Key Points
This study demonstrates allosteric RNA structure alteration resulting from an exonic variation, thereby interfering with splicing.
This study details a novel mechanism by which silent mutation distant to the 5′ splice site could still result in intron retention.
Abstract
Disease-associated silent mutations are considered to affect the accurate pre–messenger RNA (mRNA) splicing either by influencing regulatory elements, leading to exon skipping, or by creating a new cryptic splice site. This study describes a new molecular pathological mechanism by which a silent mutation inhibits splicing and leads to intron retention. We identified a heterozygous silent mutation, c.7464C>T, in exon 44 of the von Willebrand factor (VWF) gene in a family with type 1 von Willebrand disease. In vivo and ex vivo transcript analysis revealed an aberrantly spliced transcript, with intron 44 retained in the mRNA, implying disruption of the first catalytic step of splicing at the 5′ splice site (5′ss). The abnormal transcript with the retained intronic region coded a truncated protein that lacked the carboxy-terminal end of the VWF protein. Confocal immunofluorescence characterizations of blood outgrowth endothelial cells derived from the patient confirmed the presence of the truncated protein by demonstrating accumulation of VWF in the endoplasmic reticulum. In silico pre-mRNA secondary and tertiary structure analysis revealed that this substitution, despite its distal position from the 5′ss (85 bp downstream), induces cis alterations in pre-mRNA structure that result in the formation of a stable hairpin at the 5′ss. This hairpin sequesters the 5′ss residues involved in U1 small nuclear RNA interactions, thereby inhibiting excision of the pre-mRNA intronic region. This study is the first to show the allosteric-like/far-reaching effect of an exonic variation on pre-mRNA splicing that is mediated by structural changes in the pre-mRNA.
Introduction
von Willebrand factor (VWF) is a multimeric plasma glycoprotein synthesized in endothelial cells and platelet precursors and plays crucial roles in hemostasis.1 The VWF gene (VWF) is composed of 52 exons spanning ∼178 kb of the genome, and it is transcribed into an 8.8-kb messenger RNA (mRNA).2 Deficient VWF results in von Willebrand disease (VWD), which is classified as quantitative (type 1 and type 3) or qualitative (type 2).3,4 Type 1 VWD, characterized by partial reduction in VWF levels, is the most common form of the disorder.5 Inheritance of type 1 VWD is considered to be autosomal dominant; however, in ∼15% of index cases, more than a single candidate VWF variant is detected.6-8
Pre-mRNA splicing is regulated by consensus core splicing sites comprising the 5′ splice site (5′ss), the 3′ splice site (3′ss), and the branch point sequences.9 The splicing reaction is initiated by RNA-RNA base pairing of the consensus 5′ss motif and the U1 small nuclear RNA (snRNA) terminus, which is followed by excision of the 5′ end of the intron, ligation of the adjacent exons, and releasing of the intron.10-12 In addition to the core splicing signals, auxiliary splicing regulatory elements (SREs) have a critical role in recognition of exons and splicing efficiency.9,13,14 Furthermore, recent studies have indicated that pre-mRNA structure modulates the splicing machinery.10,15,16
Previous studies suggest that disease-causing silent mutations distant to the core splice sites interfere with exon recognition either by affecting auxiliary SREs, leading to exon skipping, or by creating cryptic splice sites, resulting in partial deletion in exons.9,17,18 Our current study, for the first time in the literature of human genetic mutations, presents a synonymous variant outside the core splice sites that results in intron retention. Intron retention is a rare splicing defect that has been previously reported solely as a result of mutations residing in core 5′ss or branch point motif sequences.19-21
We had previously reported a translationally silent mutation c.7464C>T (p.Gly2488) in exon 44 of VWF, 25 bp after the 3′ss and 85 bp downstream of the 5′ss, in a patient with type 1 VWD.22 In the present study, in vivo and ex vivo transcript analysis unexpectedly revealed that it inhibited splicing at the 5′ donor splice site, despite its 85-bp distance to the 5′ss, and led to an aberrant transcript with a retained intronic region in mature mRNA. The previously described mechanisms for exonic substitutions, such as contribution to cis-regulatory elements, could not provide a plausible explanation for this event. Interestingly, a combination of in silico secondary and tertiary structure analysis of the pre-mRNA clarified this long-distance influence by demonstrating allosteric modifications in RNA folding resulting from the given mutation, which inhibited accessibility of the 5′ core splice site.
Materials and methods
Patients: phenotypic analysis
An 11-year-old girl index patient (IP) diagnosed with VWD from a non-consanguineous family and her parents were included in the study. Laboratory investigation of VWF antigen (VWF:Ag), VWF binding to platelet glycoprotein Ib, factor VIII coagulant activity, and VWF multimers (1.2% [w/v] and 1.6% [w/v] agarose gels) was performed as previously described.4,22 The bleeding score was calculated on the basis of a bleeding questionnaire for type 1 VWD.23 This study was approved by the local ethics committee, and informed consent was obtained from all patients (vote 091/09).
Gene analysis
Genomic DNA was isolated from peripheral whole blood of the IP and her parents by standard methods. Mutation screening analysis was carried out by direct sequencing of exons 1 to 52, including exon/intron boundaries, 5′ and 3′ untranslated regions, and the promoter region of the VWF as described before.22 In addition, sequence analysis of the whole intron 44 of VWF was performed. The current single-nucleotide polymorphism (SNP) database was checked for the presence of unknown substitutions through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SNP, accessed March 2015). The splice-site prediction tool Human Splicing Finder version 2.4.1 (http://www.umd.be/HSF, accessed February 2014) was used to analyze the effect of the novel identified variant on splicing regulatory sites.24
The Multiplex Ligation-Dependent Probe Amplification assay (kits P011 and P012; MRC-Holland, Amsterdam, The Netherlands) was applied to detect the potential presence of VWF exon rearrangements.25
BOEC isolation
Blood outgrowth endothelial cells (BOECs) were isolated from blood of the IP, her mother, and 3 healthy individuals based on the published standardized protocols (see supplemental Methods, available on the Blood Web site).26
RNA isolation and RT-PCR assay
The total RNA was isolated from whole blood of the IP, her mother, and healthy control subjects using the Tempus Spin RNA Isolation Kit (Applied Biosystems, United Kingdom) according to the manufacturer’s instructions. In addition, RNA was extracted from cultured BOECs and platelets by using RNeasy Mini Kit (QIAGEN, Germany). The isolated mRNA was reverse transcribed (RT) to complementary DNA (cDNA), and subsequently, VWF cDNA was polymerase chain reaction (PCR)-amplified in 14 overlapping fragments containing multiple exons using the QIAGEN LongRange 2Step RT-PCR Kit according to the manufacturer’s recommendations. PCR reactions were performed in the following cycling conditions: 3 minutes at 93°C; followed by 35 cycles of 15 seconds at 93°C, 30 seconds at 55°C, and 2 minutes at 68°C; and a final extension of 2 minutes at 68°C. To allow amplification of the probable aberrant transcript with the larger size, RT-PCRs of overlapping segments 12 and 13 were repeated using the same primers but increasing the extension time of PCR cycling from 2 minutes to 6, 8, and 10 minutes. The sequence and position of the primers and the product sizes of the RT-PCR are shown in supplemental Table 1. The RT-PCR products were separated on 1% agarose gel and sequenced to identify the variations in the mRNA transcript.
Subsequent RT-PCR reactions using 4 allele-specific primer combinations were performed to ascertain whether intron 44 was retained within the mature mRNA. In the first pair, a forward primer was designed across the junction of exon 40-41, and a reverse primer was designed to target a sequence in intron 44. In the second, third, and fourth primer pairs, forward primers targeted 3 different sites in intron 44, and the reverse primer was directed at the exon 48-49 junction (supplemental Table 2).
VWF expression assessment and confocal immunofluorescence (IF) microscopy of BOECs
VWF:Ag levels in the medium and lysate of BOECs were measured as described in supplemental Methods. Furthermore, western blotting of intracellular VWF was performed to evaluate production of the precursor VWF (pre-pro-VWF) in BOECs (supplemental Methods).
In addition, BOECs were fixed and stained with immunofluorescent antibodies to visualize VWF, PECAM-1, cis- and trans-Golgi compartments, and endoplasmic reticulum (ER) (supplemental Methods). The comparative degree of colocalization for the wild-type (wt) and mutant VWF was calculated as mean Pearson correlation coefficient.27
In silico secondary and tertiary pre-mRNA structure analysis
Structural analysis of the pre-mRNA was performed to generate information on the secondary and tertiary structure corresponding to the wt and mutant variant. Secondary structure analysis was performed on the mfold Web server (http://mfold.rit.albany.edu/?q=mfold, accessed February 2014).28,29 Tertiary structure analysis was performed on 2 ab initio Web servers and on the basis of secondary structure prediction made by mfold on RNAComposer (http://rnacomposer.cs.put.poznan.pl, accessed May 2014).30 The Rosetta online platform (http://rosie.rosettacommons.org/rna_denovo, accessed April 2014) was used to generate de novo models of RNA sequences 30 nucleotides long surrounding the mutated nucleotide for both the mutant and wt sequences.31 Because the Rosetta RNA de novo platform has a size limitation, in order to investigate any distal allosteric influences on structure upstream or downstream of the mutation loci, we generated an ab initio model of a longer sequence (137 nucleotides) of pre-mRNA (including the mutated nucleotide and parts of exon 44 and intron 44 covering both the 5′ss and 3′ss) on the iFold server (http://troll.med.unc.edu/ifold, accessed June 2014).32 In another type of model generation, the secondary structure prediction for the lowest energy structure was downloaded in dot-bracket format from mfold and was used as an input file on the RNAComposer server to generate the 3-dimensional coordinates corresponding to the secondary structure prediction. Default modeling parameters were used for all Web servers. Modeling was also performed with all these servers for generating the 3-dimensional structure for the U1 snRNA sequence (only for first 69 nucleotides). Subsequently, in silico docking for the U1 snRNA structures was performed on the 3-dimensional structures of the mutant and wt structures generated from iFold on the ZDOCK server (version 2.3; http://zdock.umassmed.edu, accessed September 2014).33 Because the server applies Fanelli parameters to implement protein-protein or protein–nucleic acid docking, the RNA-RNA dock was analyzed only from a coarse-grained perspective.34 To compare the effectiveness of individual base pairs in RNA-RNA interaction, a hypothetical U1 snRNA–5′ donor splice site complex structure was reproduced by first cofolding the 2 RNAs on the RNAcofold server (http://rna.tbi.univie.ac.at/cgi-bin/RNAcofold.cgi, accessed September 2014) and subsequently generating the 3-dimensional coordinates for the 2-dimensional prediction using the RNAComposer server. The tertiary structures were visualized and analyzed, and their images were rendered with YASARA version 13.8.26.35 Supplemental Table 3 defines the sequences submitted for structure prediction with respect to the location of the mutation and the splice sites.
Results
Characterization of patients
The type 1 VWD IP with a cumulative bleeding score of 17 had a history of epistaxis, frequent bruises, prolonged bleeding from small wounds, and oral-cavity bleeding. Furthermore, she had a tendency to bleed after tooth extraction and surgery. The IP had a significantly reduced VWF:Ag of 9 IU/dL and a prolonged platelet function assay (PFA-100) closure time (Table 1). Her VWF multimer profile revealed normal multimer size distribution but a reduction in concentration of all multimers due to her low VWF.Ag level (supplemental Figure 1). One hour after desmopressin administration, the IP’s VWF:Ag level was increased from 9 to 19 IU/dL (along with an increase in VWF ristocetin cofactor activity from <6 to 19 IU/dL). These levels remained almost persistent 2 hours after treatment. However, this poor VWF response to desmopressin was not clinically sufficient in many occasions, and treatment with VWF/factor VIII concentrates was required.
Phenotypic characteristics and genetic data of the IP and her parents
| . | VWD diagnosis . | Mutation . | Age, y . | Blood group . | Closure time PFA-collagen/ADP, s . | VWF:Ag, IU/dL . | VWF:GPIb, IU/dL . | FVIII:C, IU/dL . | Multimer . |
|---|---|---|---|---|---|---|---|---|---|
| IP | Type 1 VWD | c.7464C>T | 11 | A1 | >300 | 9 | 6 | 35 | Normal multimer size distribution* |
| Mother | Has risk factor for bleeding | c.7464C>T | 45 | A1 | 149 | 49 | 46 | 92 | Normal |
| Father | Has risk factor for bleeding | No mutation | 46 | A1 | 126 | 44 | 35 | 108 | Normal |
| Normal range | — | — | — | — | 71-118 | 65-165 | 64-150 | 70-157 | — |
| . | VWD diagnosis . | Mutation . | Age, y . | Blood group . | Closure time PFA-collagen/ADP, s . | VWF:Ag, IU/dL . | VWF:GPIb, IU/dL . | FVIII:C, IU/dL . | Multimer . |
|---|---|---|---|---|---|---|---|---|---|
| IP | Type 1 VWD | c.7464C>T | 11 | A1 | >300 | 9 | 6 | 35 | Normal multimer size distribution* |
| Mother | Has risk factor for bleeding | c.7464C>T | 45 | A1 | 149 | 49 | 46 | 92 | Normal |
| Father | Has risk factor for bleeding | No mutation | 46 | A1 | 126 | 44 | 35 | 108 | Normal |
| Normal range | — | — | — | — | 71-118 | 65-165 | 64-150 | 70-157 | — |
ADP, adenosine 5′-diphosphate; FVIII:C, factor VIII coagulant activity; PFA, platelet function assay; VWF:Ag, VWF antigen; VFW:GPIb, VWF binding to platelet glycoprotein Ib; —, not applicable.
The size distribution of the multimer pattern was normal but the concentration of the all multimers was reduced.
Although both parents of the IP had no bleeding history, they showed prolonged closure time and a modestly decreased VWF:Ag level, indicating that they have a risk factor for bleeding (Table 1).
VWF gene mutations
DNA sequencing revealed a heterozygous unknown synonymous variant, c.7464C>T (p.Gly2488), in exon 44. Genetic analysis of the parents showed that the IP had inherited the unknown variant, c.7464C>T, from her mother. No other VWF mutations were detected by the sequencing of all exons and the promoter region. No exon rearrangement was found in the explored region of VWF by Multiplex Ligation-Dependent Probe Amplification analysis. The impact of the detected silent mutation (c.7464C>T) on splice enhancing and silencing motifs (ie, SREs) was evaluated with the Human Splicing Finder prediction tool, and the results are presented in supplemental Table 4.
In addition, no unknown variant was detected after sequence analysis of intron 44; only 8 SNPs that were already recorded in the SNP database were identified in intron 44 (supplemental Figure 2). The minor allele of each detected SNP had a frequency of >10% (supplemental Figure 2).
RNA analysis
Further RNA analysis was performed for assessment of the pathogenicity of the variant c.7464C>T with respect to RNA splicing. In addition, complete sequencing of the VWF cDNA was performed based on the assumption that there might be another mutation deep in an intron that could affect splicing of the second allele.
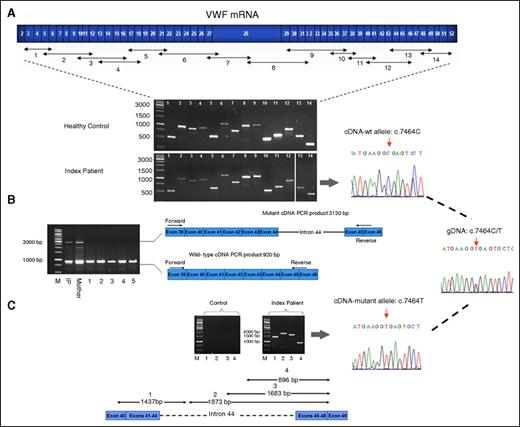
There were no differences in either the number or the size of the RT-PCR amplification products of IP VWF cDNA compared with wt on the agarose gel (Figure 1A). Moreover, the sequence analysis of the whole VWF cDNA showed no abnormality. However, surprisingly, analysis of the cDNA sequence of fragments 12 and 13 (both compromising exon 44 with the heterozygous variant c.7464C>T) demonstrated the monoallelic presentation of the wt transcript, c.7464C, in the sequence chromatogram, indicating no normal mRNA transcript from the mutant allele (Figure 1A). The presence of heterozygous exonic SNPs in the other RT-PCR fragments indicated that the mutant transcript is present but cannot be amplified, probably due to its large size, leading to preferential amplification of the wt transcript. Repetition of RT-PCR of segments 12 and 13 with an increase in extension time of PCR cycling confirmed this presumption. The RT-PCR of segment 12 showed 2 amplicons: an expected 920-bp segment corresponding to the normal transcript, and an aberrantly spliced transcript of ∼3200 bp that, in addition to the normal transcript, had an insert of ∼2200 bp (Figure 1B). Likewise, repetition of RT-PCR of fragment 13 confirmed an insert with same size (supplemental Figure 3). Further RT-PCRs with 4 pairs of allele-specific primers confirmed the retention of whole intron 44 (Figure 1C). However, the RT-PCR of the control RNAs performed with the same allele-specific primers failed, as expected (Figure 1C).
RT-PCR products on agarose gel. (A) Schematic scale of the coding region of VWF (exons 2-52) with the primer positions designed for amplification of the full-length VWF mRNA and corresponding amplicon segments. Agarose gel electrophoresis image shows the 14 overlapping RT-PCR products of VWF using total RNA from the IP’s blood as template, under thermocycling conditions with 2 minutes of extension time. The sequence chromatogram of segments 12 and 13 (both covering exon 44 carrying the silent mutation c.7464C>T) demonstrate single-peak manifestation of wt nucleotide C, c.7464C, indicating a fail in amplification of the mutant transcript. Lanes 13 and 14 were run in a separate gel but with similar running conditions. (B) RT-PCR products of segment 12 amplified with primers residing in exon 39 and exons 45/46 boundary, and with increased extension time (6 minutes) of thermocycling. RT-PCR products of RNA obtained from blood of the IP and her mother demonstrate a smaller product (920 bp) relevant to the normal transcript and an aberrant larger fragment (3130 bp) corresponding to the retained intron 44 in mRNA, whereas RT-PCRs using RNA from 5 healthy control subjects as template show only the smaller normal fragment (lanes 1-5). (C) RT-PCR amplification using allele-specific primers to confirm intron 44 retention. The primer combinations and expected amplicon sizes, if intron 44 is retained, are as follows: segment 1 (1437 bp), forward primer in exons 40/41 boundary and reverse primer targeted in intron 44, 861 nucleotides downstream of the exon 44 (lane 1); and segments 2, 3 and 4 (length 1873, 1683, and 896 bp, respectively), forward primers directed in intron 44 in 3 different positions (+787, +977, and +1764) and reverse primer in exons 48/49 boundary (lanes 2, 3, and 4). Sequence analysis of the cDNA segment 1 exhibited monoallelic presentation of the mutant variant T (c.7464T) in the sequence chromatogram, indicating that the aberrant transcript is derived solely from the mutant allele. Lane M represents the molecular weight marker (1-kb ladder).
RT-PCR products on agarose gel. (A) Schematic scale of the coding region of VWF (exons 2-52) with the primer positions designed for amplification of the full-length VWF mRNA and corresponding amplicon segments. Agarose gel electrophoresis image shows the 14 overlapping RT-PCR products of VWF using total RNA from the IP’s blood as template, under thermocycling conditions with 2 minutes of extension time. The sequence chromatogram of segments 12 and 13 (both covering exon 44 carrying the silent mutation c.7464C>T) demonstrate single-peak manifestation of wt nucleotide C, c.7464C, indicating a fail in amplification of the mutant transcript. Lanes 13 and 14 were run in a separate gel but with similar running conditions. (B) RT-PCR products of segment 12 amplified with primers residing in exon 39 and exons 45/46 boundary, and with increased extension time (6 minutes) of thermocycling. RT-PCR products of RNA obtained from blood of the IP and her mother demonstrate a smaller product (920 bp) relevant to the normal transcript and an aberrant larger fragment (3130 bp) corresponding to the retained intron 44 in mRNA, whereas RT-PCRs using RNA from 5 healthy control subjects as template show only the smaller normal fragment (lanes 1-5). (C) RT-PCR amplification using allele-specific primers to confirm intron 44 retention. The primer combinations and expected amplicon sizes, if intron 44 is retained, are as follows: segment 1 (1437 bp), forward primer in exons 40/41 boundary and reverse primer targeted in intron 44, 861 nucleotides downstream of the exon 44 (lane 1); and segments 2, 3 and 4 (length 1873, 1683, and 896 bp, respectively), forward primers directed in intron 44 in 3 different positions (+787, +977, and +1764) and reverse primer in exons 48/49 boundary (lanes 2, 3, and 4). Sequence analysis of the cDNA segment 1 exhibited monoallelic presentation of the mutant variant T (c.7464T) in the sequence chromatogram, indicating that the aberrant transcript is derived solely from the mutant allele. Lane M represents the molecular weight marker (1-kb ladder).
The same aberrant transcript was observed when the IP’s platelet RNA (data not shown) and BOEC RNA (supplemental Figure 4) were used as the template. Similarly, the mother with the same substitution (c.7464C>T) detected in exon 44 of the VWF showed the same defect in RNA splicing (Figure 1B; supplemental Figure 3).
Confocal IF microscopy of BOECs
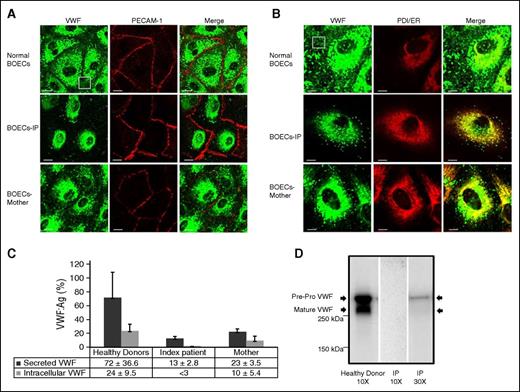
The isolated BOECs were characterized by immunofluorescent staining for the endothelial cell-specific marker, PECAM-1, and VWF (Figure 2A). Confocal IF analysis showed that in contrast to pervasive VWF staining in normal BOECs, VWF staining among BOECs isolated from the IP was less and variable. Only 64 of 100 inspected IP BOECs expressed VWF protein, which itself exhibited some degree of variability. Furthermore, VWF staining in the IP BOECs was mostly diffuse, with only a limited number of rounded WPBs compared with the VWF stored in discrete elongated WPBs within normal BOECs (Figure 2A-B). Immunofluorescent VWF staining of the mother-derived BOECs demonstrated a combination of VWF stored in WPBs and diffuse staining (Figure 2A-B). Further IF analysis illustrated that the diffuse VWF within the BOECs of the IP and her mother is colocalized with the ER marker, protein disulfide isomerase, demonstrating retention of truncated VWF proteins in the ER (Figure 2B). The mean Pearson coefficient, representing the degree of ER colocalization, was significantly higher (P < .01) for the BOECs of the IP and her mother (0.540 ± 0.114 and 0.248 ± 0.154, respectively) compared with normal BOECs (0.040 ± 0.019).
Subcellular distribution and expression of VWF in the BOECs isolated from the IP and healthy donors. (A) Characteristics of the BOECs via staining of the cell-specific markers VWF (light green) and PECAM (red) with secondary antibodies conjugated with Alexa Fluor-488 and Alexa Fluor-594, respectively. However, only 64 of 100 inspected IP BOECs emitted light green fluorescent signals, representing production of VWF protein. In the VWF-expressing IP BOECs, VWF staining is mostly diffuse, accumulating around the nucleus of the cells, whereas in normal BOECs, VWF can be seen as distinct elongated structures, indicating storage in WPBs. The BOECs isolated from the IP’s mother illustrate a combination of VWF stored in WPBs and diffuse staining. The white box points out secreting VWF strings in normal BOECs that are not visible in the IP-derived BOECs. Bars represent 20 µm. (B) Diffuse staining observed in BOECs obtained from the IP and her mother (carrying the mutation c. 7464C>T) was colocalized with protein disulfide isomerase (PDI). Staining was performed using primary antibodies anti-VWF (left channel, light green) and anti-PDI (ER marker; middle, red) and secondary antibodies conjugated with Alexa Fluor-488 and Alexa Fluor-555, respectively. Colocalization of VWF and PDI staining is illustrated in the right channel (Merge). Bars represent 10 µm. (C) Bar graph of the mean of VWF:Ag levels in the medium and lysates of BOECs obtained from the IP, her mother (3 independent experiments, N = 3), and 3 healthy donors (each 3 independent experiments, N = 9). The mean VWF:Ag was determined after 10× concentration of the collected medium and cell lysates of confluent BOECs in 75 cm2 flasks. Error bars indicate the standard deviation. (D) Western blot analysis of BOEC intracellular VWF after electrophoresis on 4% to 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Left lane shows both pre-pro-VWF and mature VWF in the lysate of the normal BOECs after 10× concentration. Middle and right lanes are representative of the IP’s BOEC lysates after 10× and 30× concentrations, respectively. WPBs, Weibel-Palade bodies.
Subcellular distribution and expression of VWF in the BOECs isolated from the IP and healthy donors. (A) Characteristics of the BOECs via staining of the cell-specific markers VWF (light green) and PECAM (red) with secondary antibodies conjugated with Alexa Fluor-488 and Alexa Fluor-594, respectively. However, only 64 of 100 inspected IP BOECs emitted light green fluorescent signals, representing production of VWF protein. In the VWF-expressing IP BOECs, VWF staining is mostly diffuse, accumulating around the nucleus of the cells, whereas in normal BOECs, VWF can be seen as distinct elongated structures, indicating storage in WPBs. The BOECs isolated from the IP’s mother illustrate a combination of VWF stored in WPBs and diffuse staining. The white box points out secreting VWF strings in normal BOECs that are not visible in the IP-derived BOECs. Bars represent 20 µm. (B) Diffuse staining observed in BOECs obtained from the IP and her mother (carrying the mutation c. 7464C>T) was colocalized with protein disulfide isomerase (PDI). Staining was performed using primary antibodies anti-VWF (left channel, light green) and anti-PDI (ER marker; middle, red) and secondary antibodies conjugated with Alexa Fluor-488 and Alexa Fluor-555, respectively. Colocalization of VWF and PDI staining is illustrated in the right channel (Merge). Bars represent 10 µm. (C) Bar graph of the mean of VWF:Ag levels in the medium and lysates of BOECs obtained from the IP, her mother (3 independent experiments, N = 3), and 3 healthy donors (each 3 independent experiments, N = 9). The mean VWF:Ag was determined after 10× concentration of the collected medium and cell lysates of confluent BOECs in 75 cm2 flasks. Error bars indicate the standard deviation. (D) Western blot analysis of BOEC intracellular VWF after electrophoresis on 4% to 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Left lane shows both pre-pro-VWF and mature VWF in the lysate of the normal BOECs after 10× concentration. Middle and right lanes are representative of the IP’s BOEC lysates after 10× and 30× concentrations, respectively. WPBs, Weibel-Palade bodies.
Expression of VWF in BOECs
To quantify total production of the VWF in BOECs, VWF:Ag levels (%) in secreted medium and lysates of the BOECs were determined. Data are presented as mean ± standard deviation. Secreted VWF from the IP-derived BOECs was considerably lower (P < .01) than healthy-donor BOECs (13% ± 2.8% vs 72% ± 36.6%). The significant reduction in secretion of VWF was not associated with a subsequent increase in intracellular accumulation. In fact, the amount of the VWF in lysates of the IP’s BOECs was below the detection limit (<3%) compared with a VWF:Ag level of 24% ± 9.5% detected in normal BOEC lysates. Furthermore, the amount of VWF in both medium and lysates of the mother’s BOECs was reduced (P < .05) compared with normal BOECs (23% ± 3.5% vs 72% ± 36.6% and 10% ± 5.4% vs 24% ± 9.5%, respectively) but they were still higher than those measured in the IP’s BOECs (P < .05) (Figure 2C).
Western blot analysis of the healthy-donor BOEC lysates (concentrated 10×) revealed two protein bands representing pre-pro-VWF and mature VWF (Figure 2D). However, no protein band was detected after electrophoresis of the same amount of the 10× IP BOEC lysates. Only weak protein bands were visualized when electrophoresis and blotting of the IP BOEC lysates were repeated with more concentration (30×).
Secondary and tertiary structure predictions of pre-mRNA
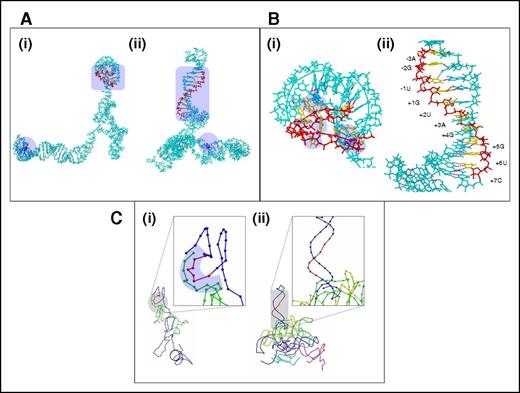
The 3-dimensional structure prediction using the ab initio servers/RNAComposer and the 2-dimensional structure prediction presented a unique summation/conclusion for the effects observed in the cDNA analysis experiments. Although the predictions performed on different servers differed from each other to variable degrees, a common conclusion derived from all the servers was that the mutation brings about gross changes in the overall structure of the pre-mRNA. The ab initio structures generated from iFold differed completely between the wt and the mutant sequences (Figure 3A). Most remarkably, the 5′ donor splice-site region forms part of a highly ordered stem-loop or hairpin structure in the mutated sequence (Figure 3B). All nucleotides that are known to bind U1 snRNA are already sequestered by a combination of canonical and noncanonical base pairs within this hairpin, which is 42 nucleotides long, stretching on to the end of the evaluated sequence (Figure 3Bii). In comparison, the wt sequence did not show any such hairpins (Figure 3Bi). One canonical base pair between the +5G (5′ donor splice-site residues) and +14C, a noncanonical base pairing between +8C (5′ donor splice-site residues) and −4G, and a backbone hydrogen bond between the phosphate group of +2U (5′ donor splice-site residues) and +8C were all observed in the region that binds to U1 snRNA in the wt sequence. Most other residues corresponding to U1 snRNA binding were unfulfilled and, therefore, in single-stranded form (Figure 3Bi). In addition, the purine/pyrimidine base atoms that usually participate in hydrogen bonding were more exposed in the wt structure and, therefore, more amenable to binding to U1 snRNA (supplemental Table 5). Interestingly, the secondary structure prediction (iFold) (Figure 4), the 3-dimensional coordinate structures generated on RNAComposer (supplemental Figure 5), and the Rosetta server structures for the mutated and wt sequences also agreed with the observations made on the 3-dimensional ab initio structures, although the correlations were not absolute. Similar to the observations made in the ab initio structures, many of the 5′ donor splice-site residues that bind to U1 snRNA were observed to be free in the wt sequence but prebound or physically constrained in a double-stranded form in the mutated sequence. However, the base pairings suggested from the secondary structure predictions and the ab initio structures varied significantly from one another.
Ab initio models of pre-mRNA and docking analysis. (A) Ab initio structures corresponding to wt (i) and mutated (ii) pre-mRNA sequences generated on iFold. The structure is depicted in stick format and shown in cyan. The mutated residue location is depicted in blue and marked with a lavender-shaded area. Hydrogen bonds are depicted as magenta dots throughout panels A-C. The backbone of the region corresponding to residues that bind to U1 snRNA is red, whereas the bases are yellow. This region is also marked by the lavender-shaded area. (B) Region corresponding to U1 snRNA binding for the wt (i) and mutated (ii) pre-mRNA models at a closer view. Color coding is as observed in panel A. The residues that bind to U1 snRNA are numbered in the mutated sequence structure model. In the wt sequence model, the unbound residues corresponding to U1 snRNA binding are marked with lavender-shaded regions. (C) Coarse-grained depiction of the dock of U1 snRNA over the wt (i) and mutated (ii) pre-mRNA sequence structure. Because the depiction is coarse-grained, the entire model is depicted only as a beaded trace. The trace is blue for the pre-mRNA sequence, with only the region corresponding to U1 snRNA binding shown in red. The different putative U1 snRNA docked structures are colored differently. Because only 1 putative dock was observed for the wt sequence, U1 snRNA is shown as light green in the dock between the U1 snRNA and wt pre-mRNA structures. The inset images provide a closer view of the U1 snRNA binding region on the pre-mRNA structures. The proximal regions in the U1 snRNA and wt pre-mRNA structures are marked by the lavender-shaded area.
Ab initio models of pre-mRNA and docking analysis. (A) Ab initio structures corresponding to wt (i) and mutated (ii) pre-mRNA sequences generated on iFold. The structure is depicted in stick format and shown in cyan. The mutated residue location is depicted in blue and marked with a lavender-shaded area. Hydrogen bonds are depicted as magenta dots throughout panels A-C. The backbone of the region corresponding to residues that bind to U1 snRNA is red, whereas the bases are yellow. This region is also marked by the lavender-shaded area. (B) Region corresponding to U1 snRNA binding for the wt (i) and mutated (ii) pre-mRNA models at a closer view. Color coding is as observed in panel A. The residues that bind to U1 snRNA are numbered in the mutated sequence structure model. In the wt sequence model, the unbound residues corresponding to U1 snRNA binding are marked with lavender-shaded regions. (C) Coarse-grained depiction of the dock of U1 snRNA over the wt (i) and mutated (ii) pre-mRNA sequence structure. Because the depiction is coarse-grained, the entire model is depicted only as a beaded trace. The trace is blue for the pre-mRNA sequence, with only the region corresponding to U1 snRNA binding shown in red. The different putative U1 snRNA docked structures are colored differently. Because only 1 putative dock was observed for the wt sequence, U1 snRNA is shown as light green in the dock between the U1 snRNA and wt pre-mRNA structures. The inset images provide a closer view of the U1 snRNA binding region on the pre-mRNA structures. The proximal regions in the U1 snRNA and wt pre-mRNA structures are marked by the lavender-shaded area.
Secondary structure prediction of pre-mRNA sequence. Secondary structure of the wt pre-mRNA sequence (A) and the mutated pre-mRNA sequence (B) predicted by mfold. Many of the 5′ donor splice site residues (5 residues: −1U, +1G, +2U, +3A, and +7C) that bind to U1 snRNA were observed to be free in the wt sequence but prebound or physically constrained in a double-stranded form in the mutated sequence. The regions corresponding to residues that bind to U1 snRNA are marked by the lavender-shaded area. The mutated residue locations are also marked by the magenta-shaded area and identified by a thick arrow. The inset images in each panel correspond to a close-up view of the region of pre-mRNA that binds to U1 snRNA in the 3-dimensional model of the wt and mutated structures generated from mfold secondary structure prediction on the RNAComposer Web server. The model structure is depicted in stick format (light green). The residues that bind to U1 snRNA are shown in blue. Hydrogen bonds are depicted as magenta dots.
Secondary structure prediction of pre-mRNA sequence. Secondary structure of the wt pre-mRNA sequence (A) and the mutated pre-mRNA sequence (B) predicted by mfold. Many of the 5′ donor splice site residues (5 residues: −1U, +1G, +2U, +3A, and +7C) that bind to U1 snRNA were observed to be free in the wt sequence but prebound or physically constrained in a double-stranded form in the mutated sequence. The regions corresponding to residues that bind to U1 snRNA are marked by the lavender-shaded area. The mutated residue locations are also marked by the magenta-shaded area and identified by a thick arrow. The inset images in each panel correspond to a close-up view of the region of pre-mRNA that binds to U1 snRNA in the 3-dimensional model of the wt and mutated structures generated from mfold secondary structure prediction on the RNAComposer Web server. The model structure is depicted in stick format (light green). The residues that bind to U1 snRNA are shown in blue. Hydrogen bonds are depicted as magenta dots.
The RNA-RNA docking of U1 snRNA (supplemental Figure 6) and our pre-mRNA ab initio structures (mutant and wt) showed an interesting consequence for the mutation upon RNA-RNA interaction. The wt structure U1 snRNA docking showed only 1 putative dock, which was localized proximal to the well-known U1 snRNA–5′ donor splice-site binding region (Figure 3Ci). The mutated structure U1 snRNA docking showed multiple putative docks, but none within the vicinity of the U1 snRNA–5′ donor splice-site binding region (Figure 3Cii). The hypothetical U1 snRNA–5′ donor splice-site complex showed interaction of the wt 5′ss consensus sequence −1 to +5 (UGUAGG) to nucleotides 4 to 9 of U1 snRNA. The nucleotides relevant to this kind of interaction have been illustrated in detail in Figure 5.
Hypothetical U1 snRNA–pre-mRNA complex structure. The main image represents the secondary structure depiction of the hypothetical U1 snRNA–pre-mRNA complex derived from the RNAcofold server. The inset images show the secondary structure and the tertiary structure generated from the same secondary structure prediction in closer detail. In the secondary structure (lower inset), the residues of U1 snRNA are shown in dark green, whereas the residues of the pre-mRNA participating in the interaction with U1 snRNA are red. The canonical base pairing is shown by lines, whereas noncanonical ones are represented by a blue dot. The tertiary structure (upper inset) is depicted in stick format. U1 snRNA is shown in light green, whereas the pre-mRNA interacting residues are red. The rest of the pre-mRNA is gray.
Hypothetical U1 snRNA–pre-mRNA complex structure. The main image represents the secondary structure depiction of the hypothetical U1 snRNA–pre-mRNA complex derived from the RNAcofold server. The inset images show the secondary structure and the tertiary structure generated from the same secondary structure prediction in closer detail. In the secondary structure (lower inset), the residues of U1 snRNA are shown in dark green, whereas the residues of the pre-mRNA participating in the interaction with U1 snRNA are red. The canonical base pairing is shown by lines, whereas noncanonical ones are represented by a blue dot. The tertiary structure (upper inset) is depicted in stick format. U1 snRNA is shown in light green, whereas the pre-mRNA interacting residues are red. The rest of the pre-mRNA is gray.
Discussion
We identified a translationally silent variation in exon 44 of VWF, c.7464C>T, in a type 1 VWD patient with severely reduced VWF:Ag levels. Subsequent RT-PCR analysis proved that the given variant impairs the efficiency of RNA splicing. Our gene analysis showed that the IP inherited this disease-causing silent mutation from her mother, with reduced VWF:Ag levels. A comparison of VWF:Ag levels of the IP (9%) with those of her mother (49%) indicates that the investigated silent mutation accounts for a reduction of only 50% of VWF:Ag levels in plasma. Furthermore, a significant reduction of total VWF biosynthesis in the IP BOECs and the absence of a clear mature VWF band on western blot analysis of cell lysates imply that production of VWF from both gene alleles is disrupted. Although genetic analyses and a full-length VWF mRNA investigation failed to detect any other mutation, a second causative variation inherited from the IP’s father impairing gene transcription or mRNA stability could not be excluded.
In vivo and ex vivo transcript analysis demonstrated that the detected single-nucleotide substitution variation localized 25 nucleotides after the 3′ss and 85 nucleotides downstream of the 5′ donor splice site interrupts the catalytic excision of intron 44 and leads to intron retention. The aberrant transcript with the intronic region insertion leads to premature termination after adding 50 amino acids; this would code a truncated protein that lacks the carboxy-terminal end of the VWF protein required for dimerization of VWF monomers in intracellular VWF processing.36,37 Confocal IF investigations confirmed accumulation of a truncated protein in the ER of the IP’s BOECs, as expected. We therefore conclude that the truncated proteins accumulated in the ER will be degraded and result in quantitative deficiencies in VWF. Intracellular retention of VWF is already known as a common mechanism for type 1 VWD.6
The occurrence of intron retention rather than exon 44 skipping implies that the given mutation does not have any impact on the accuracy of splicing at 3′ss. Therefore, it can be safely concluded that although bioinformatics predictions suggest interference with cis-SREs (supplemental Table 4), this disease-causing silent mutation would not exert its pathological influence through this mechanism. It is classically considered that disease-associated synonymous variants might interfere with exon definition by affecting cis-regulatory elements and lead to exon skipping, as opposed to our aberrantly spliced transcripts in which we observed intron retention. Therefore, we explored the possibility that the variant might have a long-distance/allosteric influence on RNA structure (more specifically, the 5′ss), thereby resulting in intron retention. Using in silico RNA secondary and tertiary structure determination, we have now shown that this is, in fact, the case for our variant. All predicted models for the mutated and wt sequences suggested a change in structure between the wt sequence and the mutated sequence. The typical RNA duplex formation U1:5′ss is mediated via canonical base pairing of the U1 snRNA 5′ terminus to the 5′ss consensus sequence, spanning the positions −3 to +6 (CAG/GURAGU).38,39 However, natural 5′ss exhibit considerable variation in different positions that lead to alternative noncanonical base-pairing registers, including bulged or shifted nucleotides.39-41 In keeping with the variations in natural 5′ss sequences, our hypothetical pre-mRNA–U1 snRNA duplex structure (generated on the RNAcofold server) showed that the residue positions between −1 to +5 of our pre-mRNA participate in interacting with U1 snRNA. All models (tertiary and secondary) suggest that the mutated sequence generates a stable long hairpin at the 5′ss that structurally constrains the residues at this site from binding to U1 snRNA. This does not happen in the wt sequences in which most residues are unbound and available for interaction with U1 snRNA. This finding was further confirmed by docking U1 snRNA with the mutated and wt pre-mRNA structures.
In line with our finding, recent studies involving genome-wide RNA structure profiling demonstrated universal secondary structure features in human transcripts, in which 5′ss were found to be less structured.42,43 In addition, a genome-wide RNA structure profiling of Arabidopsis thaliana revealed native stable structures at the 5′ss of unspliced pre-mRNAs in alternative spliced transcripts, which suggests that secondary structure inhibits the first step of the splicing.42,44 Moreover, a previous survey showed that synthetic stable hairpins sequestering 5′ss had a strong inhibitory effect on splicing.15
Although the role of RNA structure in regulation of splicing is understood, the impact of disease-associated single-nucleotide variants on pre-mRNA structure, which could putatively affect the efficiency of RNA splicing, is not adequately studied, except for few human genes such as survival motor neuron and tau genes.14,15 Nevertheless, studies such as the one reported on genome variations associated with spinal muscular atrophy disease in the survival motor neuron gene have demonstrated the influence of exonic mutations located in vicinity of the 5′ss on pre-mRNA folding.15,45,46 Our study adds to these findings by suggesting that the exonic variations not only have a localized influence but they also can mediate distant but cis changes by bringing about allosteric modifications in pre-mRNA structure and fold. This is clearly demonstrated by our variation, which is located in an exon and closer to the 3′ss but influences the 5′ss.
In conclusion, this study revealed a novel pathomolecular mechanism by which a disease-causing silent mutation exerts major allosteric changes in RNA folding, thereby impairing the splicing process. Our study, therefore, highlights the importance of investigating the putative impact of silent/exonic variations on pre-mRNA structure.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Jan Voorberg for technical advice on culturing BOECs.
This work was supported by a research grant from Grifols Deutschland GmbH (J.O. and H.Y.).
Authorship
Contribution: H.Y. designed the study, performed the experimental work, interpreted the data, and wrote the paper; A.B. designed and performed the molecular modeling and structural analysis, performed the confocal image analysis, and wrote the paper; M.S.A. performed the western blotting; J.D. evaluated and interpreted VWF multimers and reviewed and edited the manuscript; V.I. and N.M. contributed with patient data; and J.O. designed and supervised the study, evaluated phenotypic and genotypic data of patients, and reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Johannes Oldenburg, Institute of Experimental Haematology and Transfusion Medicine, University Clinic Bonn, Sigmund-Freud-Str 25, 53105 Bonn, Germany; e-mail: johannes.oldenburg@ukb.uni-bonn.de.
References
Author notes
H.Y. and A.B. contributed equally to this study.