Key Points
Cytokine-activated NK cells display distinct gene expression programs in response to cytokine withdrawal.
IL-15 sustains antitumor functions of NK cells through mTOR-governed metabolic processes.
Abstract
Treatment of hematological malignancies by adoptive transfer of activated natural killer (NK) cells is limited by poor postinfusion persistence. We compared the ability of interleukin-2 (IL-2) and IL-15 to sustain human NK-cell functions following cytokine withdrawal to model postinfusion performance. In contrast to IL-2, IL-15 mediated stronger signaling through the IL-2/15 receptor complex and provided cell function advantages. Genome-wide analysis of cytosolic and polysome-associated messenger RNA (mRNA) revealed not only cytokine-dependent differential mRNA levels and translation during cytokine activation but also that most gene expression differences were primed by IL-15 and only manifested after cytokine withdrawal. IL-15 augmented mammalian target of rapamycin (mTOR) signaling, which correlated with increased expression of genes related to cell metabolism and respiration. Consistently, mTOR inhibition abrogated IL-15–induced cell function advantages. Moreover, mTOR-independent STAT-5 signaling contributed to improved NK-cell function during cytokine activation but not following cytokine withdrawal. The superior performance of IL-15–stimulated NK cells was also observed using a clinically applicable protocol for NK-cell expansion in vitro and in vivo. Finally, expression of IL-15 correlated with cytolytic immune functions in patients with B-cell lymphoma and favorable clinical outcome. These findings highlight the importance of mTOR-regulated metabolic processes for immune cell functions and argue for implementation of IL-15 in adoptive NK-cell cancer therapy.
Introduction
Natural killer (NK)-cell–based immunotherapy is a potential therapeutic modality in patients with advanced cancers as transfer of haploidentical NK cells induces beneficial responses in patients with hematological malignancies; and leukemia clearance correlates with persistence and in vivo expansion of NK cells after infusion.1-3 Thus, sustained NK-cell activity in vivo likely represents a therapy performance-limiting factor.
The type I cytokine family members interleukin-2 (IL-2) and IL-15 are essential in controlling homeostasis of innate and adaptive immunity.4 Despite their different protein sequences, IL-2 and IL-15 bind to shared β (IL-2/IL-15Rβ) and γ (γc) subunits, but engage separate α-chain receptors (IL-2/IL-15Rα).5 Although IL-2/IL-15 receptor complexes activate similar signal transduction cascades, they display distinct activities in vivo. Although IL-2 preferentially expands regulatory T cells and CD4+ helper T cells,6 IL-15 supports development of central memory T cells7,8 and NK cells.9,10 Furthermore, sustained persistence of infused NK cells is linked to increased levels of endogenous IL-15 following treatment with high-dose cyclophosphamide and fludarabine.1 Thus, NK-cell activation with IL-15 may have beneficial effects on their postinfusion activity. The molecular mechanisms underlying distinct effects from IL-2 and IL-15 on NK-cell proliferation and persistence are, however, unknown.
Recently, studies of “translatomes” (ie, the pool of translated messenger RNAs [mRNAs]) using polysome or ribosome profiling have highlighted mRNA translation as a key mechanism controlling gene expression and influencing a wide range of functions in immune cells.11-13 Changes in translational efficiency affect protein levels without changes in steady-state mRNA levels, thereby enabling rapid adaptation to environmental changes essential for a functional immune system.13 Thus, profiling mRNA translation may be essential to efficiently link observed immune cell phenotypes to underlying gene expression programs.
Here, we uncover IL-15–mediated improved post-cytokine-withdrawal functions of NK cells associated with augmented mammalian target of rapamycin (mTOR) signaling and an IL-15–primed gene expression program. Such detailed mechanistic and functional understanding of IL-15’s impact on human NK cells supports implementation of IL-15 in adoptive NK-cell therapy.
Materials and methods
Patient gene expression data sets
We used a recent data set14 to investigate the role of IL-15 in patients (supplemental Methods, available on the Blood Web site).
Cell cultures
K562 (short tandem repeat fingerprint in supplemental Table 1) and Epstein-Barr virus (EBV)-transformed B cells were maintained in Iscove modified Dulbecco medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen). Prior to NK-cell isolation, T cells were removed by CD3 depletion (RosetteSep kit from StemCell Technologies) during Ficoll gradient centrifugation. Primary human NK cells were subsequently isolated by magnetic-activated cell sorting purification (purity >98%; Miltenyi Biotec).
Cytokine activation and expansion of NK cells
To activate NK cells, 18.3 ng/mL recombinant human IL-2 (50% effective dose <0.1 ng/mL; Peprotech) or recombinant human IL-15 (50% effective dose <0.1 ng/mL; Biological Resources Branch, National Cancer Institute) was added to 1 mL of Iscove modified Dulbecco medium supplemented with 10% human AB serum containing 2 × 106 NK cells in 24-well plates for 48 hours. To evaluate the molecular mechanisms, dimethyl sulfoxide (DMSO), the mTOR inhibitor torin-1 (1 µM; Tocris Biosciences), the STAT-3–selective inhibitor S3I-201 (100 μM; Sigma-Aldrich), or the STAT-5–selective inhibitor CAS285986-31-4 (400 µM; Merck) was added during the activation. For in vitro expansion of human primary NK cells, a clinically applicable protocol with minor modifications was used15 (see supplemental Methods).
NK-cell function
The cytolytic activity of NK cells was measured using the chromium (51Cr) release assay. Proliferation of NK cells was determined by 3H-thymidine incorporation (see supplemental Methods). To evaluate levels of soluble interferon-γ (IFN-γ) released by NK cells, culture media was harvested and quantified using enzyme-linked immunosorbent assay (MabTech).
Flow cytometry
Fluorochrome-conjugated antibodies and labeling dyes are listed in supplemental Table 2 and were applied as previously described.16 Fluorescently labeled cells were acquired in a BD LSRII flow cytometer and analyzed by FlowJo software (TreeStar; see supplemental Methods).
Oxygen and glucose consumption
Five million primary human NK cells were activated with 18.3 ng/mL IL-2 or IL-15 in a 12-well plate, in presence of DMSO, torin-1 (1 µM), or torin-1 (1 µM) plus CAS285986-31-4 (400 µM). Oxygen consumption rate (OCR) and extracellular acidity rate (ECAR) were determined using the XF cell Mito Stress Test kit and the Seahorse XF analyzer (Seahorse Bioscience; see supplemental Methods).
RNA isolation and RNAseq library generation
Cytosolic and polysome-associated RNA was collected from 2 to 4 million NK cells per donor and treated as described.17 smartSeq2 RNA-sequencing (RNAseq) libraries were prepared as described (from step 518 ). Pooled libraries were sequenced (50-bp paired-end reads generated with Illumina HiSeq 2500; see supplemental Methods).
RNAseq data analysis
RNAseq reads were mapped to the hg19 genome assembly using HISAT19 and quantified as described using default settings.20 Data were transformed using r-log, and a random variance model20 was used to identify differential expression using data from cytosolic or polysome-associated mRNA while changes in polysome-associated mRNA that were independent of changes in cytosolic mRNA levels (ie, differential translation) were identified using anota.21 Generally Applicable Gene-Set Enrichment was used to identify enrichment of genes with functions annotated by the Gene Ontology Consortium using data for all genes as input,22 whereas GOstats was used to identify enrichment in identified gene subsets (see supplemental Methods).
In vivo experiments
All experiments were approved by the ethical review board at Karolinska Institutet (ethical approval #N175/15). NOD/SCID/γ−/− (NSG) mice (bred and maintained at the Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet) were injected intraperitoneally with 1× 106 to 5 × 106 IL-2– or IL-15–expanded NK cells. Prior to injection, NK cells were labeled for 20 minutes with 12.5 µg/mL of the near infrared dye DiIC18(7) (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide [DiR; Invitrogen]). On day 4 to 5 after infusion, livers, lungs, and spleens were resected, mechanically homogenized, passed through a 70-μm nylon mesh filter, and evaluated for live-dead fixable aqua dead cell marker (Invitrogen) and anti-CD56-PeCy7 (clone HCD56; Biolegend) using a BD LSRII flow cytometer and analyzed by FlowJo (TreeStar) (see supplemental Figure 7A for gating strategy).
Statistics
Data were analyzed using Prism (GraphPad) and evaluated for normality before appropriate Student t test or nonparametric Mann-Whitney U tests were performed. Data from at least 3 donors and 3 independent experiments are presented as mean ± standard deviation (SD). Representative fluorescence-activated cell sorter (FACS) analysis histograms were selected to resemble across replicate mean values. The association between IL-15 and GZMB/PRF1 was quantified by Spearman correlation. We explored the association between IL-15 expression and risk of death using martingale residuals plots.23,24 Patients were grouped according to IL-15 expression and IL-15’s association with overall survival was assessed using a score (log-rank) test and visualized using Kaplan-Meier plots.
Results
IL-15 primes for survival and cytolytic activity in human NK cells
We reasoned that an in vitro model assessing IL-15 and IL-2 effects after their withdrawal may allow us to uncover cytokine-specific properties relevant for postinfusion performance of NK cells. At the selected concentration, activation with IL-15 or IL-2 induced a comparable increase in primary human NK-cell proliferation and cytolytic activity (P < .05; Figure 1A). However, at suboptimal cytokine concentrations (<9.15 ng/mL), IL-15 was superior to IL-2 in maintaining NK-cell proliferation (data not shown). After cytokine withdrawal, IL-15–treated NK cells maintained a higher level of cytotoxicity (P < .05; Figure 1B) and showed lower levels of apoptosis (P < .05; Figure 1C) compared with cells treated with IL-2. Furthermore, reexposure of IL-15–treated NK cells with IL-15 resulted in higher levels of CD25+/CD137+–activated NK compared with reexposure of IL-2–treated NK cells with IL-2 (data not shown). This suggests that IL-15 and IL-2 differ in their ability to sustain cytokine signaling, induce expression of cytokines and/or their receptors, and/or induce STAT-3 and/or -5 phosphorylation. Indeed, IL-15 induced expression of IL-2Rα (CD25), but not IL-15Rα or IL-2/15Rβ (P < .001), retained membrane binding of IL-15 and IL-2, increased phosphorylation of STAT-3 (Y705) and STAT-5 (Y694), and elevated expression of the antiapoptotic protein Bcl-2 compared with IL-2–activated NK cells (Figure 1D-E; supplemental Figure 1A).
IL-15 primes NK cells with improved survival and cytolytic activity following cytokine withdrawal. Primary human NK cells were isolated from fresh peripheral blood mononuclear cells (PBMCs) and activated with IL-2 or IL-15 (both at 18.3 ng/mL) for 48 hours. (A) Cytolytic capacity against NK-sensitive target K562 (effector-to-target [E:T] ratio = 5:1) and proliferation of resting or cytokine activated human NK cells were measured by chromium release assay and thymidine incorporation assay, respectively. Following cytokine activation for 48 hours, NK cells were cultivated without cytokines (cytokine withdrawal) for an additional 24 hours and tested for their (B) cytolytic activity against K562 cells or (C) viability. Flow cytometry analysis of NK cells following cytokine activation including (D) frequencies of CD25+ cells, (E) expression of membrane-bound cytokines, and cytokine receptor complexes; intracellular expression of Bcl-2; and phosphorylation of STAT-3 (Y705) and STAT-5 (Y694). Results from multiple donors (n > 5) were summarized and are presented as mean ± SD. *P < .05; ***P < .001; Mann-Whitney nonparametric U test. cyt., cytokine; n.s., not significant.
IL-15 primes NK cells with improved survival and cytolytic activity following cytokine withdrawal. Primary human NK cells were isolated from fresh peripheral blood mononuclear cells (PBMCs) and activated with IL-2 or IL-15 (both at 18.3 ng/mL) for 48 hours. (A) Cytolytic capacity against NK-sensitive target K562 (effector-to-target [E:T] ratio = 5:1) and proliferation of resting or cytokine activated human NK cells were measured by chromium release assay and thymidine incorporation assay, respectively. Following cytokine activation for 48 hours, NK cells were cultivated without cytokines (cytokine withdrawal) for an additional 24 hours and tested for their (B) cytolytic activity against K562 cells or (C) viability. Flow cytometry analysis of NK cells following cytokine activation including (D) frequencies of CD25+ cells, (E) expression of membrane-bound cytokines, and cytokine receptor complexes; intracellular expression of Bcl-2; and phosphorylation of STAT-3 (Y705) and STAT-5 (Y694). Results from multiple donors (n > 5) were summarized and are presented as mean ± SD. *P < .05; ***P < .001; Mann-Whitney nonparametric U test. cyt., cytokine; n.s., not significant.
Critically, after cytokine withdrawal, the expression levels of membrane-bound cytokines, cytokine receptors, and Bcl-2 were higher in IL-15–activated NK cells compared with IL-2–activated NK cells. In contrast, similar expression of IL-15 receptor α (IL-15Rα), phosphorylated STAT-3 (pSTAT-3), and pSTAT-5 were observed between IL-2– and IL-15–treated NK cells after cytokine withdrawal (supplemental Figure 1B). Notably, the frequencies of CD56bright/CD16neg NK cells were comparable after IL-2 or IL-15 activation. However, after cytokine withdrawal, the percentages of CD56bright NK cells increased more in IL-15– than IL-2–activated NK cells (supplemental Figure 1C). Thus, IL-15 induces NK-cell characteristics consistent with improved post-cytokine-withdrawal activity.
IL-15 and IL-2 differentially modulate steady-state mRNA levels and translational efficiencies in NK cells after cytokine withdrawal
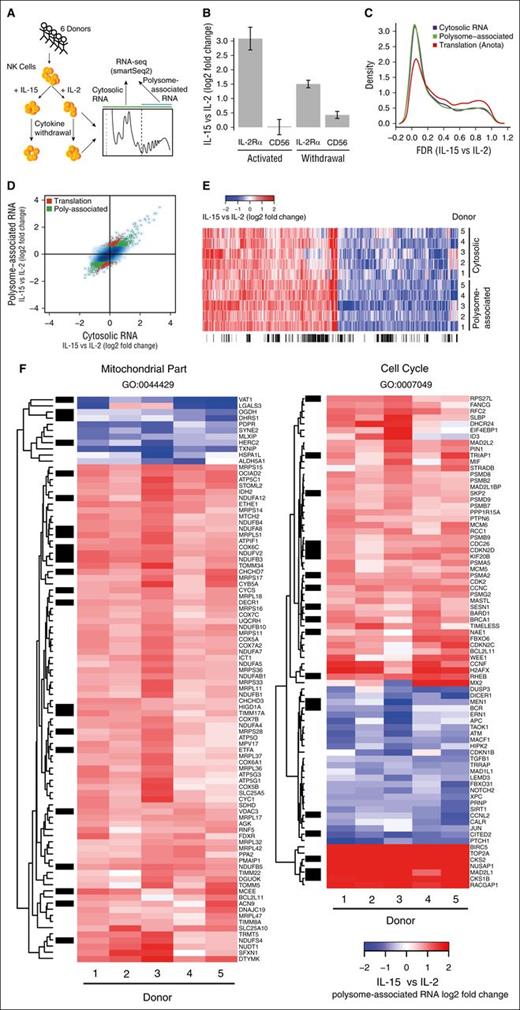
To explore why stimulation with IL-15 results in superior function of NK cells after cytokine withdrawal compared with IL-2, we assessed steady-state cytosolic mRNA levels (commonly referred to as the transcriptome although both transcription and RNA degradation modulate such mRNA levels) and the translatome (by measuring polysome-associated mRNA levels, ie, mRNAs associated with >2 ribosomes) in NK cells from 6 donors after cytokine activation and following their withdrawal using RNAseq,12 resulting in 5 complete replicates (Figure 2A). Because polysome-associated mRNA levels reflect changes in translational efficiency and steady-state mRNA levels, we used anota analysis,25 which adjusts changes in polysome-associated mRNA for those observed in steady-state mRNA to identify genes that are regulated via differential translational efficiency (hereafter “differential translation”). The data set (supplemental Table 3) showed good treatment effects as indicated by principal components analysis (supplemental Figure 2A-B) and, consistent with FACS analysis, both polysome-associated and cytosolic IL-2Rα mRNA levels were increased in IL-15–treated cells as compared with IL-2–treated cells before and after cytokine withdrawal whereas expression of CD56 remained largely unchanged (Figure 2B; supplemental Figure 2C). As differences in cytolytic ability were most notable after cytokine withdrawal, false discovery rates (FDRs) were calculated for differential expression between IL-15– and IL-2–treated NK cells under this condition using data from polysome-associated or cytosolic mRNA, or differential translation using anota analysis. Substantial congruent changes in polysome-associated and cytosolic mRNA was observed and a gene subset was regulated by differential translation (Figure 2C-D).
Abundant differential cytosolic mRNA levels and differential translation between NK cells activated with IL-15 or IL-2 following cytokine withdrawal. (A) Overview of gene expression studies. Isolation of polysome-associated mRNA (ie, associated with >2 ribosomes) and cytosolic mRNA from 6 donors and 4 conditions followed by generation of smartSeq2 RNAseq libraries. (B) Relative (IL-15 vs IL-2) polysome-associated mRNA levels for IL-2Rα and CD56 after 48 hours of cytokine activation and following withdrawal for an additional 24 hours. (C) Densities of genome-wide FDRs comparing IL-15– to IL-2–activated cells post-cytokine withdrawal using data from cytosolic or polysome-associated mRNA and translational efficiency as identified by anota. (D) Genome-wide log2 fold changes (IL-15 vs IL-2) following cytokine withdrawal using data from cytosolic or polysome-associated mRNA. mRNAs with differential polysome-association (green) or differential translational efficiency (red; by anota analysis) are indicated. (E) Heatmap showing log2 fold changes using data from cytosolic or polysome-associated mRNA for mRNAs showing differential polysome association between NK cells activated with IL-15 vs IL-2 following cytokine withdrawal. (F) Heatmap showing log2 fold changes using data from polysome-associated mRNA for differentially expressed genes belonging to the indicated gene ontology (GO) processes. (E-F) The sidebar indicates differentially translated genes identified by anota analysis (black).
Abundant differential cytosolic mRNA levels and differential translation between NK cells activated with IL-15 or IL-2 following cytokine withdrawal. (A) Overview of gene expression studies. Isolation of polysome-associated mRNA (ie, associated with >2 ribosomes) and cytosolic mRNA from 6 donors and 4 conditions followed by generation of smartSeq2 RNAseq libraries. (B) Relative (IL-15 vs IL-2) polysome-associated mRNA levels for IL-2Rα and CD56 after 48 hours of cytokine activation and following withdrawal for an additional 24 hours. (C) Densities of genome-wide FDRs comparing IL-15– to IL-2–activated cells post-cytokine withdrawal using data from cytosolic or polysome-associated mRNA and translational efficiency as identified by anota. (D) Genome-wide log2 fold changes (IL-15 vs IL-2) following cytokine withdrawal using data from cytosolic or polysome-associated mRNA. mRNAs with differential polysome-association (green) or differential translational efficiency (red; by anota analysis) are indicated. (E) Heatmap showing log2 fold changes using data from cytosolic or polysome-associated mRNA for mRNAs showing differential polysome association between NK cells activated with IL-15 vs IL-2 following cytokine withdrawal. (F) Heatmap showing log2 fold changes using data from polysome-associated mRNA for differentially expressed genes belonging to the indicated gene ontology (GO) processes. (E-F) The sidebar indicates differentially translated genes identified by anota analysis (black).
IL-15 modulates expression of genes related to the cell cycle and mitochondrial function
Because polysome-associated mRNA levels reflect changes in both cytosolic mRNA levels and differential translation, subsequent analysis was focused on data from polysome-associated mRNA while monitoring which changes were mediated via differential translation as determined by anota analysis. One thousand two hundred twelve mRNAs showed significantly different polysome-association (FDR <0.15 and fold change >1.5) with minor donor heterogeneity (Figure 2E) in IL-15– vs IL-2–treated NK cells after cytokine withdrawal (639 upregulated genes and 573 downregulated genes) out of which 350 genes (29%) were differentially translated. Gene set enrichment analysis revealed selective upregulation of genes related to mitochondrial functions (eg, electron transport chain and cellular respiration) and cell cycle (supplemental Figure 3; Figure 2F). Mitochondria-related genes were largely upregulated by IL-15 with only a few exceptions including negative regulators of metabolism such as TXNIP. In contrast, genes involved in cell cycle regulation were upregulated or downregulated. Nevertheless, several promotors for cell survival and proliferation including BIRC5 (Survivin), TOP2A, CKS2, and RACGAP1 were upregulated by IL-15 whereas antiproliferative or proapoptotic proteins such as TGFB1, ATM, and PTCH1 were downregulated. Thus, gene expression signatures are consistent with improved functional activity of NK cells after cytokine withdrawal.
IL-15 orchestrates gene expression programs in NK cells via priming
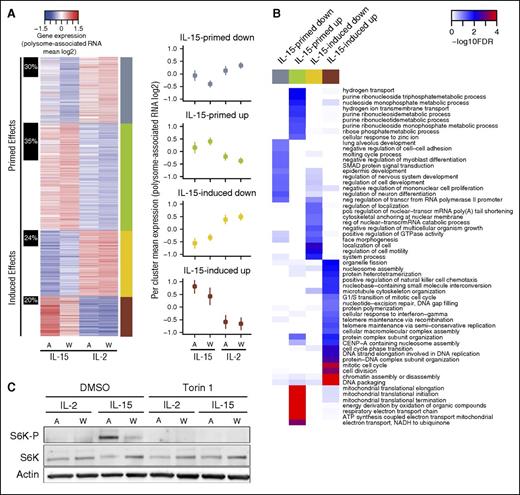
The observed differences in gene expression after cytokine withdrawal between cells activated with IL-15 or IL-2 can be the result of “cytokine-induced” (ie, altered during cytokine activation and maintained after cytokine withdrawal) or “cytokine-primed” regulation (ie, emerging following the stress-response associated with cytokine withdrawal). Indeed, clustering of identified differentially expressed genes using both pre- and post-cytokine-withdrawal data identified 4 distinct groups (Figure 3A): cytokine-primed upregulated (IL-15 vs IL-2) genes (n = 466), cytokine-primed downregulated genes (n = 286), cytokine-induced upregulated genes (n = 173), and cytokine-induced downregulated genes (n = 287). Thus, surprisingly, IL-15 primarily regulates gene expression in NK cells via cytokine priming (62% of all genes). Intriguingly, cytokine-primed genes were preferentially regulated by differential translation as compared with cytokine-induced genes (1.7-fold, Fisher exact test P = .0002). Moreover, each regulatory mode targeted a specific set of functions: cytokine-primed upregulated genes were enriched for functions related to respiration, metabolic processes, and translation; cytokine-primed downregulated genes were enriched for transcription, cell signaling, and developmental processes; cytokine-induced upregulated genes were enriched for cell cycle functions; and cytokine-induced downregulated genes were enriched for processes including cell motility, development, and cell signaling (Figure 3B). Thus, IL-15 orchestrates gene expression programs via cytokine-induced and cytokine-primed modes of regulation thereby targeting distinct cellular functions consistent with improved NK-cell activity.
IL-15 primes a stress-induced gene expression program in NK cells. (A) A heatmap of mean polysome-association (log2) per condition (A = activated, W = cytokine withdrawal) for mRNAs showing differential (IL-15 vs IL-2) polysome association after cytokine withdrawal. Clustering was performed using data from this set of mRNAs across all 4 conditions. The right sidebar indicates the identified gene subsets; the left sidebar shows proportions of genes regulated by differential translation (anota) per subset. Clusters corresponding to induced or primed modes of regulation are indicated. Across genes mean ± SD per condition and cluster is also plotted. (B) A heatmap illustrating results from enrichment analysis within subsets identified in panel A for cellular processes defined by GO. The color scale indicates significance for enrichment for GO terms identified as enriched in at least 1 subset (FDR <0.1 and odds ratio >1.5). (C) Freshly isolated human primary NK cells were activated (A) with IL-2 or IL-15 (18.3 ng/mL) for 48 hours with or without torin-1 (1 µM) and subsequently deprived of cytokines for 24 hours (W). Phosphorylation of the mTOR target S6 kinase was determined by western blot and compared with total S6K. Actin served as a loading control.
IL-15 primes a stress-induced gene expression program in NK cells. (A) A heatmap of mean polysome-association (log2) per condition (A = activated, W = cytokine withdrawal) for mRNAs showing differential (IL-15 vs IL-2) polysome association after cytokine withdrawal. Clustering was performed using data from this set of mRNAs across all 4 conditions. The right sidebar indicates the identified gene subsets; the left sidebar shows proportions of genes regulated by differential translation (anota) per subset. Clusters corresponding to induced or primed modes of regulation are indicated. Across genes mean ± SD per condition and cluster is also plotted. (B) A heatmap illustrating results from enrichment analysis within subsets identified in panel A for cellular processes defined by GO. The color scale indicates significance for enrichment for GO terms identified as enriched in at least 1 subset (FDR <0.1 and odds ratio >1.5). (C) Freshly isolated human primary NK cells were activated (A) with IL-2 or IL-15 (18.3 ng/mL) for 48 hours with or without torin-1 (1 µM) and subsequently deprived of cytokines for 24 hours (W). Phosphorylation of the mTOR target S6 kinase was determined by western blot and compared with total S6K. Actin served as a loading control.
mTOR mediates improved metabolic and cytotoxic functions following cytokine withdrawal in IL-15–activated human NK cells
Our finding that cytokine-primed upregulated genes are enriched for regulation via differential translation and mitochondria-related functions parallels recent data on translational control of mitochondrial function via selective translation of mRNAs encoding mitochondria-related proteins downstream of the mTOR pathway.25,26 Thus, it is plausible that differential activation of mTOR signaling by IL-2 or IL-15 could, at least partly, explain observed gene expression differences and associated cellular phenotypes.25,27 Consistently, IL-15 induced increased phosphorylation of the mTOR substrate S6K, as compared with IL-2 and IL-15, associated S6K phosphorylation was maintained, albeit at a lower level, after 24 hours of cytokine withdrawal (Figure 3C). We therefore investigated the impact of mTOR activity on NK-cell function using the selective active site mTOR inhibitor torin-1 during in vitro cytokine activation of primary human NK cells.
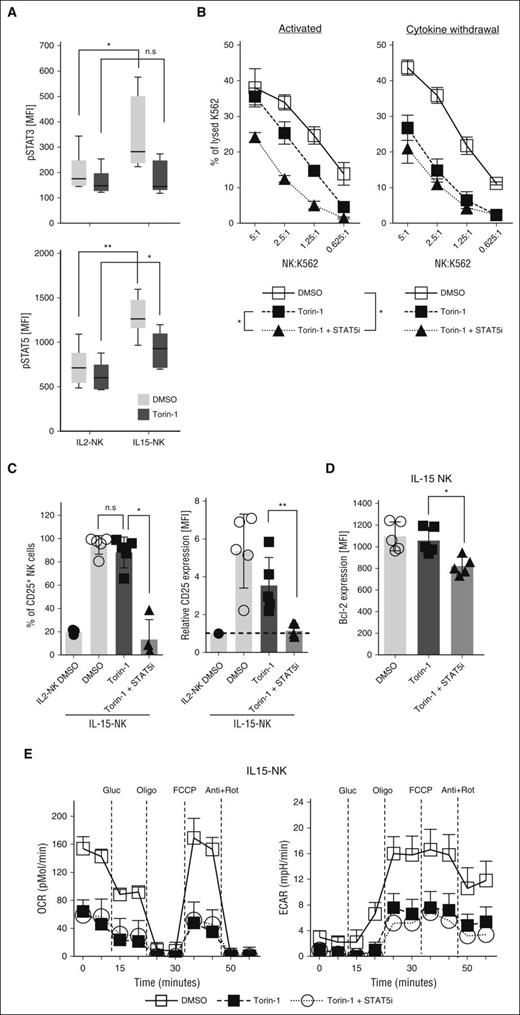
NK cells treated with IL-15 showed a higher basal and maximal cellular respiratory function compared with those treated with IL-2 (supplemental Figure 4A). Torin-1 treatment eliminated IL-15–associated S6K phosphorylation pre- and post-cytokine withdrawal (Figure 3C) and reduced respiratory activity induced by IL-15 (P < .05; Figure 4A; supplemental Figure 4B). Moreover, torin-1 reduced the expression intensity (but not frequency) of CD25 (P < .05; Figure 4B) and expression of membrane-bound IL-2 (P < .01) and IL-15Rα (P < .001; Figure 4C) in IL-15–treated NK cells. In contrast, torin-1 did not affect IL-15–associated expression of membrane-bound IL-15 or expression of Bcl-2 or IL-2/15Rβ (Figure 4C; supplemental Figure 4C). Torin-1 also reduced the cytolytic activity by NK cells activated with IL-2 (P < .001; Figure 4D) or IL-15, but for the latter only at low effector-to-target ratios. Critically, torin-1 treatment reversed the improved postwithdrawal cytolytic activity of IL-15–activated, but not IL-2–activated, NK cells at all effector-to-target ratios tested to a level observed in IL-2–activated NK cells post-cytokine withdrawal (P < .01; Figure 4D). Thus, augmented mTOR activity is essential for improved cytotoxic and metabolic activities of NK cells treated with IL-15 following cytokine withdrawal.
mTOR contributes to IL-15–associated protective and metabolic benefits in human NK cells. Freshly isolated primary human NK cells were activated with IL-2 or IL-15 for 48 hours in the presence of DMSO or the selective mTOR inhibitor torin-1 (1 μM). (A) The real-time OCR (picomoles per minute) and glycolysis rate (ECAR; milli-pH per minute) were measured using the Seahorse platform. (B) The impact of torin-1 on frequency and expression intensity of CD25 was measured on NK cells by FACS. (C) Expression levels of membrane-bound cytokines or IL-15Rα chain as determined by FACS. (D) Lysis of K562 cells mediated by IL-15–activated NK cells, in the presence of torin-1 (filled symbols), DMSO (open symbols) after direct activation or following cytokine withdrawal. Shaded symbols show lysis of K562 cells by IL-2–activated NK cells after cytokine withdrawal in the presence of DMSO. Results from multiple donors (n > 4) were summarized and presented as mean ± SD. Representative histograms were chosen based on proximity to average values. *P < .05; **P < .01; ***P < .001; Mann-Whitney nonparametric U test. FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; Gluc, glucose.
mTOR contributes to IL-15–associated protective and metabolic benefits in human NK cells. Freshly isolated primary human NK cells were activated with IL-2 or IL-15 for 48 hours in the presence of DMSO or the selective mTOR inhibitor torin-1 (1 μM). (A) The real-time OCR (picomoles per minute) and glycolysis rate (ECAR; milli-pH per minute) were measured using the Seahorse platform. (B) The impact of torin-1 on frequency and expression intensity of CD25 was measured on NK cells by FACS. (C) Expression levels of membrane-bound cytokines or IL-15Rα chain as determined by FACS. (D) Lysis of K562 cells mediated by IL-15–activated NK cells, in the presence of torin-1 (filled symbols), DMSO (open symbols) after direct activation or following cytokine withdrawal. Shaded symbols show lysis of K562 cells by IL-2–activated NK cells after cytokine withdrawal in the presence of DMSO. Results from multiple donors (n > 4) were summarized and presented as mean ± SD. Representative histograms were chosen based on proximity to average values. *P < .05; **P < .01; ***P < .001; Mann-Whitney nonparametric U test. FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; Gluc, glucose.
STAT-5 mediates mTOR-independent NK-cell phenotypes during IL-15 activation but not following withdrawal
As mTOR inhibition did not completely reduce the cytolytic ability of IL-15–activated NK cells in the presence of cytokines, additional pathways are likely involved. In contrast to reduced STAT-3 phosphorylation, STAT-5 phosphorylation was higher in IL-15–activated NK cells as compared with those activated by IL-2 also in the presence of torin-1 (P < .05; Figure 5A). As inhibition of STAT-5 or mTOR reduced STAT-3 phosphorylation (supplemental Figure 5A), we compared STAT-5 plus mTOR inhibition to mTOR inhibition alone to study the role of STAT-5 signaling (supplemental Figure 5B). Indeed, this treatment reduced the cytolytic functions of IL-15–activated NK cells at all effector-to-target ratios (P < .05; Figure 5B). Notably, concurrent inhibition of STAT-5 and mTOR did not influence the cytolytic activity of NK cells after cytokine withdrawal in comparison with torin-1 alone, indicating that IL-15–primed prolonged NK-cell activation is mTOR dependent but STAT-5 independent (Figure 5B). Moreover, STAT-5 and mTOR inhibition reduced expression of CD25 and intracellular Bcl-2 (P < .05; Figure 5C-D) but enhanced expression of common IL-2/15Rβ and membrane-bound IL-2 on IL-15–activated NK cells as compared with mTOR inhibition alone (P < .01; supplemental Figure 5C). Consistent with the role of mTOR in maintaining metabolic processes, mitochondrial and glycolytic properties of activated NK cells were not modulated by additional STAT-5 inhibition (Figure 5E). Thus, although STAT-5 affects some IL-15–associated NK-cell phenotypes, cytotoxic and metabolic improvements after cytokine withdrawal mainly depend on mTOR and not STAT-5 signaling.
STAT-5 mediates mTOR-independent signaling upon IL-15 activation. (A) Phosphorylation of STAT-3 (n = 8) or STAT-5 (n = 7) were compared between NK cells activated with IL-2 or IL-15 in the presence or absence of torin-1. Alternatively, a selective STAT-5 inhibitor (400 μM) was added in combination with torin-1 during IL-15 activation of NK cells and (B) K562 lysis was assessed after 48 hours of activation or following an additional 24 hours of cytokine withdrawal. (C) Frequencies of CD25+ cells and expression intensities of CD25 on IL-15–activated NK cells. (D) Expression levels of intracellular Bcl-2 and (E) oxygen consumption and glycolytic potential were measured in NK cells cultured under described conditions. Results from multiple donors (n > 5) were summarized and presented as mean ± SD. Representative histograms were chosen based on proximity to average values. *P < .05; **P < .01; Mann-Whitney nonparametric U test. MFI, mean fluorescence intensity.
STAT-5 mediates mTOR-independent signaling upon IL-15 activation. (A) Phosphorylation of STAT-3 (n = 8) or STAT-5 (n = 7) were compared between NK cells activated with IL-2 or IL-15 in the presence or absence of torin-1. Alternatively, a selective STAT-5 inhibitor (400 μM) was added in combination with torin-1 during IL-15 activation of NK cells and (B) K562 lysis was assessed after 48 hours of activation or following an additional 24 hours of cytokine withdrawal. (C) Frequencies of CD25+ cells and expression intensities of CD25 on IL-15–activated NK cells. (D) Expression levels of intracellular Bcl-2 and (E) oxygen consumption and glycolytic potential were measured in NK cells cultured under described conditions. Results from multiple donors (n > 5) were summarized and presented as mean ± SD. Representative histograms were chosen based on proximity to average values. *P < .05; **P < .01; Mann-Whitney nonparametric U test. MFI, mean fluorescence intensity.
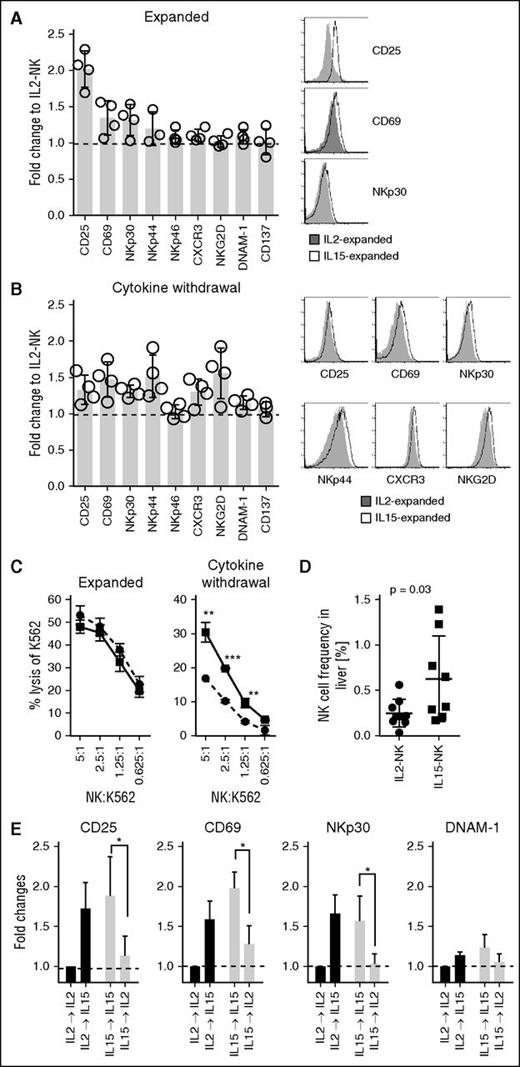
NK cells expanded with IL-15 are resistant to cytokine withdrawal
Next, we sought to investigate whether NK cells expanded in the presence of IL-15 using a clinically approved protocol would also display improved functions compared with those expanded with IL-2. Although no difference in NK-cell yield was observed (supplemental Figure 6A), expansion with IL-15 resulted in higher expression of CD25 on NK cells as compared with IL-2 (Figure 6A). After cytokine withdrawal, the expression of CD25 and several other activating receptors including CD69, NKp30, NKp44, CXCR3, and NKG2D remained elevated on IL-15–expanded NK cells compared with IL-2–expanded NK cells (Figure 6B). Although the release of IFN-γ was comparable between IL-2– and IL-15–expanded NK cells (supplemental Figure 6B), NK cells expanded with IL-15 showed higher lytic ability after cytokine withdrawal (Figure 6C). To investigate whether NK cells expanded with IL-15 would have improved persistence in vivo, NK cells were injected intraperitoneally into NSG mice. Indeed, the frequency of IL-15–expanded human NK cells was higher in livers compared with IL-2–expanded NK cells 4 to 5 days postinfusion (Figure 6D). Although no statistically significant differences (P = .27) were observed in lungs, the median frequency of NK cells was 0.16% and 0.05% for mice injected with IL-15–NK and IL-2–NK, respectively. No differences in NK-cell frequencies between mice injected with IL-15–expanded and IL-2–expanded NK cells were observed in the spleens (supplemental Figure 7B).
NK cells expanded with IL-15 are resistant to cytokine withdrawal. For expansion, 0.5 × 106 to 2 × 106 purified NK cells were cultured at a 1:10 ratio with irradiated EBV-transformed B cells in X-VIVO 20 medium supplemented with 10% heat-inactivated human AB serum, in the presence of recombinant human IL-2 (1000 IU/mL; Proleukin) or IL-15 (61 ng/mL). Fresh media supplemented with AB serum and cytokines (500 IU/mL for IL-2 or 30.5 ng/mL for IL-15) was added on day 5 and thereafter every 3 days and cells were harvested between 11 and 14 days. Expression levels of various activation markers on NK cells (A) freshly after expansion or (B) following 48 hours of cytokine withdrawal were measured by FACS. (C) Comparison of cytolytic capacity of IL2-NK (circles, dashed line) and IL15-NK (squares, solid line) cells against K562 target cells after expansion or cytokine withdrawal (48 hours). (D) NSG mice were injected (intraperitoneally) with 1 × 106 to 5 × 106 DiR-labeled IL-2– or IL-15–expanded NK cells. The liver was resected 4 to 5 days after injection of NK cells. Single-cell suspension of the liver was stained with a live-dead fixable aqua dead cell stain and anti-CD56 and thereafter acquired by flow cytometry. The frequency of NK cells calculated is based on viable CD56+/DiR+ cells. Each symbol represents 1 mouse injected with IL2-NK (n = 9) or IL15-NK (n = 8). Error bars show mean and SD and P value is calculated by a t test. (E) NK cells expanded with IL-2 or IL-15 were cultured in the same or alternative cytokines for 24 hours and expression intensities of various activating markers including CD25, CD69, NKp30, and DNAM-1 were measured by FACS. Results from 4 independent expansions were summarized and presented as mean ± SD. Representative histograms were chosen based on proximity to average values. *P < .05; **P < .01; ***P < .001; Mann-Whitney nonparametric U test.
NK cells expanded with IL-15 are resistant to cytokine withdrawal. For expansion, 0.5 × 106 to 2 × 106 purified NK cells were cultured at a 1:10 ratio with irradiated EBV-transformed B cells in X-VIVO 20 medium supplemented with 10% heat-inactivated human AB serum, in the presence of recombinant human IL-2 (1000 IU/mL; Proleukin) or IL-15 (61 ng/mL). Fresh media supplemented with AB serum and cytokines (500 IU/mL for IL-2 or 30.5 ng/mL for IL-15) was added on day 5 and thereafter every 3 days and cells were harvested between 11 and 14 days. Expression levels of various activation markers on NK cells (A) freshly after expansion or (B) following 48 hours of cytokine withdrawal were measured by FACS. (C) Comparison of cytolytic capacity of IL2-NK (circles, dashed line) and IL15-NK (squares, solid line) cells against K562 target cells after expansion or cytokine withdrawal (48 hours). (D) NSG mice were injected (intraperitoneally) with 1 × 106 to 5 × 106 DiR-labeled IL-2– or IL-15–expanded NK cells. The liver was resected 4 to 5 days after injection of NK cells. Single-cell suspension of the liver was stained with a live-dead fixable aqua dead cell stain and anti-CD56 and thereafter acquired by flow cytometry. The frequency of NK cells calculated is based on viable CD56+/DiR+ cells. Each symbol represents 1 mouse injected with IL2-NK (n = 9) or IL15-NK (n = 8). Error bars show mean and SD and P value is calculated by a t test. (E) NK cells expanded with IL-2 or IL-15 were cultured in the same or alternative cytokines for 24 hours and expression intensities of various activating markers including CD25, CD69, NKp30, and DNAM-1 were measured by FACS. Results from 4 independent expansions were summarized and presented as mean ± SD. Representative histograms were chosen based on proximity to average values. *P < .05; **P < .01; ***P < .001; Mann-Whitney nonparametric U test.
After adoptive NK-cell therapy, patients are often treated with recombinant IL-2 to sustain NK cells in vivo. To mimic this, the effect of intermediate cytokine dosage on NK cells expanded in different conditions was investigated. Short-term exposure of IL-15 substantially improved activation of IL-2–expanded NK cells, as demonstrated by enhanced expression of various surface markers (Figure 6E; supplemental Figure 6C). In contrast, the same dose of IL-2 failed to sustain activation of expanded NK cells in either condition (Figure 6E; supplemental Figure 6C). This effect was not mediated through IFN-γ (supplemental Figure 6D), but relied on STAT-5 signaling (supplemental Figure 6E).
The expression of IL-15 is associated with improved clinical outcome in patients with B-cell lymphoma
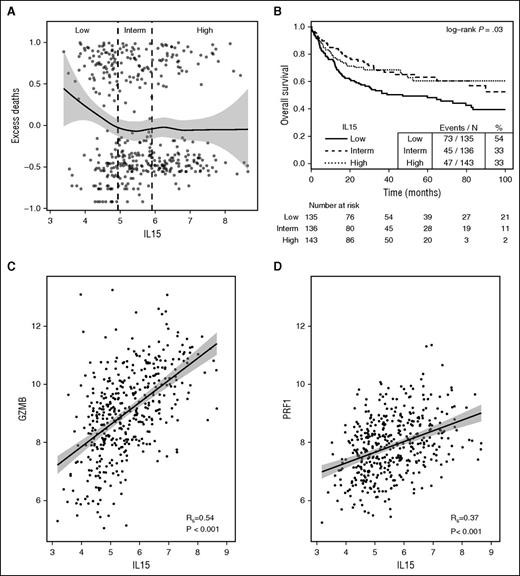
To evaluate the prognostic potential of IL-15 expression, a publicly available data set assessing steady-state mRNA levels in tissue samples from patients with B-cell lymphoma was reanalyzed.14 The effect of IL-15 expression on overall survival was explored by plotting residuals of an empty Cox model (interpreted as excess deaths23 ) against IL-15 expression. This revealed that low expression of IL-15 is associated with worse outcome (Figure 7A). This was also observed when comparing survival of 3 patient subgroups based on IL-15 expression (Figure 7B). Notably, IL-15 mRNA levels correlated with the expression of cytotoxic proteins granzyme B (R = 0.54; P < .001) and perforin 1 (R = 0.37; P < .001) (Figure 7C-D). The corresponding analysis for IL-2 could not be performed as the DNA-microarray signals for IL-2 did not exceed background levels.
IL-15 gene expression is associated with improved clinical outcome in patients with B-cell lymphomas. (A) Scatter plot of excess deaths (martingale residuals of the survival outcome in an empty Cox model) vs IL-15 gene expression (log2); tertile categories are separated by vertical dashed lines: low (<4.9), intermediate (4.9-5.9), high (>5.9). A loess curve with 95% confidence bands (gray) is indicated. (B) Kaplan-Meier plot for overall survival of patients categorized according to tertiles of IL-15 expression. (C) Correlation between IL-15 and Granzyme B (GZMB) or (D) Perforin 1 (PRF1) mRNA levels visualized using scatter plots and fitted linear model estimates with 95% confidence bands. Rs, Spearman rank correlation coefficient.
IL-15 gene expression is associated with improved clinical outcome in patients with B-cell lymphomas. (A) Scatter plot of excess deaths (martingale residuals of the survival outcome in an empty Cox model) vs IL-15 gene expression (log2); tertile categories are separated by vertical dashed lines: low (<4.9), intermediate (4.9-5.9), high (>5.9). A loess curve with 95% confidence bands (gray) is indicated. (B) Kaplan-Meier plot for overall survival of patients categorized according to tertiles of IL-15 expression. (C) Correlation between IL-15 and Granzyme B (GZMB) or (D) Perforin 1 (PRF1) mRNA levels visualized using scatter plots and fitted linear model estimates with 95% confidence bands. Rs, Spearman rank correlation coefficient.
Discussion
Persistence of transferred NK cells, commonly activated with IL-2, is associated with clinical responses in patients with hematological malignancies.1,28 Various cytokines including IL-7, IL-15, IL-12, IL-18, and IL-21 have been assessed for improving NK-cell activity.29 IL-15 has been of particular interests as it is essential for NK-cell development and mice lacking IL-15,9 IL-15Rα,30 IL-2Rβ,31 or γc32 subunits showed reduced NK-cell frequency. In contrast, NK cells in IL-2–deficient mice remained functional.32,33 Nevertheless, several studies indicate that IL-15 does not provide NK cells with proliferative or cytolytic advantages compared with IL-2.33,34 We confirm that short-term IL-2– or IL-15–activated or expanded NK cells show comparable activities. IL-15 has been shown to augment intracellular signaling and recruit antiapoptotic proteins, which could support functional durability of NK cells.35,36 This agrees with studies showing that IL-15–activated human NK cells are functionally resistant to steroid inhibition37 and sustain their superior in vivo proliferation.1,35,38
In T cells, IL-15 promotes survival via upregulating Bcl-239,40 and antioxidant proteins41,42 and limiting proapoptotic caspase-3.43 In addition, we show that the expression of activating NK-cell receptors is upregulated on IL-2–expanded NK cells after short-term incubation with IL-15, possibly contributing to the result that, upon injection into immunodeficient mice, the frequency of IL-15–expanded NK cells in the liver was higher as compared with IL-2–expanded NK cells. Thus, our findings support implementation of IL-15 into current adoptive cellular therapy protocols, either during the expansion phase or infused at low doses after NK-cell transfer.
To date, a few studies have explored in vivo administration of IL-15. The study by Conlon et al concluded that IV infusion of 0.3 μg/kg per day for 12 consecutive days was the maximum-tolerated dose and resulted in a 10-fold increase in NK-cell numbers.10 Similarly, subcutaneous or IV intermittent administration of IL-15 was shown to be safe and to expand NK cells in nonhuman primates.44,45 Thus, these reports and our data showing that IL-15 can reactivate NK cells after cytokine withdrawal suggest the use of low-dose intermittent treatment with IL-15.
Because of potential organ-detrimental effects of inappropriately activated immune cells, it is likely essential that gene expression circuits in immune cells are governed by several regulatory levels including transcription, mRNA stability, and mRNA translation.12,13 This also implies that for a more complete understanding of immune cell functions each of these regulatory levels needs to be assessed. Although measurement of steady-state mRNA levels (ie, transcriptomes) estimates the combined regulatory impact from transcription and mRNA stability, such studies will not capture regulation occurring via differential translation. Studies of translatomes are therefore powerful tools to enable a more complete understanding of how immune cell functions are controlled. Studying the translatome is likely particularly important when phenotypes involving modulated mTOR signaling are included as mTOR primarily regulates gene expression via translational control.46 This is consistent with the result that ∼30% of the observed differences in gene expression between IL-2– and IL-15–activated NK cells, which leads to differential mTOR activation, were mediated via differential translation but also underlines that other mechanisms acting at the level of transcription or mRNA stability largely shape the proteome in IL-15–activated NK cells. The time point we selected following cytokine withdrawal is relatively late (24 hours) whereas most studies of the effects of mTOR on gene expression are performed at earlier time points (2-12 hours) and the proportion of genes regulated via different mechanisms is likely to change over time.
It was recently demonstrated that activation with IL-15 or IL-2 resulted in similar steady-state mRNA profiles in murine CD8+ T cells.47 In contrast, we show striking steady-state mRNA differences and differential translation between IL-15– and IL-2–activated human NK cells. Intriguingly, by comparing cells during activation and after cytokine withdrawal, we discovered 2 modes for how IL-15 orchestrates gene expression programs. While IL-15 directly induces expression changes in a distinct set of genes, it also primes NK cells to respond differently to cytokine withdrawal as compared with IL-2. Although both modes of regulation (ie, induced and primed) involve alterations in steady-state mRNA levels and translation, priming is to a larger extent mediated via translation. This is consistent with posttranscriptional programs being essential for efficient adaptation to changes in the environment, such as stress. Moreover, induced and primed modes of regulation coordinate distinct cellular functions where priming appears important for superior postwithdrawal capabilities by stimulating selective expression of, for example, mitochondria-related genes.48
One of the key pathways that targets mRNA translation and regulates cell proliferation and metabolism is the mTOR pathway.25,46,49,50 Although the precise mechanisms remain elusive,51 recent findings provide evidence that IL-15 activates multifaceted metabolic activities during maturation and survival of NK cells via mTOR.27,51 Indeed, we found that selective inhibition of mTOR decreases recruitment of survival signals and abolishes metabolic and functional advantages of IL-15–activated NK cells. These results suggest that NK-cell metabolic activity associates with antitumor immunity.48 Consistently, Keppel et al recently demonstrated that IL-15 potentiates glycolytic functions on human NK cells which are essential for their effector functions.52 Furthermore, Chang et al showed that tumor-infiltrating lymphocytes that regained glycolytic potential after checkpoint blockade therapy are more efficient in eliminating tumor cells.53 In this context, it would be of interest to monitor NK-cell activity in patients treated with mTOR inhibitors, such as rapalogs. Although such data are scarce, Sarkaria et al reported, in agreement with herein presented data, robust suppression of NK, T, and B cells in patients with glioblastoma multiforme after treatment with a combination of temsirolimus with chemoradiation.54 Taken together, high glycolytic capacity appears to enable NK cells to sustain their in vivo performance.
IL-2 and IL-15 are potent in eliciting signaling transduction through STAT-3 and STAT-5.4,5 mTOR has been reported to phosphorylate STAT-3 at S727, which increases STAT-3 transcriptional activity,55,56 and, under some conditions of augmented mTOR signaling, also Y70557 which was assessed in this study. We identify enhanced phosphorylation of STAT-3 and -5 in human NK cells by IL-15 prior to cytokine withdrawal and, consistent with the ability of mTOR to phosphorylate STAT-3 at Y705, mTOR inhibition reduced STAT-3 Y705 but not STAT-5 phosphorylation. Indeed, mTOR-independent STAT-5 activation was necessary for some NK-cell functions during cytokine activation but not following cytokine withdrawal. Although combined inhibition of STAT-5 and mTOR abolishes some favorable phenotypes, no additional impact was observed on metabolic or cytotoxic activities.
In conclusion, this study adds to our understandings about establishment and maintenance of tumor-reactive NK cells and support clinical implementation of IL-15 for adoptive NK-cell therapy. More broadly, our studies suggest that a large aspect of cytokine-mediated gene expression programs and downstream cellular functions, including antitumor capacity, are overlooked if postactivation conditions are omitted. This is likely not limited to NK cells and should hence be considered in similar studies of immune cells.
The data reported in this article have been deposited at the Gene Expression Omnibus database (accession number GSE77808).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff at the research facility at the Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet. The authors also thank Axel Liljencrantz, Department of Oncology-Pathology Karolinska Institutet, for assistance with cell culture.
This work was supported by the Swedish Cancer Society (#CAN 2012/474 and #CAN 2015/421), the Swedish Childhood Cancer Foundation (#PR2014-0093), the Swedish Foundation for International Cooperation in Research and Higher Education (#IB2014-5690), the Cancer Research Foundations of Radiumhemmet (#141272) (A.L.); and the Swedish Cancer Society (CAN 2013/737), the Swedish Research Council (2013-3055), Wallenberg Academy Fellow Program, and the Strategic Research Programme in Cancer (STRATCAN) (O.L.).
Authorship
Contribution: Y.M. designed and performed research, analyzed data, and wrote the paper; V.v.H. designed and performed research, contributed vital new reagents and analytical tools, analyzed data, and wrote the paper; X.Z. and L.M. performed research and contributed vital new reagents; E.W. designed and performed research; J.L. contributed vital new analytical tools, analyzed data, and wrote the paper; K.W. designed and performed research and analyzed data; S.L. and S.M. performed research; O.L. designed research, contributed vital new reagents and analytical tools, analyzed data, and wrote the paper; R.K. designed research, contributed vital new reagents, and wrote the paper; and A.L. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ola Larsson, Department of Oncology-Pathology, Karolinska Institutet, SciLifeLab, 17165, Solna, Sweden; e-mail: ola.larsson@ki.se; Rolf Kiessling, Department of Oncology-Pathology, Cancer Center Karolinska, R8:01, Karolinska University Hospital, Karolinska Institutet, 17176 Stockholm, Sweden; e-mail: rolf.kiessling@ki.se; and Andreas Lundqvist, Department of Oncology-Pathology, Cancer Center Karolinska, R8:01, Karolinska University Hospital, Karolinska Institutet, 17176 Stockholm, Sweden; e-mail: andreas.lundqvist@ki.se.
References
Author notes
Y.M. and V.v.H. contributed equally to this study.

![Figure 1. IL-15 primes NK cells with improved survival and cytolytic activity following cytokine withdrawal. Primary human NK cells were isolated from fresh peripheral blood mononuclear cells (PBMCs) and activated with IL-2 or IL-15 (both at 18.3 ng/mL) for 48 hours. (A) Cytolytic capacity against NK-sensitive target K562 (effector-to-target [E:T] ratio = 5:1) and proliferation of resting or cytokine activated human NK cells were measured by chromium release assay and thymidine incorporation assay, respectively. Following cytokine activation for 48 hours, NK cells were cultivated without cytokines (cytokine withdrawal) for an additional 24 hours and tested for their (B) cytolytic activity against K562 cells or (C) viability. Flow cytometry analysis of NK cells following cytokine activation including (D) frequencies of CD25+ cells, (E) expression of membrane-bound cytokines, and cytokine receptor complexes; intracellular expression of Bcl-2; and phosphorylation of STAT-3 (Y705) and STAT-5 (Y694). Results from multiple donors (n > 5) were summarized and are presented as mean ± SD. *P < .05; ***P < .001; Mann-Whitney nonparametric U test. cyt., cytokine; n.s., not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/11/10.1182_blood-2016-02-698027/4/m_1475f1.jpeg?Expires=1770252240&Signature=goekdBPipSRe8wr-HAvdq5NEzEUjDgglrOIp4RU1Yf9A-bdOCI5kVT4D1R4BHqG10ycenkhzP3fGtxNf~S12Ir3EHh6jqhRINXKwfxf39kE2qZhSV3spvnb9rQsWrUUYxgJcO4zRe6WIgV6FFaj6j2Bde0BwTOIPVvrI0ar3MJrzdds7sRbh9~S~oSGHy44uhh~M-FB2Z74VrbD7ipzuc6d2iolZrpF0N6RhqTIgJgnc67EkB8nNis23ZUx5xBdWOJCYvdlS3UweAaQ3W5y-A9ySROfCSQ7z~McCFVu7PJtev3g4LiHupZWgQS8CIX2jTDNxWj0G0~kFCmQ-tOWjeQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)