To the editor:
Mutations in genes involved in telomere maintenance are implicated in a broad range of degenerative disorders collectively known as telomere diseases.1,2 Their clinical presentations extend from severe childhood syndromes such as dyskeratosis congenita (DC) to later-onset, phenotypically restricted disorders including aplastic anemia (AA), myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), and idiopathic pulmonary fibrosis.3,4 Identifying cryptic germ line telomere disease in adults presenting with isolated AA/MDS/AML carries important therapeutic implications,5,6 mandating rigorous clinical evaluation and targeted genetic testing. The finding of variants of unknown significance, especially in patients without syndromic features, presents a diagnostic and management conundrum.
Telomerase reverse transcriptase (TERT) and telomerase RNA component (TERC) constitute the core telomerase enzyme that replenishes telomeric DNA repeats (TTAGGG) to maintain cellular self-renewal.7,8 A moderate reduction in TERC levels is sufficient to cause telomere diseases.9-12 Several mutations in the TERC gene that affect transcription, template function, and/or stability of TERC have been defined,13-18 but newly identified, potentially pathogenic variants must be distinguished from polymorphisms that do not impair telomerase. Direct evaluation of the impact of TERC variants on telomerase in patient cells would be ideal, but is confounded by the premature senescence in culture of cells carrying short telomeres19,20 and also because most human somatic cells do not express TERT.21
Here, we characterize a previously undescribed variant in the conserved region 4 (CR4)/CR5 domain of TERC, encoding adenine in place of guanine at position 319, in a family with a high suspicion of telomere disease. By reprogramming patient cells into induced pluripotent stem cells (iPSCs), which have reactivated endogenous TERT expression and can be expanded to limitless quantities, we demonstrate a novel mechanism of telomere disease, namely impaired binding of TERC to TERT in vivo.
Patient research enrollment, genetic evaluation, and experimental methods are described in supplemental Methods (available on the Blood Web site).
The proband was a previously healthy 36-year-old man with an incidental finding of moderate thrombocytopenia during routine evaluation. He had otherwise normal hematologic parameters and physical findings, and a bone marrow examination showed normal cellularity and adequate megakaryocytes. Further evaluation revealed hepatosplenomegaly and portal hypertension, and a liver biopsy showed fibrosis and nodular regenerative changes. He was deemed to have liver disease of unclear etiology and secondary thrombocytopenia. Several months later, the proband’s previously healthy 34-year-old sister presented with pancytopenia and was found to have refractory anemia with excess blasts, type 1 (Figure 1A-B). She was treated with DNA hypomethylating agents but progressed to AML with complex cytogenetics, and after attaining remission with chemotherapy was referred for allogeneic bone marrow transplantation. A detailed history and examination revealed her brother’s liver abnormalities, their father’s death from pulmonary fibrosis, and subtle stigmata of DC in her and extended family members, raising suspicion for telomere disease (Figure 1A). Telomere length testing in her and the proband showed mean lymphocyte telomere length at most in the first percentile for age (Figure 1C). Genetic testing revealed a heterozygous TERC c.319G>A variant of unknown significance (Figure 1D), but no mutations in TERT, DKC1, NHP2, NOP10, TINF2, or TCAB1.
A novel variant in the CR4/5 region of TERC in a family with telomere disease. (A) Pedigree of the family; the proband is indicated with an arrow. Affected individuals are shown in gray and symptoms are indicated. Mutations in the individuals genotyped are shown. IPF, idiopathic pulmonary fibrosis; Skin, pigmentation abnormalities; TP, thrombocytopenia. (B) Bone marrow histology. Top, Hypercellular, fibrotic bone marrow with dyspoiesis (hematoxylin and eosin [H&E]). Bottom, CD34 staining shows increased percentage of CD34+ cells (scale bar, 20 µm). (C) Telomere length measurement by flow cytometry fluorescence in situ hybridization (flow-FISH) in lymphocytes in the proband and his sister. (D) Top, Schematic of telomerase RNP. The location of the variant in the TERC CR4/CR5 region is indicated with an arrow. Bottom, Sequencing of TERC in the proband shows the c.319G>A variant. (E) Telomeric repeat amplification protocol (TRAP) assay for telomerase activity in VA13 cells transfected with TERT and WT TERC, G319A TERC, or control plasmid. Threefold dilutions of input cell extract. Internal control (IC) amplification standard is indicated. Relative telomerase activities are shown in the graph. (F) Top, Immunoblot of TERT and actin protein levels. Bottom, Northern blot of TERC RNA from VA13 cells transfected with TERT plus WT TERC, G319A TERC, or control plasmid. Ethidium bromide staining of 28S ribosomal RNA (rRNA) is used as a loading control.
A novel variant in the CR4/5 region of TERC in a family with telomere disease. (A) Pedigree of the family; the proband is indicated with an arrow. Affected individuals are shown in gray and symptoms are indicated. Mutations in the individuals genotyped are shown. IPF, idiopathic pulmonary fibrosis; Skin, pigmentation abnormalities; TP, thrombocytopenia. (B) Bone marrow histology. Top, Hypercellular, fibrotic bone marrow with dyspoiesis (hematoxylin and eosin [H&E]). Bottom, CD34 staining shows increased percentage of CD34+ cells (scale bar, 20 µm). (C) Telomere length measurement by flow cytometry fluorescence in situ hybridization (flow-FISH) in lymphocytes in the proband and his sister. (D) Top, Schematic of telomerase RNP. The location of the variant in the TERC CR4/CR5 region is indicated with an arrow. Bottom, Sequencing of TERC in the proband shows the c.319G>A variant. (E) Telomeric repeat amplification protocol (TRAP) assay for telomerase activity in VA13 cells transfected with TERT and WT TERC, G319A TERC, or control plasmid. Threefold dilutions of input cell extract. Internal control (IC) amplification standard is indicated. Relative telomerase activities are shown in the graph. (F) Top, Immunoblot of TERT and actin protein levels. Bottom, Northern blot of TERC RNA from VA13 cells transfected with TERT plus WT TERC, G319A TERC, or control plasmid. Ethidium bromide staining of 28S ribosomal RNA (rRNA) is used as a loading control.
Given the implications of diagnosing telomere disease in this family, we undertook a detailed evaluation of the TERC G319A variant. The guanine at this position in the TERC CR4/CR5 domain is highly conserved among vertebrates22 (Figure 1D; supplemental Figure 1), and human genetic variation at this position is not evident in the Exome Aggregation Consortium database, suggesting an important albeit undefined role. We tested the function of TERC G319A in a commonly used cell-based assay.16-18,23 When we ectopically expressed normal or G319A TERC along with TERT in VA13 cells, which are TERT and TERC negative, we found that G319A TERC was capable of reconstituting telomerase activity, but at levels ∼40% lower than normal TERC (Figure 1E). Levels of ectopically expressed TERC and TERT were similar across experimental samples (Figure 1F). We regarded these results as suggestive but inconclusive that the TERC G319A variant is pathological, given the modest decrease in telomerase activity and the unclear mechanism by which it would compromise TERC function.
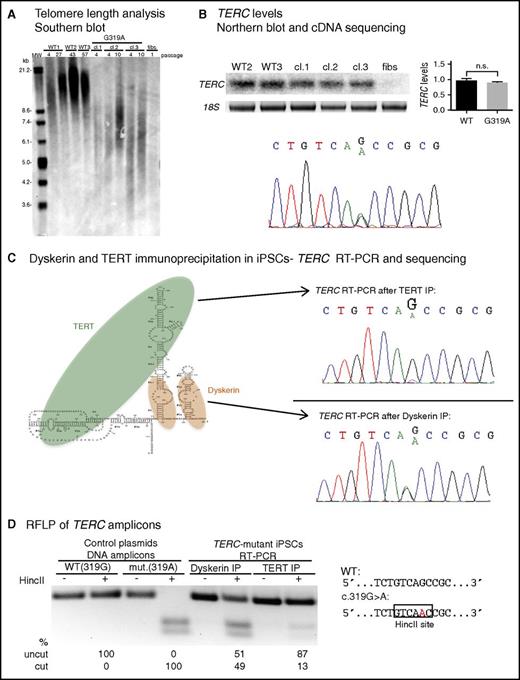
We turned our attention to patient cells by culturing fibroblasts from the proband’s bone marrow. To overcome senescence and limitations of cell numbers, and to study the function of TERC G319A in TERT-expressing cells, we reprogrammed the fibroblasts using a lentiviral vector encoding OCT4, SOX2, KLF4, and MYC, and isolated several iPSC clones (supplemental Figure 2). TERC het.G319A iPSCs showed short telomere lengths compared with wild-type (WT) iPSCs (Figure 2A; supplemental Figure 3), but displayed continuous self-renewal in culture. We found no differences in overall TERC levels in normal vs TERC het.G319A iPSCs (Figure 2B). By sequencing complementary DNA (cDNA) from patient iPSCs, we observed that transcripts from both the normal and G319A TERC alleles were equally represented (Figure 2B). These data not only indicate telomere maintenance defects in TERC het.G319A patient cells, but also that neither transcription nor posttranscriptional accumulation of TERC is compromised by the G319A variant.
The TERC G319A mutation impairs TERC:TERT interaction. (A) Telomere restriction fragment length (TRF) analysis of normal (WT) iPSCs, 3 independent TERC-mutant iPSC clones (cl), and TERC-mutant fibroblasts. (B) Top left, Representative Northern blot of TERC RNA from WT, TERC-mutant iPSCs, and TERC-mutant fibroblasts. Top right, Relative TERC levels are shown in the graph. Bottom, Sequencing of TERC cDNA from TERC-mutant iPSCs. (C) Left, Schematic of TERC RNA in complex with TERT and dyskerin. Right, Sequencing of TERC RT-PCR amplicons isolated from TERT RNP (top) and dyskerin RNP (bottom) isolated by immunoprecipitation (IP) from TERC-mutant iPSCs. (D) Restriction fragment length polymorphism (RFLP) of TERC amplicons from dyskerin or TERT IP of patient iPSCs (lanes 5-8). The HincII site created by the G319A mutation is depicted on the right. Lanes 1-4 show digest patterns of amplicons from WT (319G) and mutant (319A) plasmid DNA. Products were separated by agarose gel electrophoresis and band intensity was quantified using ImageLab. n.s., not significant.
The TERC G319A mutation impairs TERC:TERT interaction. (A) Telomere restriction fragment length (TRF) analysis of normal (WT) iPSCs, 3 independent TERC-mutant iPSC clones (cl), and TERC-mutant fibroblasts. (B) Top left, Representative Northern blot of TERC RNA from WT, TERC-mutant iPSCs, and TERC-mutant fibroblasts. Top right, Relative TERC levels are shown in the graph. Bottom, Sequencing of TERC cDNA from TERC-mutant iPSCs. (C) Left, Schematic of TERC RNA in complex with TERT and dyskerin. Right, Sequencing of TERC RT-PCR amplicons isolated from TERT RNP (top) and dyskerin RNP (bottom) isolated by immunoprecipitation (IP) from TERC-mutant iPSCs. (D) Restriction fragment length polymorphism (RFLP) of TERC amplicons from dyskerin or TERT IP of patient iPSCs (lanes 5-8). The HincII site created by the G319A mutation is depicted on the right. Lanes 1-4 show digest patterns of amplicons from WT (319G) and mutant (319A) plasmid DNA. Products were separated by agarose gel electrophoresis and band intensity was quantified using ImageLab. n.s., not significant.
Prior studies indicate that, in addition to the TERC template region, the CR4/CR5 domain serves as an independent site to which TERT binds in the telomerase holoenzyme.24,25 We hypothesized that, despite its location outside of the P6b and P6.1 stem loops in CR4/CR5 (Figure 1D) which are known to be required for TERT association,24-26 the TERC G319 residue might also be important for binding to TERT. We exploited our ability to generate millions of patient iPSCs and the heterozygosity of the TERC variant to determine the relative association of normal vs G319A TERC with TERT, in the same cell. We immunoprecipitated TERC-containing ribonucleoprotein (RNP) complexes from patient iPSCs using either anti-dyskerin or anti-TERT antibodies, and extracted and analyzed the associated RNAs (supplemental Figure 4). Sequencing of the reverse transcription polymerase chain reaction (RT-PCR) products of dyskerin-bound RNA from patient iPSCs showed equal recovery of the G319A and normal TERC species (Figure 2C). In contrast, when we pulled down TERT RNPs, we found a decreased proportion of G319A vs normal TERC (Figure 2C; supplemental Figure 5). We took advantage of the gain of an HincII restriction site in the mutant TERC cDNA sequence to quantitate the relative association of G319A vs normal TERC with TERT, and found it to be diminished by ∼75% (Figure 2D). These results demonstrate the TERC G319A mutation compromises telomerase function via decreasing binding of TERC to TERT in vivo.
In the era of clinical genome sequencing, determining the pathogenicity of variants of unknown significance is an important challenge, often with immediate implications for patient diagnosis and counseling. Patient iPSCs allowed us to overcome limitations of a heterologous cell-based assay widely used to assess TERC and TERT variants, and compare the functions of normal vs variant TERC RNA using a biochemical approach. Our results reveal a sequence in the TERC CR4/CR5 domain that is important for binding TERT, which when disrupted in a heterozygous state in humans is sufficient to cause telomere disease. A guanine residue at positions orthologous to nucleotide 319 of human TERC is highly conserved but has escaped attention in prior work, possibly due to its location outside of the CR4/CR5 stem loops and absence in the extensively studied medaka (Oryzias latipes) telomerase RNA.27,28 Validating the TERC G319A mutation in patient iPSCs allows us to proceed with genetic testing and counseling of older and younger family members at risk. In summary, our results illustrate the utility of patient-derived iPSCs for evaluating genetic variants and defining molecular mechanisms of disease in patients with bone marrow failure syndromes.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank the patients and family for participation in research. The authors also thank Aidan Moriarty for technical assistance.
This work was supported by funding from grant R01 DK107716-01 (National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases), Pedals for Pediatrics (Dana-Farber Cancer Institute), and the Translational Research Program (Boston Children’s Hospital) (S.A.), and the Scientific and Technological Research Council of Turkey (B.B.).
Contribution: B.B. and S.A. designed experiments, analyzed data, and wrote the paper; B.B. performed experiments; C.M.B., C.S.C., and S.A. examined the patients and/or provided patient information and materials; C.S.C. consented the patients to the research study; and M.D.F. analyzed histology and provided images.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Suneet Agarwal, Boston Children's Hospital, CLS 3002, 3 Blackfan Circle, Boston, MA 02115; e-mail: suneet.agarwal@childrens.harvard.edu.

![Figure 1. A novel variant in the CR4/5 region of TERC in a family with telomere disease. (A) Pedigree of the family; the proband is indicated with an arrow. Affected individuals are shown in gray and symptoms are indicated. Mutations in the individuals genotyped are shown. IPF, idiopathic pulmonary fibrosis; Skin, pigmentation abnormalities; TP, thrombocytopenia. (B) Bone marrow histology. Top, Hypercellular, fibrotic bone marrow with dyspoiesis (hematoxylin and eosin [H&E]). Bottom, CD34 staining shows increased percentage of CD34+ cells (scale bar, 20 µm). (C) Telomere length measurement by flow cytometry fluorescence in situ hybridization (flow-FISH) in lymphocytes in the proband and his sister. (D) Top, Schematic of telomerase RNP. The location of the variant in the TERC CR4/CR5 region is indicated with an arrow. Bottom, Sequencing of TERC in the proband shows the c.319G>A variant. (E) Telomeric repeat amplification protocol (TRAP) assay for telomerase activity in VA13 cells transfected with TERT and WT TERC, G319A TERC, or control plasmid. Threefold dilutions of input cell extract. Internal control (IC) amplification standard is indicated. Relative telomerase activities are shown in the graph. (F) Top, Immunoblot of TERT and actin protein levels. Bottom, Northern blot of TERC RNA from VA13 cells transfected with TERT plus WT TERC, G319A TERC, or control plasmid. Ethidium bromide staining of 28S ribosomal RNA (rRNA) is used as a loading control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/16/10.1182_blood-2016-04-710160/4/m_2089f1.jpeg?Expires=1765956974&Signature=p-tXLMeKMoziz5MZUHIHVjzYxjfS1tlI-tLuK73DtvwfuGefmn8KlM~UWcMlOo4qi9e7aAsuYADEt1wa967q~aHVbirxmjuxa5jHI2YGoTRvGvqhXril6IbJLm8F-E3DW90oZ~JD6JhDv7QuxAADAS7owTsKpLX0IrY7xHNrGQYyf-BXEwZAS5~fxNj9PiAoBirEQI1aRxjKSRfLUvIl80Ie4pVw666ZbHjZNblY7kGof8NTlL7pPrWJuuwgk0wN0HJo3IpMhrCxnncSeEvkiYFh7TJfN5QsYwrnIV66J8qualXXVCskyjGoWjClyX6vBPcxBFd0tEoy4EgtSc7llw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal