Key Points
A high frequency of diverse activating mutations in costimulatory/TCR-related signaling genes occurs in AITL and other TFH-derived PTCL.
Deregulated TCR activation may play a role in the pathogenesis of TFH-derived PTCL, paving the way for developing novel targeted therapies.
Abstract
Angioimmunoblastic T-cell lymphoma (AITL) and other lymphomas derived from follicular T-helper cells (TFH) represent a large proportion of peripheral T-cell lymphomas (PTCLs) with poorly understood pathogenesis and unfavorable treatment results. We investigated a series of 85 patients with AITL (n = 72) or other TFH-derived PTCL (n = 13) by targeted deep sequencing of a gene panel enriched in T-cell receptor (TCR) signaling elements. RHOA mutations were identified in 51 of 85 cases (60%) consisting of the highly recurrent dominant negative G17V variant in most cases and a novel K18N in 3 cases, the latter showing activating properties in in vitro assays. Moreover, half of the patients carried virtually mutually exclusive mutations in other TCR-related genes, most frequently in PLCG1 (14.1%), CD28 (9.4%, exclusively in AITL), PI3K elements (7%), CTNNB1 (6%), and GTF2I (6%). Using in vitro assays in transfected cells, we demonstrated that 9 of 10 PLCG1 and 3 of 3 CARD11 variants induced MALT1 protease activity and increased transcription from NFAT or NF-κB response element reporters, respectively. Collectively, the vast majority of variants in TCR-related genes could be classified as gain-of-function. Accordingly, the samples with mutations in TCR-related genes other than RHOA had transcriptomic profiles enriched in signatures reflecting higher T-cell activation. Although no correlation with presenting clinical features nor significant impact on survival was observed, the presence of TCR-related mutations correlated with early disease progression. Thus, targeting of TCR-related events may hold promise for the treatment of TFH-derived lymphomas.
Introduction
Angioimmunoblastic T-cell lymphoma (AITL), one of the most common peripheral T-cell lymphomas (PTCLs),1,2 comprises CD4+ neoplastic T cells with a T-follicular helper (TFH) immunophenotype, and an important reactive microenvironment.3 In addition, a subset of PTCL not otherwise specified (PTCL-NOS) exhibiting a TFH-like immunophenotype and overlapping characteristics with AITL are believed to be part of the same disease spectrum.4,5 The prognosis of AITL remains poor, with a 5-year overall survival of ∼30%.1,6 Although autologous stem cell transplantation may improve the response to conventional first-line therapy, early disease progression makes it frequently impossible.7 Thus, pinpointing new druggable targets for developing specific therapies in TFH-derived lymphomas remains critical.
Recurrent mutations in the epigenetic regulators TET2, IDH2, and DNMT3A have been detected in AITL and TFH-like PTCLs8,9 but are not likely sufficient to drive lymphomagenesis.10,11 A highly recurrent dominant-negative G17V mutation in the RHOA GTPase was recently discovered in as much as 70% of AITL and TFH-like PTCLs.12-14 RHOA is activated downstream of T-cell receptor (TCR) engagement in mature T cells and is linked to cytoskeleton reorganization after T-cell activation.12,13 Although their exact role in human T-cell transformation remains uncertain, RHOA mutations tend to co-occur with TET2 mutations, suggesting that they represent a second event in multistep AITL lymphomagenesis.15 Finally, AITL also bears a variety of other less recurrent genetic alterations.12,13,16
In B-cell lymphomas, constitutive B-cell receptor (BCR) signaling via somatic mutation-induced or antigen-mediated activation of key BCR components leads to neoplastic B-cell survival and expansion.17 Several lines of evidence suggest that TCR activation might be similarly relevant to PTCL pathogenesis. For example, NPM1-ALK chimeric proteins induce an intracellular cascade of events substituting to TCR signaling in anaplastic large cell lymphoma.18 The ITK-SYK fusion present in rare PTCLs likewise drives lymphomagenesis by triggering antigen-independent activation of TCR signaling pathways in mice.19 Mutations or gene fusions in costimulatory/TCR signaling genes as CD28, FYN, PLCG1, or CARD11 have also been reported in several PTCLs.12,20-25 However, alterations in the TCR signaling pathway have not yet been extensively studied in TFH-related lymphomas. Here, we focused on investigating mutations in the costimulatory/TCR signaling cascade in a series of AITL and TFH-like PTCLs using a targeted deep-sequencing approach. Besides frequent RHOA mutations, we demonstrated recurrent activating and virtually mutually exclusive mutations in costimulatory/TCR pathway components in 49% of the cases. Notably, we found several mutations in PLCG1 and CARD11 that we characterized as functionally activating in vitro. Integrated analysis with gene expression profiles indicated that TCR-mutated samples were enriched for molecular signatures, reflecting higher T-cell activation and proliferation. Clinically, TCR-mutated patients receiving anthracyclin-based chemotherapy showed an increased early progression risk compared with patients without such mutations. These results indicate the potential of using drugs that reduce TCR signaling for treating these lymphomas.
Methods
Patients and tumor samples
Discovery cohort.
Eleven paired tumoral and normal samples from 10 clinically annotated AITL patients were subject to whole-genome sequencing (WGS, n = 8) or whole-exome sequencing (WES, n = 3) (supplemental Table 1, available on the Blood Web site).
Extended cohort.
Targeted deep sequencing (TDS) was performed on DNA extracted from frozen tissue biopsies of an extended cohort of 85 previously untreated patients (72 AITL, 13 TFH-like PTCL-NOS). In 3 patients, paired samples were analyzed. These cases and associated clinical annotations (supplemental Table 2) were collected in the framework of the Tenomic LYSA consortium. Diagnoses were confirmed by expert hematopathologists. Criteria used to define TFH-like PTCL-NOS were previously reported.8 A subset of these patients was analyzed for TET2, IDH2, and DNMT3A mutations in previous studies.8,9 The study was approved by the local ethics committee (CPP Ile de France IX 08-009).
WGS, WES, and TDS
Libraries were constructed for WGS following a standard protocol.26 100-bp paired-end reads were generated using an Illumina HiSequation 2000 Sequencer (version 3 chemistry). Reads were mapped to the NCBI Build 36.1 reference genome using the Burrows-Wheeler Aligner alignment tool version 0.5.7, algorithm aln. Polymerase chain reaction duplicates were eliminated using Picard version 1.38. Indels and single-nucleotide variants (SNVs) were called using Strelka version 1.0.14, removing false-positive calls internally. Variants were independently identified for each tumor-germline pair, using the germline as reference. GRCh37 coordinates of the filtered indels and SNVs were obtained using LiftOverVCF. Variants were annotated with VEP, and their read counts were obtained using in-house scripts. Copy number variations (CNVs) were identified using HMMcopy. Commonly deleted or amplified regions from WGS were visually identified using the Integrative Genome Viewer Software (IGV, https://www.broadinstitute.org/igv/). Deleted loci and genes having SNVs and/or CNVs in at least 2 patients are listed in supplemental Tables 3 and 4.
TDS of 69 genes (supplemental Table 5) was performed on the extended cohort. Systematic sequencing errors were removed by adding 2 randomly selected blood samples from 4 healthy volunteers to each run. Libraries were prepared using the True-Seq Custom Amplicon kit (Illumina) and sequenced on a MiSeq machine (Illumina, 1000× mean depth, mean coverage 94%). Variants were called using MiSeq reporter using default settings and were visually inspected on IGV. All synonymous variants and known SNPs with an allelic frequency >0.1% in public databases (dbSNP131, 1000 Genomes Project, 5000 Exomes project) were filtered out. We excluded variants found in normal samples, in ambiguously mapped regions from captured pseudogenes, and in regions of low complexity. Candidate variants were independently validated in a second round of TDS: we selectively amplified the amplicons covering the positions of candidate mutations (IonAmpliseq kit, Life Technologies) and sequenced them on an Ion PGM System (Life Technologies; 1305× mean depth, mean coverage 91.64%). Variants were called using default settings of the IonReporter software and reviewed with IGV. The final variants list (supplemental Table 6) was generated applying the following filtering criteria: (a) variants called by both platforms; (b) variants with minor allele frequency (MAF) <0.4; and (c) variants already described as somatic in the COSMIC database. Filter (b) was introduced to take into account the low tumor content and the unavailability of matched normal samples. The threshold was defined based on the MAF range of well-characterized AITL mutations, such as in RHOA and CD28.12-14,25 Variants were annotated using GeneTalk (http://www.gene-talk.de/).
Cloning and functional validation
A detailed description is provided in the supplemental Material. Briefly, we performed site-directed mutagenesis of RHOA, PLCG1, and CARD11 mutations on cloned cDNAs into pcMV6-MycFlag (RHOA, PLCG1) (Origene, C-terminal tag) and pcDNA3-HA (CARD11) (N-terminal tag)27 vectors. RHOA variants were functionally assessed in vitro with rhotekin pull-down assays12-14 and SRE (Serum Responsive Element) (Promega, Madison, WI) luciferase reporter assay using HEK293T cells.
PLCG1 and CARD11 variants were functionally tested by fluorescence resonance energy transfer (FRET)-based determination of their ability to induce MALT1 protease activity in HEK293T cells28 and in luciferase-based assays using a NFAT reporter in HEK293T cells (forPLCG1) or a NF-κB reporter in Jurkat cells deficient in CARD1129 (kindly provided by Dr. Xin Lin) (for CARD11) .
Western blot analysis
The following primary antibodies were used in western blotting: anti-HA (Covalab Biotechnology), anti-Myc (clone 4A6, Millipore), anti-CARD11 (clone 1D12, Cell Signaling), and anti-MALT128 and anti-Actin (ab3280, Abcam). Secondary antibodies were purchased from Jackson Laboratories.
Gene expression and enrichment analysis
Gene expression profiles generated by hybridization on Affymetrix U133 Plus 2.0 were available for the extended cohort.3 Rotation testing using mean ranks (ROMER, R package limma) was used to determine the gene set signature enrichments in TCR_Mut compared with TCR_WT patients. A total of 304 signatures, including 23 of 50 hallmark signatures and 9 signatures from the curated and immunogenic signature collection of the Molecular Signatures Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb), all signatures of interest in lymphoid biology (Staudt, lymphochip.nih.gov/signaturedb/), and 2 manually annotated sequences linked to TCR signaling, were tested.
Clinical correlations and survival analysis
Frequency differences of categorical variables were analyzed using the Fisher exact test, whereas differences in continuous variables were tested with the Wilcoxon rank-sum test. Overall survival (OS) was defined as time from diagnosis to death or last follow-up, whereas progression-free survival (PFS) was defined as time from treatment initiation to death or progression or last follow-up. Analyses of OS and PFS were performed using the Kaplan-Meier method, and hazard ratio differences were computed with the log-rank test.
Results
Whole-exome/genome sequencing reveals alterations in T-cell signaling genes in AITL
Eleven tumor samples from 10 AITL patients were subject to WGS or WES and compared with germline DNA (supplemental Figure 1; supplemental Table 1). On average, 91% of the genome had >10× coverage, with 68% having >30×. A mean of 27 SNVs per patient (range 11-50) was observed in coding regions, except in a sample from a relapsing patient with a chromothripsis pattern of structural variation (supplemental Figure 2) and 225 SNVs. In one patient refractory to first-line therapy, WGS performed on tumor biopsies at diagnosis and after 5 cycles of chemotherapy revealed a similar profile of alterations, with identical TET2 and ANKRD62 SNVs in both samples. CNV analysis revealed multiple loci deleted in a number of patients, with 8 minimal common regions (supplemental Table 3), mostly in 3 patients. A total of 39 genes had SNVs and/or CNVs in at least 2 patients (supplemental Table 4). Apart from RHOA G17V and TET2 mutations, 9 of 39 (23%) altered genes were related to TCR (n = 7), Toll-like receptor (n = 1), or Wnt (n = 1) signaling. Other mutations were found in genes involved in the cell cycle (n = 5), autophagy (n = 2), or other pathways (n = 6). Six additional genes relevant to TCR or JAK/STAT signaling (CTNNB1, VAV1, MTOR, CREBBP, PIK3C2G, JAK1) were altered in one patient each. Collectively, T-cell signaling alterations were found in 7 of 10 patients.
Frequent mutations of RHOA and other TCR-related genes are confirmed in an extended cohort of TFH-derived lymphomas
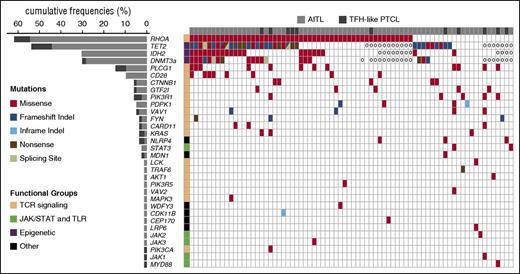
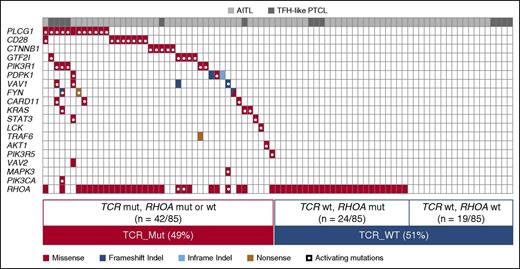
We designed a panel of 69 genes for TDS, partly based on findings in the discovery cohort (supplemental Table 5; supplemental Figure 1) for application on our extended cohort (supplemental Table 2). After orthogonal cross-validation and conservative filtering (supplemental Table 6; supplemental Figure 3), we validated 70 variants in 29 genes (Figure 1), including missenses (61), frameshifts (5), nonframeshift deletions (2), and stop-gain mutations (2). Mutations were detected in 70 of 85 samples (82%, mean mutations per sample = 1.8). RHOA and other TCR signaling–related genes accounted for 19 of 29 (65.5%) mutated genes (Figures 1 and 2).
Mutational landscape of nodal TFH-derived lymphomas. The results of targeted deep sequencing of 69 genes in 72 AITL (light gray) and 13 TFH-like PTCL (dark gray) are presented. Ten cases (8 AITL and 2 TFH-like PTCL) with no mutations detected are not represented. TET2, DNMT3A, and IDH2 mutations available for a subset of the cases reported in previous studies8,9 are also shown. Case-mutation pairs for which data are not available are indicated by a 0. Mutated genes (rows) are arranged by decreasing order of mutation frequency. Patients (columns) are arranged from left to right based on their mutational status following gene ranking.
Mutational landscape of nodal TFH-derived lymphomas. The results of targeted deep sequencing of 69 genes in 72 AITL (light gray) and 13 TFH-like PTCL (dark gray) are presented. Ten cases (8 AITL and 2 TFH-like PTCL) with no mutations detected are not represented. TET2, DNMT3A, and IDH2 mutations available for a subset of the cases reported in previous studies8,9 are also shown. Case-mutation pairs for which data are not available are indicated by a 0. Mutated genes (rows) are arranged by decreasing order of mutation frequency. Patients (columns) are arranged from left to right based on their mutational status following gene ranking.
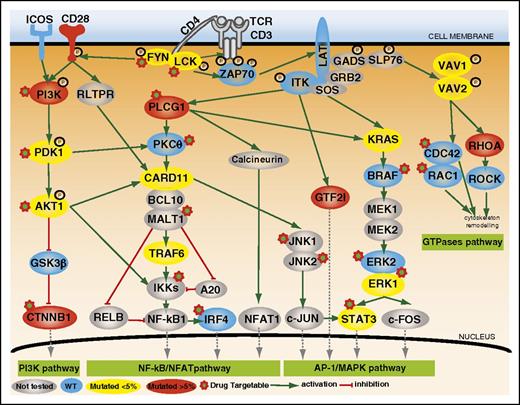
Mutations of TCR signaling–related genes in nodal lymphomas of TFH origin. The intracellular pathways after TCR ligation and costimulatory activation were reconstructed using the Ingenuity pathway analysis (IPA) tools, the KEGG database, and other references. Four main pathways are individualized, from left to right: (1) PI3K pathway after CD28/TCR-dependent FYN phosphorylation and ultimately resulting in CTNNB1 translocation into the nucleus; (2) NF-κB/NFAT pathway proximally initiated by ITK-dependent PLCG1 activation and resulting in NFAT1, NF-κB, and IRF4 activation; (3) AP-1/MAPK pathway that comprises ITK-dependent GTF2I activation, MALT1-induced JNKs activation, and PLCG1-GRB2/SOS–induced MAPK components activation; and (4) GTPase-dependent pathway, including RHOA, responsible for cytoskeleton remodeling upon costimulatory/TCR activation. The main positive interactions are indicated by solid green arrows, whereas inhibitory effects are indicated in red. The TCR signaling elements are depicted in yellow or red if the coding genes were mutated in <5% or ≥5% cases, respectively. The most frequently mutated genes (PLCG1, CD28, PI3K components, CTNNB1, and GTF2I) were part of costimulatory, NF-κB/NFAT, PI3K, and AP-1/MAPK intracellular signaling pathways. Proteins corresponding to WT genes are indicated in blue, and genes that were not sequenced are in gray. ERK1, ERK2, JNK1, JNK2, and PDK1 are protein names for MAPK1, MAPK3, MAPK8, MAPK9, and PDPK1 genes, respectively.
Mutations of TCR signaling–related genes in nodal lymphomas of TFH origin. The intracellular pathways after TCR ligation and costimulatory activation were reconstructed using the Ingenuity pathway analysis (IPA) tools, the KEGG database, and other references. Four main pathways are individualized, from left to right: (1) PI3K pathway after CD28/TCR-dependent FYN phosphorylation and ultimately resulting in CTNNB1 translocation into the nucleus; (2) NF-κB/NFAT pathway proximally initiated by ITK-dependent PLCG1 activation and resulting in NFAT1, NF-κB, and IRF4 activation; (3) AP-1/MAPK pathway that comprises ITK-dependent GTF2I activation, MALT1-induced JNKs activation, and PLCG1-GRB2/SOS–induced MAPK components activation; and (4) GTPase-dependent pathway, including RHOA, responsible for cytoskeleton remodeling upon costimulatory/TCR activation. The main positive interactions are indicated by solid green arrows, whereas inhibitory effects are indicated in red. The TCR signaling elements are depicted in yellow or red if the coding genes were mutated in <5% or ≥5% cases, respectively. The most frequently mutated genes (PLCG1, CD28, PI3K components, CTNNB1, and GTF2I) were part of costimulatory, NF-κB/NFAT, PI3K, and AP-1/MAPK intracellular signaling pathways. Proteins corresponding to WT genes are indicated in blue, and genes that were not sequenced are in gray. ERK1, ERK2, JNK1, JNK2, and PDK1 are protein names for MAPK1, MAPK3, MAPK8, MAPK9, and PDPK1 genes, respectively.
RHOA mutations include a novel activating K18N variant
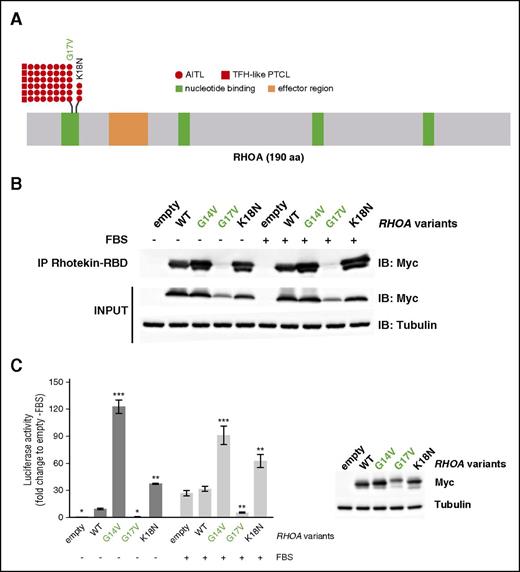
RHOA was the most frequently mutated gene among those analyzed (51/85; 60% of patients), with a similar prevalence in AITL and TFH-like PTCL. Consistent with previous reports,12,13 the G17V variant was found in most of the cases. We also identified a novel K18N mutation (variant frequency [VF] [5.5%; 7.4%; 10%]) in 3 AITL patients (Figure 3A; Table 1).30-44 Both mutations occur in the highly conserved GTP binding site of RHOA. We tested Myc-tagged G14V (constitutively active45 ), G17V (dominant-negative), K18N, and wild-type (WT) RHOA in a pull-down assay using Rhotekin-RBD (ρ binding domain) beads (Figure 3B). Although RHOA G17V already characterized as dominant-negative12-14 did not bind RBD beads, even in the presence of fetal bovine serum, compared with WT, RHOA K18N showed a marked increase of RBD beads binding similarly to RHOA G14V (Figure 3B). Moreover, unlike RHOA G17V that did not activate and even repressed transcription from the SRE under serum activation, RHOA K18N markedly enhanced its transcription (Figure 3C).
RHOA mutations in AITL and TFH-like PTCL. (A) Overview of the RHOA protein structure, showing G17V (42 AITL, 6 TFH-like PTCL) and the novel K18N (3 AITL) variants that target the highly conserved GTP/GDP binding site of RHOA. (B) Protein blot analysis of GTP-bound RHOA-Myc in rhotekin pull-down assay from HEK293T cells expressing indicated RHOA constructs. IP, immunoprecipitation. Representative of 6 independent experiments. (C) SRE (serum-responsive element) luciferase reporter assay monitoring the activity of RHOA K18N mutant, compared with WT, G14V, or G17V mutants, previously characterized as activating and dominant-negative, respectively. Cells were stimulated (light gray) or not (dark gray) with fetal bovine serum for 6 hours. Data are represented as mean ± standard error of the mean (SEM) from 4 independent experiments. Significant differences in activation activity were determined using 2-way analysis of variance (ANOVA) with repeated measurement (**P ≤ .01; ***P ≤ .001 compared with WT). Representative western blot from a luciferase assay experiment. Ectopic myc-tagged RHOA expression is revealed by anti-Myc. Anti-Actin blotting serves as loading control.
RHOA mutations in AITL and TFH-like PTCL. (A) Overview of the RHOA protein structure, showing G17V (42 AITL, 6 TFH-like PTCL) and the novel K18N (3 AITL) variants that target the highly conserved GTP/GDP binding site of RHOA. (B) Protein blot analysis of GTP-bound RHOA-Myc in rhotekin pull-down assay from HEK293T cells expressing indicated RHOA constructs. IP, immunoprecipitation. Representative of 6 independent experiments. (C) SRE (serum-responsive element) luciferase reporter assay monitoring the activity of RHOA K18N mutant, compared with WT, G14V, or G17V mutants, previously characterized as activating and dominant-negative, respectively. Cells were stimulated (light gray) or not (dark gray) with fetal bovine serum for 6 hours. Data are represented as mean ± standard error of the mean (SEM) from 4 independent experiments. Significant differences in activation activity were determined using 2-way analysis of variance (ANOVA) with repeated measurement (**P ≤ .01; ***P ≤ .001 compared with WT). Representative western blot from a luciferase assay experiment. Ectopic myc-tagged RHOA expression is revealed by anti-Myc. Anti-Actin blotting serves as loading control.
Characteristics of the 70 variants identified by targeted deep sequencing of TCR, JAK/STAT, and TLR-related genes in 85 TFH-derived PTCL samples
| Functional groups . | Gene . | Amino acid change . | Mutation type . | Domain . | New/reported . | Effect . | References . |
|---|---|---|---|---|---|---|---|
| Costimulatory and proximal TCR signaling | CD28 | D124V | Missense | Extracellular | AITL, ATLL | Gain of function (F) | 20,25 |
| CD28 | D124E | Missense | Extracellular | AITL, ATLL | Gain of function (F) | 20,25 | |
| CD28 | T195P | Missense | Cytoplasmic | AITL, ATLL | Gain of function (F) | 20,25,30 | |
| LCK | N446K* | Missense | Kinase | New | Probably gain of function (P) | ||
| LCK | P447R* | Missense | Kinase | New | Probably gain of function (P) | ||
| FYN | Q527X | Stop gain | ATLL | Probably gain of function (P) | 20 | ||
| FYN | 525_525del† | Frameshift deletion | New | Probably gain of function (P) | |||
| FYN | S186L† | Missense | SH2 | New | Probably gain of function (P) | ||
| FYN | K108fs* | Frameshift deletion | New | Probably loss of function (P) | |||
| FYN | E107S* | Missense | SH3 | New | Probably loss of function (P) | ||
| NK-kB/NF-AT pathway | PLCG1 | E47K | Missense | PH 1 | ATLL | Gain of function (F) | 20 |
| PLCG1 | R48W | Missense | PH 1 | ATLL, SS | No effect (F) | 20,24 | |
| PLCG1 | D342G | Missense | PI-PLC X-box | ATLL, MF | Gain of function (F) | 20 | |
| PLCG1 | S345F† | Missense | PI-PLC X-box | ATL, MF, SS, ATLL | Gain of function (F) | 20,-22,24 | |
| PLCG1 | S520F† | Missense | PH 2; first part | MF, SS, ATLL | Gain of function (F) | 20,21,24 | |
| PLCG1 | E730K | Missense | SH2 2 | New | Gain of function (F) | ||
| PLCG1 | G869E | Missense | Near SH3 | New | Gain of function (F) | ||
| PLCG1 | E1163K | Missense | C2 | ATLL, SS | Gain of function (F) | 20,24 | |
| PLCG1 | D1165H | Missense | C2 | ATLL, SS | Gain of function (F) | 20,24 | |
| PLCG1 | D1165G | Missense | C2 | ATLL | Gain of function (F) | 20 | |
| CARD11 | F902C | Missense | ATLL | Gain of function (F) | 20 | ||
| CARD11 | S547T | Missense | New | Gain of function (F) | |||
| CARD11 | F176C | Missense | Coiled coil | New | Gain of function (F) | ||
| TRAF6 | Q347X | Stop gain | Coiled coil /MATH | New | NA | ||
| PI3K pathway | PIK3R1 | K141R | Missense | RHO-GAP | New | Probably gain of function (P) | |
| PIK3R1 | Q475P | Missense | iSH2 | New | Probably gain of function (P) | ||
| PIK3R1 | T576A | Missense | iSH2 | New | Probably gain of function (P) | ||
| PIK3R1 | G680S | Missense | SH2 2 | New | Probably gain of function (P) | ||
| PIK3R1 | V704M | Missense | SH2 2 | New | Probably gain of function (P) | ||
| PIK3R5 | A259V | Missense | Carcinoma (liver) | Probably gain of function (P) | 31 | ||
| PIK3CA | L1001P | Missense | PI3K/PI4K | New | Probably gain of function (P) | ||
| PDPK1 | 19_20del | Inframe deletion | New | NA | |||
| PDPK1 | 151_152del | Frameshift deletion | Protein kinase | New | Probably loss of function (P) | ||
| PDPK1 | R324Q | Missense | Protein kinase | New | Probably gain of function (P) | ||
| PDPK1 | P340Q | Missense | Protein kinase | New | Probably gain of function (P) | ||
| AKT1 | G294R | Missense | Protein kinase | New | Probably gain of function (P) | ||
| CTNNB1 | T41A† | Missense | Phospho by GSK3b | Hepatocellular carcinoma, acute lymphoblastic leukemia, breast cancer, Wilms’ tumor | Gain of function (F) | 32 | |
| CTNNB1 | H36P | Missense | Hepatocellular carcinoma, acute lymphoblastic leukemia, breast cancer, Wilms’ tumor | Gain of function (F) | 32 | ||
| CTNNB1 | S45F† | Missense | Hepatocellular carcinoma, acute lymphoblastic leukemia, breast cancer, Wilms’ tumor | Gain of function (F) | 32 | ||
| CTNNB1 | K335T | Missense | Hepatocellular carcinoma | Gain of function (F) | 33 | ||
| AP-1/MAPK pathway | KRAS | I36M | Missense | Hematopoeitic neoplasms | Gain of function (F) | 34 | |
| KRAS | A18D | Missense | GTP binding | Lymphoma | Gain of function (F) | 34 | |
| KRAS | G13D | Missense | GTP binding | Solid tumors | Gain of function (F) | 34 | |
| MAPK3 | R278Q | Missense | Protein kinase | New | Probably gain of function (P) | ||
| STAT3 | E616G | Missense | SH2 | Lymphoma | Gain of function (F) | 26,35 | |
| STAT3 | E616K | Missense | SH2 | Lymphoma | Gain of function (F) | 26,35 | |
| GTF2I | D317E | Missense | New | Probably gain of function (P) | |||
| GTF2I | N340S | Missense | New | Probably gain of function (P) | |||
| GTF2I | R523S | Missense | GTF2I-like | New | Probably gain of function (P) | ||
| GTF2I | L607F | Missense | GTF2I-like | New | Probably gain of function (P) | ||
| GTF2I | R702Q | Missense | New | Probably gain of function (P) | |||
| GTPases pathway | RHOA | K18N | Missense | GTP binding | New | Gain of function (F) | |
| RHOA | G17V | Missense | GTP binding | AITL, ATLL | Dominant negative (F) | 12,-14,20,36 | |
| VAV1 | 151_158del | Frameshift deletion | New | Probably loss of function (P) | |||
| VAV1 | 778_783del | Frameshift deletion | New | Probably gain of function (P) | |||
| VAV1 | D797G | Missense | SH3 2 | New | Probably gain of function (P) | ||
| VAV1 | Y826S | Missense | SH3 2 | New | Probably gain of function (P) | ||
| VAV2 | Y214C | Missense | DH | Gastric carcinoma | Probably dominant negative (P) | ||
| JAK/STAT and TLR pathways | JAK1 | D831E | Missense | Kinase | New | Probably gain of function (P) | |
| JAK2 | V617F | Missense | Myeloproliferative disorders | Gain of function (F) | 37,,,,,-43 | ||
| JAK3 | A699V | Missense | Kinase | Lymphoma, carcinoma | Gain of function (F) | 37,,,,,-43 | |
| MYD88 | S219C | Missense | TIR | DLBCL, lymphocytic leukemia | Probably gain of function (P) | 44 |
| Functional groups . | Gene . | Amino acid change . | Mutation type . | Domain . | New/reported . | Effect . | References . |
|---|---|---|---|---|---|---|---|
| Costimulatory and proximal TCR signaling | CD28 | D124V | Missense | Extracellular | AITL, ATLL | Gain of function (F) | 20,25 |
| CD28 | D124E | Missense | Extracellular | AITL, ATLL | Gain of function (F) | 20,25 | |
| CD28 | T195P | Missense | Cytoplasmic | AITL, ATLL | Gain of function (F) | 20,25,30 | |
| LCK | N446K* | Missense | Kinase | New | Probably gain of function (P) | ||
| LCK | P447R* | Missense | Kinase | New | Probably gain of function (P) | ||
| FYN | Q527X | Stop gain | ATLL | Probably gain of function (P) | 20 | ||
| FYN | 525_525del† | Frameshift deletion | New | Probably gain of function (P) | |||
| FYN | S186L† | Missense | SH2 | New | Probably gain of function (P) | ||
| FYN | K108fs* | Frameshift deletion | New | Probably loss of function (P) | |||
| FYN | E107S* | Missense | SH3 | New | Probably loss of function (P) | ||
| NK-kB/NF-AT pathway | PLCG1 | E47K | Missense | PH 1 | ATLL | Gain of function (F) | 20 |
| PLCG1 | R48W | Missense | PH 1 | ATLL, SS | No effect (F) | 20,24 | |
| PLCG1 | D342G | Missense | PI-PLC X-box | ATLL, MF | Gain of function (F) | 20 | |
| PLCG1 | S345F† | Missense | PI-PLC X-box | ATL, MF, SS, ATLL | Gain of function (F) | 20,-22,24 | |
| PLCG1 | S520F† | Missense | PH 2; first part | MF, SS, ATLL | Gain of function (F) | 20,21,24 | |
| PLCG1 | E730K | Missense | SH2 2 | New | Gain of function (F) | ||
| PLCG1 | G869E | Missense | Near SH3 | New | Gain of function (F) | ||
| PLCG1 | E1163K | Missense | C2 | ATLL, SS | Gain of function (F) | 20,24 | |
| PLCG1 | D1165H | Missense | C2 | ATLL, SS | Gain of function (F) | 20,24 | |
| PLCG1 | D1165G | Missense | C2 | ATLL | Gain of function (F) | 20 | |
| CARD11 | F902C | Missense | ATLL | Gain of function (F) | 20 | ||
| CARD11 | S547T | Missense | New | Gain of function (F) | |||
| CARD11 | F176C | Missense | Coiled coil | New | Gain of function (F) | ||
| TRAF6 | Q347X | Stop gain | Coiled coil /MATH | New | NA | ||
| PI3K pathway | PIK3R1 | K141R | Missense | RHO-GAP | New | Probably gain of function (P) | |
| PIK3R1 | Q475P | Missense | iSH2 | New | Probably gain of function (P) | ||
| PIK3R1 | T576A | Missense | iSH2 | New | Probably gain of function (P) | ||
| PIK3R1 | G680S | Missense | SH2 2 | New | Probably gain of function (P) | ||
| PIK3R1 | V704M | Missense | SH2 2 | New | Probably gain of function (P) | ||
| PIK3R5 | A259V | Missense | Carcinoma (liver) | Probably gain of function (P) | 31 | ||
| PIK3CA | L1001P | Missense | PI3K/PI4K | New | Probably gain of function (P) | ||
| PDPK1 | 19_20del | Inframe deletion | New | NA | |||
| PDPK1 | 151_152del | Frameshift deletion | Protein kinase | New | Probably loss of function (P) | ||
| PDPK1 | R324Q | Missense | Protein kinase | New | Probably gain of function (P) | ||
| PDPK1 | P340Q | Missense | Protein kinase | New | Probably gain of function (P) | ||
| AKT1 | G294R | Missense | Protein kinase | New | Probably gain of function (P) | ||
| CTNNB1 | T41A† | Missense | Phospho by GSK3b | Hepatocellular carcinoma, acute lymphoblastic leukemia, breast cancer, Wilms’ tumor | Gain of function (F) | 32 | |
| CTNNB1 | H36P | Missense | Hepatocellular carcinoma, acute lymphoblastic leukemia, breast cancer, Wilms’ tumor | Gain of function (F) | 32 | ||
| CTNNB1 | S45F† | Missense | Hepatocellular carcinoma, acute lymphoblastic leukemia, breast cancer, Wilms’ tumor | Gain of function (F) | 32 | ||
| CTNNB1 | K335T | Missense | Hepatocellular carcinoma | Gain of function (F) | 33 | ||
| AP-1/MAPK pathway | KRAS | I36M | Missense | Hematopoeitic neoplasms | Gain of function (F) | 34 | |
| KRAS | A18D | Missense | GTP binding | Lymphoma | Gain of function (F) | 34 | |
| KRAS | G13D | Missense | GTP binding | Solid tumors | Gain of function (F) | 34 | |
| MAPK3 | R278Q | Missense | Protein kinase | New | Probably gain of function (P) | ||
| STAT3 | E616G | Missense | SH2 | Lymphoma | Gain of function (F) | 26,35 | |
| STAT3 | E616K | Missense | SH2 | Lymphoma | Gain of function (F) | 26,35 | |
| GTF2I | D317E | Missense | New | Probably gain of function (P) | |||
| GTF2I | N340S | Missense | New | Probably gain of function (P) | |||
| GTF2I | R523S | Missense | GTF2I-like | New | Probably gain of function (P) | ||
| GTF2I | L607F | Missense | GTF2I-like | New | Probably gain of function (P) | ||
| GTF2I | R702Q | Missense | New | Probably gain of function (P) | |||
| GTPases pathway | RHOA | K18N | Missense | GTP binding | New | Gain of function (F) | |
| RHOA | G17V | Missense | GTP binding | AITL, ATLL | Dominant negative (F) | 12,-14,20,36 | |
| VAV1 | 151_158del | Frameshift deletion | New | Probably loss of function (P) | |||
| VAV1 | 778_783del | Frameshift deletion | New | Probably gain of function (P) | |||
| VAV1 | D797G | Missense | SH3 2 | New | Probably gain of function (P) | ||
| VAV1 | Y826S | Missense | SH3 2 | New | Probably gain of function (P) | ||
| VAV2 | Y214C | Missense | DH | Gastric carcinoma | Probably dominant negative (P) | ||
| JAK/STAT and TLR pathways | JAK1 | D831E | Missense | Kinase | New | Probably gain of function (P) | |
| JAK2 | V617F | Missense | Myeloproliferative disorders | Gain of function (F) | 37,,,,,-43 | ||
| JAK3 | A699V | Missense | Kinase | Lymphoma, carcinoma | Gain of function (F) | 37,,,,,-43 | |
| MYD88 | S219C | Missense | TIR | DLBCL, lymphocytic leukemia | Probably gain of function (P) | 44 |
Genes are organized by functional groups.
AITL, angioimmunoblastic T-cell lymphoma; ATLL, adult T-cell lymphoma/leukemia; DLBCL, diffuse large B-cell lymphoma; F, functional validation; MF, mycosis fungoides; P, literature- and models-based; SS, Sézary syndrome.
Gene variants found in the same patient and in the same allele.
Gene variants found in the same patient but in different alleles.
Mutations in other TCR signaling–related genes are variably recurrent and diverse
CD28 and TCR-proximal signaling genes.
Three different CD28 mutations—T195P, D124V, and D124E—all described previously13,25,30 were identified in 4, 2, and 2 AITL patients, respectively (8/85, 9.4%) (Figure 4A). All TFH-like PTCL were CD28-WT. T195P and D124V mutations are known to enhance TCR/CD28-induced NF-κB activity in vitro.25,30 Three patients (2 AITL, 1 TFH-like PTCL; 3.5%) harbored mutations in FYN, a TCR- and CD28-proximal kinase component (Figure 4B). Mutations in the SH2 domain (S186L) and the absence of phosphorylated tyrosine 531 (T524fs or Q527X mutations) probably confer enhanced kinase activity by disrupting their inhibitory interaction.12 The 2 amino acid substitutions in the tyrosine kinase domain of LCK observed in one AITL patient is expected to enhance its kinase activity.46 We did not find mutations in ICOS, ZAP70, LAT, or ITK.
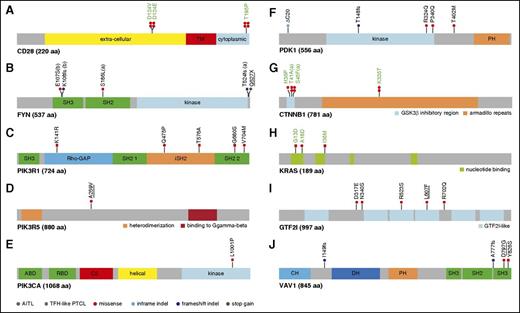
Mapping of variants in TCR signaling genes mutated in at least 3 patients. (A) CD28: the D124V/E variants involve the extracellular part of the receptor, whereas T195P lies in the intracellular, C-terminal, domain between the 2 domains, allowing interaction with PIK3R1 (YVKM sequence) or GRB2/VAV (PRRP sequence) proteins. (B) FYN: the five mutations indicated occurred in three patients; 1 patient harbored 2 mutations (a); 2 mutations in adjacent positions in the SH3 domain (b) were observed on the same allele in another patient. (C-E) PI3K subunits: when appropriate, cellular stimuli are present, the nSH2 and cSH2 domains of PIK3R1 bind phosphorylated tyrosines (YXXM motif) in activated receptors (CD28) and adapter proteins, thereby activating the PIK3CA (p110a) catalytic subunit without releasing the PIK3R1 (p85a) interaction with p110a through their iSH2 and ABD domains, respectively. The K141R missense affects the ρ-GAP domain, whereas iSH2 and the second SH2 domains of PIK3R1 bore 2 missenses each (Q475P, T576A and G680S, V704M, respectively). The A259V mutation, described as somatic in COSMIC, affects a linker region of PIK3R5. Finally, the L1001P point mutation affects the PIK3CA kinase domain. (F) PDK1: one inframe INDEL, one frameshift INDEL, and 3 missense affect PDK1 protein. (G) CTNNB1: 3 previously described activating missenses affect the GSK3β inhibitory domain (exon 3), whereas the K335T activating mutation affects the armadillo repeats region. (H) KRAS: 3 missense mutations alter 2 N-terminal nucleotide binding regions. (I) GTF2I: 5 missense mutations affect the GTF2I transcription factor, among which two are found in different GTF2I-like domains. (J) VAV1: 2 frameshift deletions and 2 missenses affect VAV1, three being localized in the C-terminal SH3 domain of the protein. In all figure panels, previously described activating mutations are in green boldface, and mutations previously described but not functionally tested are underlined. PDK1 is the protein name of PDPK1 gene.
Mapping of variants in TCR signaling genes mutated in at least 3 patients. (A) CD28: the D124V/E variants involve the extracellular part of the receptor, whereas T195P lies in the intracellular, C-terminal, domain between the 2 domains, allowing interaction with PIK3R1 (YVKM sequence) or GRB2/VAV (PRRP sequence) proteins. (B) FYN: the five mutations indicated occurred in three patients; 1 patient harbored 2 mutations (a); 2 mutations in adjacent positions in the SH3 domain (b) were observed on the same allele in another patient. (C-E) PI3K subunits: when appropriate, cellular stimuli are present, the nSH2 and cSH2 domains of PIK3R1 bind phosphorylated tyrosines (YXXM motif) in activated receptors (CD28) and adapter proteins, thereby activating the PIK3CA (p110a) catalytic subunit without releasing the PIK3R1 (p85a) interaction with p110a through their iSH2 and ABD domains, respectively. The K141R missense affects the ρ-GAP domain, whereas iSH2 and the second SH2 domains of PIK3R1 bore 2 missenses each (Q475P, T576A and G680S, V704M, respectively). The A259V mutation, described as somatic in COSMIC, affects a linker region of PIK3R5. Finally, the L1001P point mutation affects the PIK3CA kinase domain. (F) PDK1: one inframe INDEL, one frameshift INDEL, and 3 missense affect PDK1 protein. (G) CTNNB1: 3 previously described activating missenses affect the GSK3β inhibitory domain (exon 3), whereas the K335T activating mutation affects the armadillo repeats region. (H) KRAS: 3 missense mutations alter 2 N-terminal nucleotide binding regions. (I) GTF2I: 5 missense mutations affect the GTF2I transcription factor, among which two are found in different GTF2I-like domains. (J) VAV1: 2 frameshift deletions and 2 missenses affect VAV1, three being localized in the C-terminal SH3 domain of the protein. In all figure panels, previously described activating mutations are in green boldface, and mutations previously described but not functionally tested are underlined. PDK1 is the protein name of PDPK1 gene.
NF-κB/NFAT pathway.
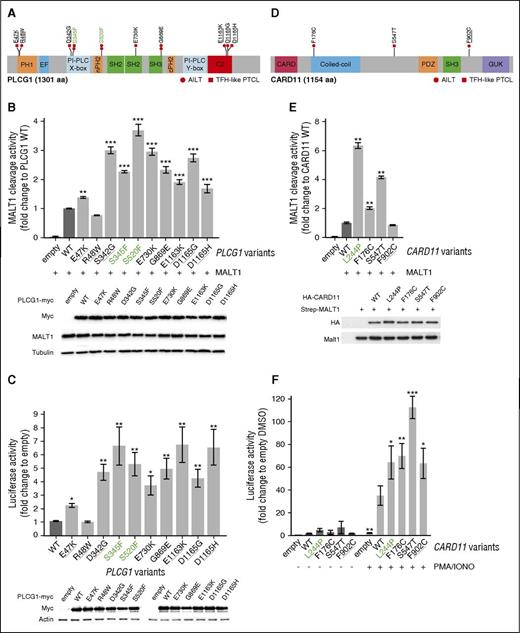
Ten different missense mutations spanning the coding region of PLCG1 (VF [2.1%; 38%]) were identified in 12 of 85 patients (8 AITL, 4 TFH-like PTCL; 14.1%) (Figure 5A). Apart from previously reported variants in adult T-cell leukemia/lymphoma (ATLL) or in other PTCL,20-24 we identified 2 novel variants (E730K and G869E). We generated all mutant constructs and tested their activity against WT PLCG1 in a FRET-based reporter assay of MALT1 protease activity27,28 (Figure 5B) and in a NFAT luciferase reporter assay (Figure 5C). Both experiments confirmed that the S345F and S520F variants were activating.21 All 6 variants in the PI-PLC, SH2, SH3, and C2 domains were also activating, increasing the FRET signal by 1.7- to threefold and the luciferase expression by four- to fivefold. Of the 2 PH1 domain variants, R48W had no effect in both assays, whereas E47K increased NFAT reporter activity by 2.1-fold and FRET signal by 1.5-fold.
Mapping and functional analysis of PLCG1 variants and CARD11 variants. (A) Schematic representation of PLCG1 protein with mapping of the 10 missense mutations identified in AITL (circles) or TFH-like PTCL (squares) cases. Previously described activating mutations are in green boldface and mutations previously described but not functionally tested are underlined. (B) Monitoring of PLCG1-mediated MALT1 activation via a FRET-based reporter assay. Data are represented as mean ± SEM from 3 independent experiments. Significant differences in activation activity were determined using 1-way ANOVA (**P ≤ .01; ***P ≤ .001). Representative western blot from a MALT1 activation experiment. PLCG1 expression is revealed by anti-Myc tag blotting, whereas MALT1 expression is shown by anti-MALT1 antibody. (C) NFAT luciferase reporter assay monitoring activity of PLCG1 mutants, compared with previously reported activating mutants (green). Data are represented as mean ± SEM from 7 independent experiments. Significant differences in activation activity were determined using 1-way ANOVA (*P ≤ .05; **P ≤ .01). Representative western blot from a luciferase assay experiment. Ectopic myc-tagged PLCG1 expression is revealed by anti-Myc. Anti-Actin blotting serves as loading control. (D) Schematic representation of CARD11 protein with mapping of the 3-point mutations found in 2 AITL (circles) and 1 TFH-like PTCL-NOS (square) patients. A previously described mutation is underlined. (E) Monitoring of CARD11-mediated MALT1 activation via a FRET-based reporter assay. Data are represented as mean ± SEM from 6 independent experiments. Significant differences in activation activity were determined using 1-way ANOVA (*P ≤ .05; **P ≤ .01). The known activating L244P variant (green) was used as a positive control for the experiment. Representative western blot from a MALT1 activation experiment. CARD11 expression is revealed by anti-HA tag blotting, whereas MALT1 expression is shown by anti-MALT1 antibody. (F) NF-κB luciferase reporter assay in Jurkat cells deficient for CARD11 monitoring activity of CARD11 mutants, compared with previously reported activating mutants (green). Data are represented as mean ± SEM from 4 independent experiments. Significant differences in activation activity were determined using 2-way ANOVA (*P ≤ .05; **P ≤ .01; ***P ≤ .001 compared with WT PMA/IONO).
Mapping and functional analysis of PLCG1 variants and CARD11 variants. (A) Schematic representation of PLCG1 protein with mapping of the 10 missense mutations identified in AITL (circles) or TFH-like PTCL (squares) cases. Previously described activating mutations are in green boldface and mutations previously described but not functionally tested are underlined. (B) Monitoring of PLCG1-mediated MALT1 activation via a FRET-based reporter assay. Data are represented as mean ± SEM from 3 independent experiments. Significant differences in activation activity were determined using 1-way ANOVA (**P ≤ .01; ***P ≤ .001). Representative western blot from a MALT1 activation experiment. PLCG1 expression is revealed by anti-Myc tag blotting, whereas MALT1 expression is shown by anti-MALT1 antibody. (C) NFAT luciferase reporter assay monitoring activity of PLCG1 mutants, compared with previously reported activating mutants (green). Data are represented as mean ± SEM from 7 independent experiments. Significant differences in activation activity were determined using 1-way ANOVA (*P ≤ .05; **P ≤ .01). Representative western blot from a luciferase assay experiment. Ectopic myc-tagged PLCG1 expression is revealed by anti-Myc. Anti-Actin blotting serves as loading control. (D) Schematic representation of CARD11 protein with mapping of the 3-point mutations found in 2 AITL (circles) and 1 TFH-like PTCL-NOS (square) patients. A previously described mutation is underlined. (E) Monitoring of CARD11-mediated MALT1 activation via a FRET-based reporter assay. Data are represented as mean ± SEM from 6 independent experiments. Significant differences in activation activity were determined using 1-way ANOVA (*P ≤ .05; **P ≤ .01). The known activating L244P variant (green) was used as a positive control for the experiment. Representative western blot from a MALT1 activation experiment. CARD11 expression is revealed by anti-HA tag blotting, whereas MALT1 expression is shown by anti-MALT1 antibody. (F) NF-κB luciferase reporter assay in Jurkat cells deficient for CARD11 monitoring activity of CARD11 mutants, compared with previously reported activating mutants (green). Data are represented as mean ± SEM from 4 independent experiments. Significant differences in activation activity were determined using 2-way ANOVA (*P ≤ .05; **P ≤ .01; ***P ≤ .001 compared with WT PMA/IONO).
Point mutations in CARD11, which encodes a scaffolding protein downstream of PLCG1 required for CD28/TCR-induced NF-κB activation,28 were found in 2 AITL and 1 TFH-like PTCL (3.5%). The F176C variant affects the coiled-coil domain of the protein, commonly mutated in diffuse large B-cell lymphoma (DLBCL).27 The S547T and F902C variants map to linker regions with undefined structure (Figure 5D). In a FRET-based assay,28 F176C and S547T enhanced MALT1 proteolytic activity by two- and fourfold, respectively, whereas the F902C mutant, previously reported in ATLL patients,20 had no detectable effect (Figure 5E). When transfected into Jurkat cells deficient for CARD11,29 we found that, compared with WT CARD11, all 3 variants induced enhanced NF-κB reporter activity in response to phorbol myristate acetate/ionomycin stimulation (Figure 5F).
PI3K pathway.
Six patients (3 AITL, 3 TFH-like PTCL; 7%) showed mutations in PI3K genes encoding the regulatory subunits PIK3R1 (n = 5), PIK3R5 (n = 1), or the catalytic subunit PIK3CA (n = 1) (Figure 4C-E). These mutations likely enhance the catalytic subunit activity or increase PIK3R1 binding to CD28.31 Five AITL patients (5.9%) had mutations in PDPK1 (PDK1), a master serine/threonine kinase with multiple targets including AKT. Three missense mutations were found on or near its kinase domain, suggesting an activating effect47 (Figure 4F). Four different CTNNB1 mutations, known to occur in a variety of carcinomas and Wilms’ tumors, were identified in 5 patients (4 AITL, 1 TFH-like PTCL; 5.9%) (Figure 4G). All have been previously characterized as activating or stabilizing variants that induce persistent signaling and increased proliferation, and have been linked to both poor treatment response and frequent tumor relapse in various tumors.32,33
AP-1/MAPK pathway.
Eleven patients (8 AITL, 3 TFH-like PTCL; 13%) had mutually exclusive activating mutations in the MAPK pathway, which regulate the activity of the AP-1 transcription factor family. Activating KRAS34 and STAT326,35 mutations were detected in 3 and 2 patients, respectively (Figure 4H). We also found 5 missense mutations in GTF2I (TFII-I)48 (Figure 4I), which are likely activating as those reported in thymic epithelial tumors.49
GTPases pathway.
Apart from highly recurrent RHOA mutations, 4 patients (3 AITL, 1 TFH-like PTCL; 4.7%) harbored previously undescribed frameshift deletions or missense mutations in VAV1, a guanosine exchange factor for RAC1, CDC42, and RHOA (Figure 4J). Two missense mutations were on the second SH3 domain, where a D797 residue mutation was previously shown to activate VAV1-mediated transformation via cell-cell contact deregulation.50
Analysis of sequential biopsies.
Analysis of paired samples in 4 refractory or relapsed patients (supplemental Table 8) showed that the mutations identified at diagnosis were also present in the second biopsy, with overall very similar VF. Within the limit of the panel of genes examined, 2 patients presented one additional mutation at relapse (CTNNB1 and VAV1), at somewhat lower VF than those present at diagnosis. Although this analysis is limited to a few pairs of samples, it indicates that the mutational heterogeneity that characterizes many AITL samples appears to be preserved and is essentially not modified by the treatments administered.
Activating mutations in TCR signaling–related genes correlate with molecular signatures reflecting higher T-cell activation and with response to therapy
Sixty-six of 85 patients harbored mutations in TCR signaling genes. To assess the biological and clinical impact of the tumor mutational status, we focused on patients with mutations in TCR signaling–related genes other than RHOA, designated hereafter as “TCR_Mut” (42/85 cases, 49%), and compared them with those with only RHOA or no detectable mutation, designated “TCR_WT” (43/85 cases, 51%) (Figure 6).
Mutual exclusivity of TCR signaling variants. The mutational status of TCR-related genes is represented for the 85 patients of the extended cohort. Genes other than RHOA are ranked by decreasing mutation frequency and show an essentially mutually exclusive mutation pattern. In total, 49% of cases were mutated in 1 or several TCR-related gene(s) other than RHOA (hereafter considered as TCR_Mut), 28.5% were mutated in RHOA only, and 22.5% harbored no mutation in any of the genes tested (collectively considered as TCR_WT).
Mutual exclusivity of TCR signaling variants. The mutational status of TCR-related genes is represented for the 85 patients of the extended cohort. Genes other than RHOA are ranked by decreasing mutation frequency and show an essentially mutually exclusive mutation pattern. In total, 49% of cases were mutated in 1 or several TCR-related gene(s) other than RHOA (hereafter considered as TCR_Mut), 28.5% were mutated in RHOA only, and 22.5% harbored no mutation in any of the genes tested (collectively considered as TCR_WT).
Mutations in TCR-related genes other than RHOA were virtually nonoverlapping, and the vast majority (47/56 distinct variants, 84%) were either functionally validated (n = 24) or predicted (n = 23) to be biologically gain-of-function mutants (Figure 6). Consequently, nearly all TCR_Mut patients (39/42 patients, 93%) harbored at least 1 activating mutation.
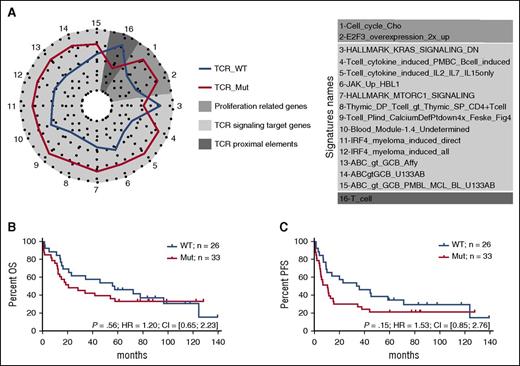
There were no significantly differentially expressed genes between TCR_Mut and TCR_WT samples (supplemental Figure 4). However, the molecular signatures of TCR_Mut were significantly enriched in 15 gene sets compared with TCR_WT by enrichment analysis (Figure 7A; supplemental Table 7; supplemental Figure 5), reflecting the activation of signaling pathways like PI3K, NF-κB,17 IRF4, and JAK-STAT, as well as the upregulation of calcium-signaling target genes. This points toward TCR signaling activation in TCR_Mut PTCLs. TCR_Mut samples also showed enrichment in cell-cycle signatures reflecting increased proliferative activity. Finally, there was a trend for TCR_Mut samples to have downregulated proximal TCR signaling genes (signature “T_cell”) such as TCR (TCRα, TCRβ; CD3δ, CD3γ), signaling molecules (LAT, TRIM, SAP, FYB, ZAP70), and cell surface markers (adjusted P = .10), a feature consistent with sustained T-cell activation.51
Biological significance and clinical relevance of TCR signaling–related mutations. (A) Spider plot representation of gene sets differentially enriched in patients with or without mutations in genes related to TCR signaling (TCR_Mut vs TCR_WT). Genes tested in the enrichment analysis were selected from signatures relevant in T- and B-cell differentiation and activation. Statistical significance of the enrichment was reached for gene sets 1 to 15 (P < .05); for gene set 16, marginal significance was observed (P = .1). (B-C) Overall survival (B) and progression-free survival (C) of patients with (red) or without (blue) mutations in TCR signaling–related genes. Analyses are restricted to the 59 patients treated with anthracyclin-based chemotherapy. Mutated patients show a trend toward a shorter PFS (11 vs 36 months) than WT patients (P = .15).
Biological significance and clinical relevance of TCR signaling–related mutations. (A) Spider plot representation of gene sets differentially enriched in patients with or without mutations in genes related to TCR signaling (TCR_Mut vs TCR_WT). Genes tested in the enrichment analysis were selected from signatures relevant in T- and B-cell differentiation and activation. Statistical significance of the enrichment was reached for gene sets 1 to 15 (P < .05); for gene set 16, marginal significance was observed (P = .1). (B-C) Overall survival (B) and progression-free survival (C) of patients with (red) or without (blue) mutations in TCR signaling–related genes. Analyses are restricted to the 59 patients treated with anthracyclin-based chemotherapy. Mutated patients show a trend toward a shorter PFS (11 vs 36 months) than WT patients (P = .15).
Then, when comparing the clinical features and outcome of TCR_Mut and TCR_WT patients, no significant differences in main clinical characteristics at presentation (supplemental Table 9) or in OS (18 vs 40 months median OS; 5-year OS 20% vs 29%; P = .37) were found (supplemental Figure 6). Strikingly, however, among the 59 patients (49 AITL and 10 TFH-like PTCLs) who received anthracyclin-based induction chemotherapy, 11 of 33 (33%) TCR_Mut patients vs 2 of 26 (8%) TCR_WT (P = .02, supplemental Table 9) showed early progression, relapse, or no response to treatment within the first 6 months after diagnosis. This, however, did not translate into a statistically significant difference in PFS at 5 years (21% vs 24% for TCR_Mut vs TCR_WT patients; P = .15) (Figure 7B-C).
Discussion
In this study, we confirmed the high prevalence of RHOA mutations in TFH-derived PTCL, with most samples bearing the dominant-negative G17V variant. Interestingly, we identified a new K18N variant in 3% of the patients that presented activating features in in vitro assays. This intriguing finding of distinct RHOA mutations, with apparently opposite functional properties in the same disease, was also recently documented in ATLL.36,52 Further studies are warranted to understand how these variants may contribute to the pathogenesis of PTCLs.
Apart from RHOA alterations, we identified activating and virtually mutually exclusive mutations in diverse TCR signaling genes in half of TFH-derived lymphomas. Individually, the frequencies of these mutations were variable, with the 5 most mutated genes (PLCG1, CD28, PIK3 elements, GTF2I, CTNNB1) being altered in 14% to 5% of the patients. CD28 and FYN mutations in AITL were previously described in a small number of cases.12-14 Our study significantly expands these previous results and strongly supports a role of activated TCR signaling in the pathogenesis of TFH-derived PTCLs, drawing strong parallels with the role of BCR signaling in B-cell lymphomas.
Mutations in genes related to TCR costimulation and signaling were recently reported in other PTCL entities, like cutaneous T-cell lymphomas, PTCL-NOS, and ATLL.20-24,53 Nonetheless, mutations in specific genes are variably recurrent in distinct entites,24,25 and for a given gene, the distribution of the mutations and their relative prevalence are heterogeneous. For instance, PLCG1 and CARD11 mutations are highly prevalent (up to 18% and 15% of the cases, respectively) in cutaneous T-cell lymphoma, in which conversely CD28 mutations are not found.20 CARD11 mutations are more frequent in ATLL (24%) and Sézary syndrome (10.9%) than in this series of TFH-derived lymphomas. Whether the observed mutations are sufficient to induce constitutive activation of the respective pathways, and how their effect may be dependent or influenced by antigen-driven or other stimulatory signals remains unknown. Interestingly, some of the TCR-related genes may be altered through point mutations or small indels as well as by major structural rearrangements, amplifications, and deletions.20,23,24 Collectively, these findings strongly suggest that genetic alterations affecting TCR signaling operate as a common pathogenic mechanism in several PTCL entities.
Our analysis also explored the JAK/STAT and TLR pathways. Mutation-induced activation of the JAK/STAT pathway, highly prevalent in myeloid neoplasms, was recently identified as a major oncogenic mechanism in several T-cell leukemias and lymphomas derived from innate immune cells.20,37-43,53 Our results suggest, however, that JAK/STAT mutations do not have the same importance in the pathogenesis of TFH-derived lymphomas, because only 4 of 85 patients bore a single activating mutation each in JAK1, JAK2, JAK3, or MYD88 (Table 1).44 Except for JAK2 V617F, the 3 others were observed in PLCG1-mutated cases.
Our data further support that AITL and TFH-like PTCL are closely related at the molecular level5 and share common oncogenic mechanisms. We and others have reported the occurrence of TET2, DNMT3A, and RHOA mutations in a large proportion of both AITL and TFH-like PTCL patients.5,8,9,12-14 This study extends the molecular overlap across TFH-derived neoplasms to genetic alterations in various TCR-signaling. Interestingly, CD28 mutations were exclusively identified in AITL patients, consistent with recent reports.25,30 Similarly, IDH2 mutations were rarely detected in PTCL cases,9,54 suggesting that the distribution of some genetic features might be relevant in distinguishing AITL from other TFH-derived nodal lymphomas, which otherwise share many common features.5
In the proposed multistep model of pathogenesis for TFH-derived PTCL, mutation-induced epigenetic deregulation, possibly arising in early hematopoietic precursors, may promote the emergence of premalignant cells, which requires additional genetic events to acquire a definitively malignant phenotype.13,55 It has been thus suggested that RHOA alterations occur as a second event in TET2- and/or DNMT3A-mutated cells.13,55,56 The prevalence of mutations in TET2, DNMT3A, and IDH2 analyzed in a subset of our cases was 52% (34/65 cases), 29% (17/56 cases), and 30% (21/71 cases), respectively.8,9 These mutations were not mutually exclusive, and in fact tended to co-occur with each other, as previously reported.12,13 We also found a tendency for RHOA mutations to associate with epigenetic alterations, because RHOA mutations were detected in 31 of 41 (76%) cases mutated in TET2, DNMT3A, and/or IDH2, but in only 8 of 23 (35%) cases without epigenetic mutations (P = .003). Because the proportion of neoplastic cells in AITL is typically low and variable from case to case,3 the comparison of VF of different mutated genes was performed for each individual sample. In 13 cases harboring mutations in both TET2 and/or DNMT3, and RHOA or IDH2 with available VF, the TET2/DNMT3A VF was significantly higher than for RHOA or IDH2 (supplemental Figure 7A). There was no obvious association of RHOA mutations with other TCR-related genes, because 27 of 51 (53%) RHOA-mutated and 15 of 34 (44%) RHOA-WT cases were mutated in other TCR-related genes (P = .51). In 27 cases mutated in both RHOA and one or several genes of the TCR, JAK/STAT, or TLR pathways, no significant difference in VF means was observed between RHOA and other genes (supplemental Figure 7B).
Although our results warrant confirmation in cohorts of patients treated on prospective clinical trials, they suggest that the mutational status of TCR-related genes may have important clinical implications, predicting early treatment failure with anthracyclin-based chemotherapy in TFH-related PTCL patients. Importantly, several activating mutations found in PI3K or NF-κB pathways could be targeted by idelalisib or the proteasome inhibitor bortezomib, respectively. It is of interest to determine their efficacy in TCR_Mut patients,12,21 possibly in combination with demethylating agents.57 Thus, similar to the importance of targeting BCR signaling in B-cell lymphomas with the BTK inhibitor ibrutinib,17 characterization of the TCR mutational status might open new avenues to design specific and more effective therapies. In clinical practice, given the high number of genes involved and the diversity of mutations found, targeted deep sequencing with high depth/coverage appears to be the method of choice for selecting patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Catherine Chapuis (Pathology, Lausanne) and Caroline Communaux from the LYSA-Pathology for their technical assistance; K. Harshman and the Lausanne Genomic Technology Facility for their technical support; and Céline Villenet and Sabine Quief from the plate-forme de génomique fonctionnelle et structurale, Lille University, for the WES experiments.
This study was supported by grants received from the Plan Cancer (Belgium), the Ligue du Cancer (Switzerland), the Institut National du Cancer (INCa AAP PLBIO13-085, INCa-DGOS 2010-085, and INCa-Plan Cancer 2013), and the MEDIC foundation.
Authorship
Contribution: D.V., R.D.M., M.T., R.D.G., L.d.L., and P.G. conceived and designed the study; D.V. and M.P.D.D. developed the methodologies; D.V., M.P.D.D., J.B., B.F., A.M., C.B., F.L., M.J., M.T., J.I., A.R., M.F., and B.B., acquired the data; D.V., M.P.D.D., E.M., F.L., M.J., O.M., O.T., C.H., M.D., O.A.B., J.-P.J., and J.G. analyzed and interpreted the data; D.V., M.P.D.D., F.L., E.M., L.d.L., and P.G. wrote the manuscript; V.F. was the administrative, technical, and material support; and L.d.L. and P.G. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laurence de Leval, CHUV Institut de Pathologie, Rue du Bugnon 25, 1011 Lausanne, Switzerland; e-mail: laurence.deleval@chuv.ch.
References
Author notes
D.V. and M.P.D.D. are considered co-first authors.
P.G. and L.d.L. are considered co-last authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal