECAM-1 IgD1-D2 across species. The alignment was generated with the ESPript server48 and modified manually. Conserved residues are boxed and highlighted in red. Secondary structures were defined by the DSSP program49,50 based on the crystal structure of PECAM-1. Disulfide linkages, N-linked glycosylation sites, the epitope for mAb PECAM-1.3, and the IgD1-D2 boundary are indicated. The interfacial residues are marked with different shapes, with color codes corresponding to Figures 4 and 5. The F-G bridge region and the swapped G strand are indicated with blue bars.
Sequence alignment of PECAM-1 IgD1-D2 across species. The alignment was generated with the ESPript server48 and modified manually. Conserved residues are boxed and highlighted in red. Secondary structures were defined by the DSSP program49,50 based on the crystal structure of PECAM-1. Disulfide linkages, N-linked glycosylation sites, the epitope for mAb PECAM-1.3, and the IgD1-D2 boundary are indicated. The interfacial residues are marked with different shapes, with color codes corresponding to Figures 4 and 5. The F-G bridge region and the swapped G strand are indicated with blue bars.
At the relatively dilute conditions used for size-exclusion chromatographic purification, IgD1-D2 molecules eluted as a monomeric species (supplemental Figure 1A), but chemical crosslinking of IgD1-D2 revealed concentration-dependent formation of dimeric species (supplemental Figure 1B), raising the possibility that the strand-swapped cis dimer observed in the IgD1-D2 crystal may exist in solution. However, whether the same dimer is also present in full-length PECAM-1 will require further investigation.
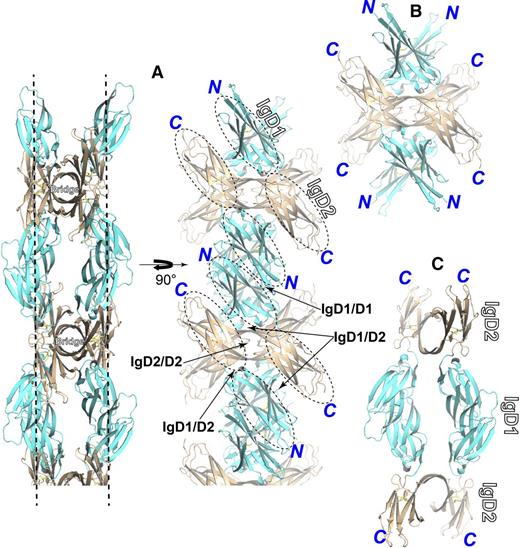
Domain interfaces observed in the crystal lattice contacts
As dissected in Figure 3, in the IgD1-D2 crystal lattice, 2 strand-swapped dimers cross at the bridge region to form a tetrameter of IgD1-D2, which enables antiparallel IgD1/D2 contacts (Figure 3A-B). The tetramers then form an array through the antiparallel IgD1/D1 contacts (Figure 3A). The tetramer arrays are orthogonal to each other to build the lattice packing (supplemental Figure 5). Thus, the 2 copies of IgD1-D2 monomer form 2 layers of domain contacts in each array of tetramers (Figure 3A), which define the homophilic interdomain interfaces of IgD1/D1, IgD1/D2, and IgD2/D2, in addition to the intrachain interface of IgD1/D2 (Figure 3A).
Molecular assembly and interfaces present in the crystal lattice. IgD1 and IgD2 are colored in cyan and wheat, respectively. The assemblies were dissected and shown separately. (A) A representative array of IgD1-D2 molecular packing in the crystal lattice. The 2 molecules of the strand-swapped dimer form 2 layers of lattice contacts marked with dashed lines. One layer of the contacts is shown by rotating the molecule 90° along the y-axis with the interfaces indicated by the arrows. (B) The assembly of a tetramer formed by 2 strand-swapped dimers crossed at the bridge region. (C) The assembly of a tetramer formed by IgD1/D1 interactions.
Molecular assembly and interfaces present in the crystal lattice. IgD1 and IgD2 are colored in cyan and wheat, respectively. The assemblies were dissected and shown separately. (A) A representative array of IgD1-D2 molecular packing in the crystal lattice. The 2 molecules of the strand-swapped dimer form 2 layers of lattice contacts marked with dashed lines. One layer of the contacts is shown by rotating the molecule 90° along the y-axis with the interfaces indicated by the arrows. (B) The assembly of a tetramer formed by 2 strand-swapped dimers crossed at the bridge region. (C) The assembly of a tetramer formed by IgD1/D1 interactions.
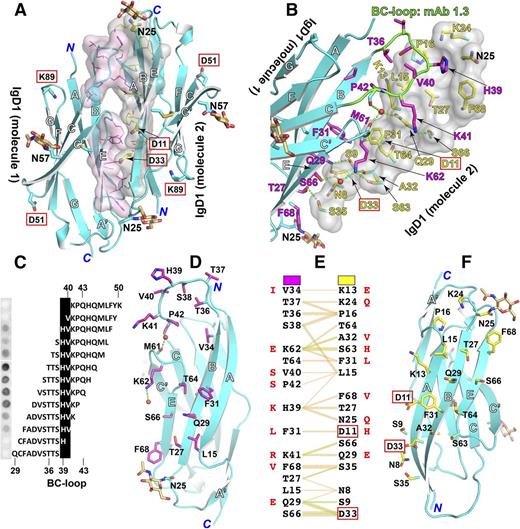
The IgD1/D1 homophilic interface
Structural analysis of IgD1/D1 homophilic interface using PDBsum51 revealed a total buried interface area of 1608 Å2 formed exclusively by the outer faces of the β sheets composed of strands A, B, and E (Figure 4A). Three regions of IgD1 molecule 1 participate in this interface: the BC loop, strands B and E, and the C′E loop (Figure 4A-B). These interact with a relatively flat interface formed by strands A, B, and E of IgD1 molecule 2 (Figure 4A-B). The BC loop of IgD1 molecule 1 packs against a long elongated region formed by residues Lys13, Leu15, Pro16, Lys24, Asn25, Thr27, Gln29, Ser66, and Phe68 within the A, B, and E strands and the A′B loop of IgD1 molecule 2 (Figure 4A-B). The epitope for mAb PECAM-1.3, known to completely block PECAM-1 homophilic interactions,6 resides within the BC loop (Figure 4B), and fine epitope mapping using a series of overlapping peptides demonstrated that His39, which is at the center of the BC loop, is the critical residue for mAb PECAM-1.3 binding (Figure 4C). In support of this, His39 is present within the BC loop of human and canine PECAM-1, both of which bind mAb PECAM-1.3, but not in the other species shown in Figure 2, which do not. The residues in strands B and E and the C′E loop of IgD1 molecule 1 pack against residues Asn8, Ser9, Asp11, Phe31, Ala32, Asp33, Ser35, Ser63, Thr64 within strands A, B, and E of IgD1 molecule 2 (Figure 4B). Finally, the common L98V polymorphic residue that has been implicated as a risk factor for cardiovascular disease52 is buried well inside the IgD1 structure, and does not contribute to the PECAM-1/PECAM-1 homophilic interface.
Homophilic interactions of PECAM-1 IgD1. (A) The IgD1/D1 interface. Interfacial residues defined by PDBsum51 are shown as sticks, with transparent surfaces and carbons in magenta for PECAM-1 IgD1-D2 molecule 1 or yellow for PECAM-1 IgD1-D2 molecule 2. Glycans at N25 and N57 are shown as sticks with carbon in wheat color. Oxygens and nitrogens are red and blue, respectively. Residues that are important for homophilic binding as identified by previous mutagenesis studies are boxed. (B) Close-up of the IgD1/D1 interface. One of the IgD1 molecules is shown as a transparent surface. The epitope of mAb PECAM-1.3 is highlighted with carbons in green. Waters are red small spheres. Hydrogen bonds are dashed in green. (C) Epitope mapping of mAb PECAM-1.3 bound to a series of overlapping peptides spanning PECAM-1 amino acids 29-50. (D-F) A split view of the IgD1/D1 interface as shown in panel B. The interacting residues are listed in panel F with hydrogen bonds and nonbonded contacts shown in green and orange, respectively. The equivalent residues that differ in mouse PECAM-1 are shown in red. The interfacial residues are also indicated in Figure 2.
Homophilic interactions of PECAM-1 IgD1. (A) The IgD1/D1 interface. Interfacial residues defined by PDBsum51 are shown as sticks, with transparent surfaces and carbons in magenta for PECAM-1 IgD1-D2 molecule 1 or yellow for PECAM-1 IgD1-D2 molecule 2. Glycans at N25 and N57 are shown as sticks with carbon in wheat color. Oxygens and nitrogens are red and blue, respectively. Residues that are important for homophilic binding as identified by previous mutagenesis studies are boxed. (B) Close-up of the IgD1/D1 interface. One of the IgD1 molecules is shown as a transparent surface. The epitope of mAb PECAM-1.3 is highlighted with carbons in green. Waters are red small spheres. Hydrogen bonds are dashed in green. (C) Epitope mapping of mAb PECAM-1.3 bound to a series of overlapping peptides spanning PECAM-1 amino acids 29-50. (D-F) A split view of the IgD1/D1 interface as shown in panel B. The interacting residues are listed in panel F with hydrogen bonds and nonbonded contacts shown in green and orange, respectively. The equivalent residues that differ in mouse PECAM-1 are shown in red. The interfacial residues are also indicated in Figure 2.
In all, 16 residues contribute to the 814-Å2 interfacial area of IgD1 molecule 1 (Figure 4D), whereas 18 residues contribute to the 794-Å2 interfacial area of IgD1 molecule 2 (Figure 4F). Specific residue interactions across the interface are listed in Figure 4E and are marked in Figure 2. Mouse and human PECAM-1 have substantially different residues at the interface (Figure 4E), consistent with the previous observation that mouse and human PECAM-1 IgD1 do not bind each other.7 Nonbonded contacts predominate in the formation of the homophilic interface (Figure 4E). Five residues within IgD1 molecule 1 and 6 residues within IgD1 molecule 2 form 7 potential direct hydrogen bonds (Figure 4B,E). Two water-mediated hydrogen-bonded networks are also present at the interface: one involving the carbonyl oxygens of Pro42 and Met61 and the side-chain nitrogen of Gln29, and another involving the carboxyl oxygen of Asp33 and the ε-nitrogen of Lys62 (Figure 4B). The antiparallel homophilic interactions are not symmetric. The BC loop of IgD1 molecule 1, but not molecule 2, contributes to the interface. Similarly, amino acids Asp11 and Asp33 of IgD1 molecule 2, but not IgD1 molecule 1, figure prominently in the interface (Figure 4D,F).
The IgD1/D1 homophilic interface determined here is consistent with extensive mutagenesis analyses of the molecular requirements for PECAM-1/PECAM-1 interactions. A large series of recombinant PECAM-1 proteins bearing single amino acid substitutions was used to identify 5 charged amino acids within IgD1, Asp11, Asp33, Lys50, Asp51, and Lys89 that when mutated individually dramatically reduced the homophilic-binding ability of PECAM-1.8 These residues are located in the opposite surfaces in our IgD1 structure (red boxes in Figure 4A), indicating that both surfaces contribute to homophilic interactions. Asp11 and Asp33, both of which disrupt PECAM-1–mediated homophilic interactions,8 figure prominently in the homophilic interface. Interestingly, mutation of Asp33, which is located at the center of a major interface, had a more dramatic effect on homophilic binding than did mutation of Asp11,8 which is closer to the edge of the interface (Figure 4B,F). Asp33 is completely conserved among different species, whereas Asp11 is present only in human PECAM-1 (Figure 2). In contrast, mutagenesis of Lys13, Lys41, or Lys62, all of which are located at the edge of the homophilic-binding interface (Figure 4B), had no detectable effect on homophilic binding.8
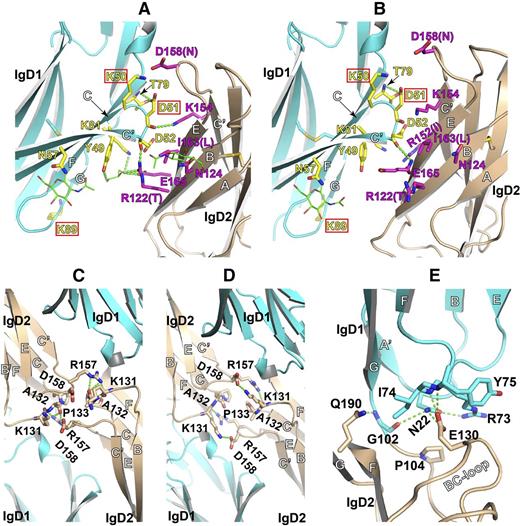
Trans homophilic contacts between adjacent IgD1/D2 and IgD2/D2 domains
The interchain interface between IgD1 and IgD2 has a buried surface area of ∼550 Å2 that consists of the outer surfaces of IgD1 β strands F, C, and C′ on the opposite side of the IgD1/D1 homophilic interface, and the outer surface of IgD2 β strands B and E (Figure 5A-B). Less than 10 residues participate in the IgD1/D2 interface, compared with 34 residues that contribute to the IgD1/D1 interface (Figures 4 and 5A). The 2 copies of the IgD1/D2 interface formed by the 2 molecules in the asymmetric unit are not identical; in one interface, shown in Figure 5A, the carbonyl oxygen of Asp51, located in the CC′ loop of IgD1, forms a hydrogen bond with the ε-nitrogen of Lys154 within IgD2. A second hydrogen bond exists between the carbonyl oxygen of Asp52 in IgD1 and the ω-nitrogen of Arg122 in IgD2. There are also indirect hydrogen bonds mediated by a glycerol molecule that lies between residues Arg122 and Glu165 of IgD2 and Tyr49 and Lys81 of IgD1. Asp52 also participates in the interface of the second copy (Figure 5B), where its carbonyl oxygen hydrogen bonds with the backbone nitrogen of Glu165 in IgD2. Despite its presence at the IgD1/D2 interface, mutation of Asp52 to Ala had little effect on the homophilic-binding activity,8 consistent with the nonfunctional role of its side chain within the interface. The side chain of Asp51 forms hydrogen bonds with Thr79 within IgD1 in both copies of PECAM-1 in the asymmetric unit (Figure 5A-B), and mutation of this residue to Arg has a pronounced negative effect on homophilic binding.8 Interestingly, Lys89, mutation of which completely disrupts PECAM-1 homophilic binding,8,9 is located within an FG loop that merely flanks the interface between IgD1 and IgD2, and is not directly involved in their interaction (Figure 5A-B). The Lys50, mutation of which has less effect on homophilic binding than the Lys89 mutation, is also located at the edge of the IgD1/D2 interface (Figure 5A-B).
Trans homophilic contacts between adjacent IgD1/D2 and IgD2/D2 domains. (A-B) Interchain IgD1/D2 interfaces present in the crystal lattice formed by the 2 PECAM-1 IgD1-D2 molecules in the asymmetric unit. (C-D) Interchain IgD2/D2 interfaces in the crystal lattice formed by the 2 PECAM-1 IgD1-D2 molecules asymmetric unit. (E) The intrachain interface between IgD1 and IgD2. Interfacial residues are shown as sticks with red oxygens and blue nitrogens. Carbohydrates are shown as sticks with green carbons. Hydrogen bonds are dashed in green. Residues that are important for homophilic binding, as identified by previous mutagenesis studies, are boxed. The equivalent residues that differ in mouse PECAM-1 are shown in parentheses. The interfacial residues are also indicated in Figure 2.
Trans homophilic contacts between adjacent IgD1/D2 and IgD2/D2 domains. (A-B) Interchain IgD1/D2 interfaces present in the crystal lattice formed by the 2 PECAM-1 IgD1-D2 molecules in the asymmetric unit. (C-D) Interchain IgD2/D2 interfaces in the crystal lattice formed by the 2 PECAM-1 IgD1-D2 molecules asymmetric unit. (E) The intrachain interface between IgD1 and IgD2. Interfacial residues are shown as sticks with red oxygens and blue nitrogens. Carbohydrates are shown as sticks with green carbons. Hydrogen bonds are dashed in green. Residues that are important for homophilic binding, as identified by previous mutagenesis studies, are boxed. The equivalent residues that differ in mouse PECAM-1 are shown in parentheses. The interfacial residues are also indicated in Figure 2.
In addition to its scaffolding function, a small homophilic interface between adjacent IgD2 domains is also present in the crystal lattice (Figure 5A), and is defined by interactions between loops C′E and BC (Figure 5C-D). Similar to the IgD1/D2 interface, the 2 copies of the IgD2/D2 interfaces are not identical (Figure 5C-D). Although the pyrrolidine rings of Pro133 face each other to form a hydrophobic interaction at the center of the interface in both copies, the side chains of Lys131, Arg157, and Asp158 are present in different rotamers, and as a result they form different hydrogen bonds in the 2 copies of the IgD2/D2 interface (Figure 5C-D). Arg157 and Asp158 are not conserved in other species. Arg157 is missing in mice, and the equivalent position of Asp158, which is also close to the IgD1/D2 interface (Figure 5A-B), is an Asn in all other species (Figure 2).
The intrachain domain boundary of IgD1 and IgD2
Five intrachain hydrogen bonds contribute to the domain boundary between IgD1 and IgD2 (Figure 5E). Conserved Glu130 within the BC loop of IgD2 lies at the center of the interface, and hydrogen bonds to 3 residues within IgD1; the backbone nitrogens of Ile74 and Tyr75 within the EF loop, and the side-chain nitrogen of Asn22 in the A′B loop (Figure 5E). The side chain of Asn22 is further stabilized through hydrogen bonding to the side chain of Arg73. The side chain of Gln190 in strand G hydrogen bonds to the carbonyl oxygen of Gly102. The boundary residue, Gly102, is well conserved between species (Figure 2), and also hydrogen bonds to the side-chain nitrogen of Asn22 through its carbonyl oxygen. These hydrogen bond interactions maintain the rigidity of the intrachain IgD1/IgD2 junction.
Carbohydrate residues at the IgD1/D1 and IgD1/D2 interface
Human PECAM-1 IgD1-D2 is predicted to contain 3 N-linked glycosylation sites: 2 within IgD1 at Asn25 and Asn57, and 1 within IgD2 at Asn124.1 Our crystal structure confirmed the presence of these glycosylation sites. Interestingly, each of them is located at a homophilic-binding interface. The N-acetylglucosamine (NAG) residue attached to Asn25 is well resolved in the crystal structure within the loop between β strands A′ and B at the IgD1/D1 homophilic-binding interface (Figure 4A). The glycan attached to Asn57 is similarly well resolved in the crystal structure, and is located in the C′ β strand of IgD1 near the IgD1/D2 interface (Figures 4A, 5A-B). The electron density of the NAG attached to Asn124 within IgD2 is not only less well resolved in the crystal structure, but also is located at the interface of human IgD1 and IgD2 (Figure 5A). It should be noted that our IgD1-D2 constructs were produced in insect cells, which have simpler N-glycans consisting only of terminal mannose residues,53 whereas native PECAM-1 expressed in mammalian cells contains larger and more complex N-glycans, many of which terminate with sialic acid residues. Sialic acids have recently been shown to contribute to homophilic binding of murine PECAM-1.21,22
Discussion
In circulating blood cells, PECAM-1 primarily functions as an inhibitory receptor that limits tyrosine kinase-mediated cellular activation events.54,55 In contrast, PECAM-1–mediated inhibitory signaling appears to be much less important in endothelial cell monolayers, where up to 2 × 106 molecules engage homophilically,9,56 resulting in junctionally enriched cell surface receptors that function to sense flow,57-59 maintain vascular integrity,11-15 regulate leukocyte transendothelial migration,19,20,60,61 and send antiapoptotic prosurvival signals into the cell.16,17,62,63 The structural features of PECAM-1 responsible for maintaining the PECAM-1/PECAM-1 homophilic contacts necessary for carrying out these activities, however, have heretofore remained unresolved. The present study was designed to provide atomic-level detail to the architecture of the interface that forms the most abundant component of the endothelial cell intercellular junction.
mAb blocking studies demonstrated 20 years ago that PECAM-1 IgD1 is required for PECAM-1 homophilic binding,6,47,64 findings that were verified using a series of mouse/human PECAM-1 chimeras7 and deletion mutants.6 Soluble IgD1 also binds endothelial cells and inhibits leukocyte transendothelial migration.19 The IgD1/D1 interface determined in our crystal structure, which is consistent with the mutagenesis studies discussed in the “Results,” provide a structural basis for IgD1-mediated PECAM-1 homophilic interactions. One caveat of our structure, however, is that the Lys89 within IgD1, mutation of which completely disrupts homophilic binding,8,9 does not appear to directly contribute to the homophilic-binding interface. Preliminary molecular docking analysis suggests that electrostatic interactions between negatively charged sialic acid moieties and positively charged Lys89 could play an important role in higher-order binding interactions between concentrated PECAM-1 oligomers present at endothelial junctions (data not shown). In fact, the side chain of Lys89 and the N-linked glycan of Asn25 are facing each other at one end of the IgD1/D1 interface (Figure 4A), indicative of a potential role for Lys89 in stabilizing the homophilic interface.
Although IgD1 by itself is capable of homophilic binding, previous studies have implicated IgD2 as also making an important, though mechanistically unclear, structural contribution. First, IgD2 has been shown to be required for homophilic interactions between PECAM-1–transfected L cells.7 Second, binding of PECAM-1/IgG to PECAM-1–transfected L cells is nearly twofold less efficient if murine IgD2 is substituted for its human counterpart.6 Finally, the mAb hec7, which recognizes a complex epitope composed of IgD1+D2,47 is as effective at inhibiting homophilic binding of both isolated PECAM-1 molecules6 and PECAM-1–expressing cells18 as are IgD1-specific mAbs.6 The IgD2-specific mAbs L133.1 and 5.6E have similar blocking effects.65 Taken together, these findings implied that IgD2 either functions as a scaffold to position IgD1 to maximize IgD1/D1 contacts or contributes directly to the homophilic-binding interface. Both appear to be operative. In addition to the rigid intrachain interface between IgD1 and IgD2 that governs the orientation of IgD1, IgD2 also forms ∼550 Å2 contacts with adjacent IgD1 domains in trans, the importance of which is highlighted by the observation that mutation of Asp 51 (a perfectly conserved residue in IgD1 that lies in the center of the IgD1/D2 homophilic interface) to Arg significantly disrupts homophilic binding.8 IgD2 also forms a small trans homophilic interface with IgD2 on adjacent molecules. Taken together, these data provide a structural explanation for the previous biochemical and cell biological observations that implicated IgD2 as having a role in the adhesive properties of PECAM-1.
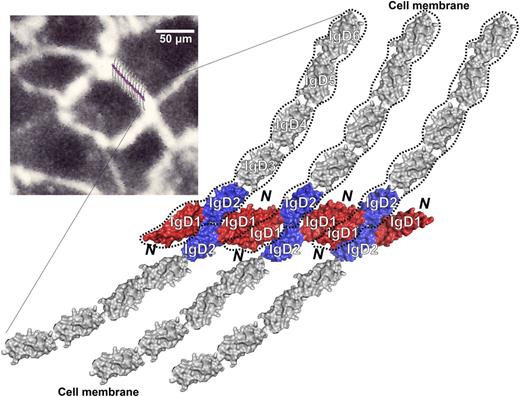
Another caveat of our structure is whether the strand-swap present in the IgD1-D2 dimer is also present in full-length PECAM-1 and physiologically functional. In addition to its well-known ability to form trans homophilic interactions between leukocytes and endothelial cells, and at endothelial cell-cell junctions, previous studies have shown that PECAM-1 forms cis dimers within the plane of the plasma membrane.24,25 The concentration-dependent dimerization observed by our chemical crosslinking assay raised the possibility that the strand-swapped cis dimer might exist in full-length PECAM-1, especially where it becomes highly concentrated at endothelial cell-cell junctions.9,56 Strand-swapping has been observed in other immunoglobulin-like cell adhesion molecules such as E-cadherin66 and NCAM2,67 in which a swapping of A strands within their homophilic-binding domains directly mediates homophilic interactions. The dimerization of IgD1-D2 in our crystal structure may have been facilitated by the swapping of strand G, which has been seen in the crystal structures of other members of the IgSF, none of which mediate cell adhesion, including CTLA-4,68 CD47,69 a llama VHH domain,70 and B7-H3.71 Although the functional roles of these strand-swapped dimers have not been elucidated, strand-swapping clearly facilitated crystal lattice packing in each of these cases. We observed an array formed by the strand-swapped IgD1-D2 dimer in the crystal lattice in which 2 layers of IgD1-D2 interfaces are formed simultaneously (Figure 3A). However, in full-length PECAM-1, the presence of IgD3-D6 could topologically constrain the array of IgD1-D2 dimers, making 2 layers of interfaces unlikely to form. The possibility that trans homophilic interactions formed by tetramers of IgD1-D2 in which only IgD1/D1 interactions are involved (Figure 3B-C) is not consistent with mutagenesis studies that have shown that both IgD1/D1 and IgD1/D2 interactions are important in PECAM-1 homophilic interactions. Finally, we cannot exclude the possibility that the strand-swapping seen in our IgD1-D2 crystal was simply due to the point at which our construct was truncated. Structural studies designed to examine the presence of the strand-swapped region in full-length PECAM-1 are in progress. Nevertheless, the IgD1-D2 interfaces defined in the crystal lattice allow us to propose a reasonable model of PECAM-1–mediated homophilic binding at the cell-cell junction in which adjacent PECAM-1 monomers form the large interfaces present between adjacent IgD1/D1, IgD1/D2, and IgD2/D2 domains (Figure 6).
Model of PECAM-1 homophilic interactions that form at the endothelial cell-cell junction. Both IgD1 and IgD2 play important roles in creating, and then solidifying, homophilic PECAM-1/PECAM-1 interactions. Note the concentration-dependent interfaces that form between IgD1/D1, IgD1/D2, and IgD2/D2 in closely apposed PECAM-1 molecules. Inset, Human umbilical vein endothelial cells immunostained with mAb PECAM-1.3, with the proposed homophilic PECAM-1/PECAM-1 model superimposed to generate a schematic of the junctional structure that may exist at endothelial cell-cell borders. PECAM-1 molecules are not to scale.
Model of PECAM-1 homophilic interactions that form at the endothelial cell-cell junction. Both IgD1 and IgD2 play important roles in creating, and then solidifying, homophilic PECAM-1/PECAM-1 interactions. Note the concentration-dependent interfaces that form between IgD1/D1, IgD1/D2, and IgD2/D2 in closely apposed PECAM-1 molecules. Inset, Human umbilical vein endothelial cells immunostained with mAb PECAM-1.3, with the proposed homophilic PECAM-1/PECAM-1 model superimposed to generate a schematic of the junctional structure that may exist at endothelial cell-cell borders. PECAM-1 molecules are not to scale.
Isolated PECAM-1 monomers exhibit surprisingly little affinity for each other, and despite its expression on the surface of circulating blood cells, PECAM-1 is unable to mediate platelet-platelet, platelet-leukocyte, or leukocyte-leukocyte adhesive interactions. In contrast, PECAM-1 molecules highly enriched at endothelial cell-cell junctions1-3,72 are able to contribute importantly to the ability of endothelial cells to maintain a permeability barrier under conditions of vascular stress.11-15 Figure 6 presents a molecular model, based on the structural studies described herein, of the aggregate of homophilic contacts that PECAM-1 makes with adjacent PECAM-1 molecules in trans, and depicts the large 1600-Å2 interface present between adjacent IgD1 domains (shown in red) that is additionally reinforced by IgD1/D2 (red/blue) and IgD2/D2 (blue/blue) contacts that form as a consequence of the large number of PECAM-1 molecules that become concentrated at endothelial cell-cell borders.9,73 Such extensive contacts likely enable PECAM-1 to maintain vascular integrity, and to resist mechanical force under conditions of fluid shear stress.74 Future planned studies should allow us to examine the mechanism by which other regions of the extracellular domain are able to affect the affinity modulation of PECAM-1 leading to adhesion strengthening of endothelial cell-cell junctions.26
Sequence alignment (Figure 2) shows that both IgD1 and IgD2 are well conserved evolutionarily, especially in regions that form β strands, suggesting that the overall structural folds observed in human PECAM-1 IgD1-D2 are likely to be evolutionarily well conserved. Because of the high degree of sequence similarity between mouse and human PECAM-1, early pharmacological studies designed to examine the ability of PECAM-1 to inhibit transendothelial leukocyte migration used soluble human recombinant PECAM-1 in a variety of murine models of inflammation. They all failed. Binding studies using a series of mouse/human chimeras later revealed that human and mouse PECAM-1 are unable to interact homophilically.7 An atomic-level explanation for the failure of human and mouse PECAM-1 to interact is provided in Figure 4E, which shows no fewer than 18 amino acid differences between human and mouse PECAM-1 at the IgD1/IgD1 homophilic-binding interface. Differences in the IgD1/IgD2 interface also likely account for the failure of mouse and human PECAM-1 to interact, as Arg122, Asp158, Ile163, and Arg152 within IgD2 (Figure 5A-B) are each different in the mouse (Figure 2). Finally, α2,6 sialic-modified glycans have been reported to be required for homophilic binding of murine PECAM-1.21,22 Human PECAM-1 IgD1 contains 2 NxL motifs, one at position 25 and another at position 57, whereas IgD2 contains an N-linked glycosylation site at position 124. Though Asn57 and Asn124 are completely evolutionarily conserved in higher mammals (Figure 2), residue 25 is a Gln in mice and rats, highlighting still another important difference in the nature of the contact sites used by murine vs human PECAM-1 to mediate homophilic binding. Such molecular differences need to be taken into account when using animal models to explore the therapeutic potential of this novel cell adhesion and signaling molecule to affect the function of blood and vascular cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Francis Peterson and Brian Volkman (Medical College of Wisconsin) for their valuable guidance during the early stages of this investigation, Dr Debra K. Newman (Blood Research Institute, BloodCenter of Wisconsin) for her critical reading of the manuscript, and the Mass Spectrometry Core of the Medical College of Wisconsin.
This work was supported by grant HL40926 from the National Heart, Lung, and Blood Institute of the National Institutes of Health (P.J.N.).
Authorship
Contribution: C.P., D.Z., and P.L. performed the research and analyzed data; and P.J.N. and J.Z. designed the research, analyzed data, and wrote the manuscript with the assistance of all of the other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jieqing Zhu, Blood Research Institute, BloodCenter of Wisconsin, PO Box 2178, Milwaukee, WI 53201; e-mail: jieqing.zhu@bcw.edu; or Peter J. Newman, Blood Research Institute, BloodCenter of Wisconsin, PO Box 2178, Milwaukee, WI 53201; e-mail: peter.newman@bcw.edu.
References
Author notes
C.P. and D.Z. contributed equally to this study.
The laboratories of P.J.N. and J.Z. contributed equally to this study.