Key Points
Graft-versus-graft alloreactivity after dUCBT involves recognition of mismatched HLA class II alleles by allele-specific CD4+ effector T cells.
Alloreactive donor CD4+ T cells may recognize recipient leukemia if mismatched for individual HLA class II alleles.
Abstract
Although double umbilical cord blood transplantation (dUCBT) in adult patients may be associated with less graft failure compared with single UCBT, hematopoietic recovery generally originates from a single cord blood unit (CBU). CBU predominance is still incompletely understood. We recently showed that blood CD4+ T-cell numbers rapidly increase after dUCBT, and early CD4+ T-cell chimerism predicts for graft predominance. Given the frequent HLA class II allele mismatches between CBUs in dUCBT, we hypothesized that alloreactive HLA class II–specific CD4+ T cells from the “winning” CBU may contribute to rejection of the “loser” CBU. We evaluated whether CD4+ T cells originating from the predominant (PD)-CBU would recognize HLA class II allele mismatches, expressed by the nonengrafting (NE)-CBU. Alloreactive effector CD4+ T cells toward 1 or more mismatched HLA class II alleles of the NE-CBU were detected in 11 of 11 patients, with reactivity toward 29 of 33 (88%) tested mismatches, and the strongest reactivity toward DR and DQ alleles early after dUCBT. Mismatched HLA class II allele-specific CD4+ T cells recognized primary leukemic cells when the mismatched HLA class II allele was shared between NE-CBU and patient. Our results suggest that cytotoxicity exerted by CD4+ T cells from the PD-CBU drives the rapid rejection of the NE-CBU, whose alloreactive effect might also contribute to graft-versus-leukemia.
Introduction
Alternative donors and stem cell sources are increasingly being used for allogeneic hematopoietic stem cell transplantation (HSCT) in patients with high-risk hematologic diseases.1-3 Results with single umbilical cord blood transplantation (sUCBT) in adults show similar outcomes to transplants from unrelated donors in terms of overall survival.4 However, the incidence of graft failure may be higher in recipients of sUCBT, especially if the total nucleated cell dose (TNC) is <2.5 × 107/kg body weight.4,5
Double UCBT (dUCBT) was developed to increase the cell dose and appeared to be associated with less graft failure in adult patients, but with similar recovery kinetics of leukocytes and platelets compared with sUCBT.6 Importantly, following a conditioning regimen without antithymocyte globulin, hematopoietic recovery after dUCBT generally originates from only a single cord blood unit (CBU), designated as the predominant (PD)-CBU.7 Single donor chimerism is readily documented within 3 months after transplant in the majority of patients,6,8,9 but actually may occur as early as 7 to 14 days after transplantation.7,10
Predictability of graft predominance could have clinical value, but to date, no consistent correlations have been found yet between graft predominance and a number of graft characteristics.6-8,11,12 Graft predominance has been suggested to be T cell mediated, but this mechanism is still incompletely understood. A correlation between higher doses of CD3+ T cells and viable hematopoietic progenitor cells (HPCs) with CBU predominance has been suggested.13,14 In addition, an alloreactive graft-versus-graft mechanism was hypothesized by Gutman et al as an important underlying mechanism for CBU predominance based on alloreactive interferon γ (IFNγ)–secreting CD8+ T cells.15
We recently reported that blood CD4+ T-cell numbers show a remarkable increase shortly after dUCBT and that early CD4+ T-cell chimerism predicts for graft predominance.7,16 Furthermore, antigen-naïve T cells in UCB require HLA class II on antigen-presenting cells and CD4 T-cell involvement for effective CD4 and CD8 T-cell immune reconstitution.17
Given the many HLA class II mismatches that generally exist between the 2 CBUs, we hypothesized that HLA class II–specific CD4+ T cells from the PD-CBU may mediate an alloreactive response toward the nonengrafting (NE)-CBU. As a consequence, mismatched HLA class II allele-specific CD4+ T cells would recognize primary leukemic cells from the patient in case the mismatched allele is expressed by the patient and thereby contribute to a graft-versus-leukemia (GVL) effect.
To investigate the above hypotheses, we evaluated whether CD4+ T cells obtained shortly after transplantation would recognize HLA class II allele mismatches expressed by the NE-CBU and primary leukemic cells (PLCs). As this investigation is challenged by limited availability of peripheral blood lymphocytes during the early phase of immune reconstitution, we developed a set of unique tools to propagate small amounts of patient T cells in vitro and to dissect immune reactivity of these T cells toward mismatched HLA class II alleles.
Methods
Patients and CBUs
Patients had undergone transplantation according to the Dutch multicenter, dUCBT trial protocol HOVON106 (EudraCTnr: 2008-000053-35).7,16 The trial protocol was approved by the Dutch Central Committee on Research Involving Human Subjects and was conducted according to the principles of the Declaration of Helsinki.
During the selection of the CBUs,7,16 HLA matching was performed at the split antigen level for HLA-A and -B and at the high-resolution level for HLA-DRB1. The minimal match grade required was 4/6 between individual units, as well as between units and recipient. Additional high-resolution typing for HLA-A, -B, -C, -DQB1, and -DPB1 was performed retrospectively.
Blood sampling and chimerism analysis
Peripheral blood (PB) samples were collected at 1, 2, 3, 6, 9, and 12 months after dUCBT for chimerism analysis, as well as isolation and cryopreservation of PB mononuclear cells (PBMCs).
Chimerism analysis, based on quantitative amplification of informative short tandem repeat (STR) regions by polymerase chain reaction (PCR),18 was performed on fresh unseparated PB cells, isolated peripheral blood T cells, and unseparated bone marrow at day +32. Complete single donor T-cell chimerism was defined as >95% donor T cells of a single CBU donor and <5% recipient or second donor T cells.
Flow cytometry
The monoclonal antibodies (mAbs) anti-CD3 peridinin chlorophyll (PerCP), anti-CD4 phycoerythrin (PE), anti-CD137 allophycocyanin (APC), anti-CD134 PE, anti-CD279 (PD1) APC, anti-CD107a PE, and anti-IFNγ PE were obtained from Becton Dickinson Biosciences (BD; San Jose, CA). Anti-CD4 fluorescein isothiocyanate mAb was from Beckman Coulter (Miami, FL). During data acquisition on a fluorescence-activated cell sorter Canto flow cytometer (BD), list mode data from 50 000 T cells, defined as CD3+, SSClow events, were collected. Analysis was performed by sequential selection of the cells of interest and quadrant statics using FCS Express 4 software (BD) (supplemental Figure 3A, available on the Blood Web site).
Cell lines and primary leukemic cells
A library of stimulator cells expressing a single HLA class II allele was generated by retroviral transduction of HLA class II–negative HeLa cells with different combinations of HLA-DRA1/B1-5, HLA-DQA1/B1, or HLA-DPA1/B1 molecules, as previously described19 (supplemental Table 1). HeLa cells were grown in Dulbecco’s modified Eagle medium (DMEM) medium (Lonza, Verviers, Belgium) supplemented with 10% fetal bovine serum (Greiner Bio-one, Alphen a/d Rijn, The Netherlands), 2 mM l-glutamin (Lonza), and 1% penicillin/streptomycin (Lonza), which will be referred to as complete DMEM hereafter.
Primary leukemic cells were isolated from patient PB at study entry and cryopreserved in liquid nitrogen. After thawing cells were rested overnight in complete DMEM prior to testing.
HLA class II allele-specific T-cell propagation
Assessment of the CD4+ T-cell HLA class II alloreactivity is challenged by the limitations of both very low T-cell numbers early after dUCBT and low frequencies of allo-reactive T cells. T cells were successfully expanded by means of in vitro HLA class II allele-specific stimulation, as previously described,20 which generated sufficient T-cell numbers to allow dissection of the immune reactivity of these T cells toward mismatched HLA class II alleles (supplemental Information).
T-cell propagation was performed in complete Iscove modified Dulbecco medium (Invitrogen, Grand Island, NY) containing 6% human serum (pool of 5 male healthy donors; Sanquin Blood Supply, Amsterdam, The Netherlands). Post-dUCBT PBMCs were cocultured with irradiated (40 Gy) HeLa cells transduced with specific HLA class II mismatch alleles of the NE-CBU (ratio 2:1) and with sequential addition of interleukin 7 (IL-7; from day 2 at 10 ng/mL; Miltenyi), IL-15 (from day 2 at 1 ng/mL; Miltenyi), and IL-2 (from day 3, escalating from 15 to 360 IU/mL; Chiron, Amsterdam, The Netherlands) and cultured for 14 to 21 days. Subsequently, these propagated T-cell cultures were subjected to assessment of HLA class II allele-specific T-cell alloreactivity.
Assessment of HLA class II allele-specific alloreactivity
Patient PBMCs after dUCBT or propagated T cells were cocultured in complete Iscove modified Dulbecco medium with HeLa cells transduced with a specific mismatched HLA class II molecule derived from the NE-CBU (at a ratio of 1:1) and at 2 to 20 hours followed by flow cytometry analysis of the T cells for measures of antigen-specific recognition. We assessed upregulation of T-cell activation antigens CD137, CD134, and CD297 (PD-1) (in a 20-hour assay) and of effector markers, such as the degranulation marker CD107 (2-hour assay) or IFNγ production (6-hour assay) within the CD4+ and CD4− (≈CD8+) T-cell subsets.
The measure of upregulation of T-cell activation antigens, typically CD137, was expressed as fold increase (FI), which reflected the proportion of T cells expressing a particular marker after an HLA class II allele HeLa stimulation divided by the proportion of T cells expressing that marker following stimulation with the control (empty) HeLa cell (of which the minimum value was set to 1). A T-cell response was defined positive at an FI exceeding 3.
Statistical analysis
The Student t test was used to compare datasets. All tests were 2 sided, and P < .05 was considered statistically significant.
Results
Characteristics of patient samples and HLA mismatches between CBUs
Patient samples were obtained from participants in the HOVON106 study.7,16 Eleven patients could be studied in-depth, as they met the following criteria: (1) successful engraftment; (2) ultimately single donor T-cell chimerism, (3) availability of cryopreserved PBMC samples early after dUCBT; and (4) HLA class II mismatch between PD- and NE-CBU for which a HLA class II–transduced HeLa cell was available. Patient characteristics are presented in Table 1. From these patients, 19 PBMC samples taken between 1.1 and 6.7 months (median, 2.1 months) after dUCBT were analyzed. PBMC sample characteristics are presented in Table 2. Single unit T-cell chimerism was confirmed in 15 samples by STR PCR, and for the remaining 4 samples, single unit chimerism was shown in unfractionated blood or bone marrow specimens. At the time of sampling, the absolute numbers of CD3+ T cells in blood were very low (ie, 0.207 × 109/L; median; range, 0.03-0.699 × 109/L; Figure 1A), of which the majority (ie, median 74%; range, 41-96%) were CD4+ T cells and the proportion of CD4+ T cells (expressed as fraction of CD3+ T cells) exceeded the upper normal limit in 41% (7 of 17) post-dUCBT PBMC samples (Figure 1B).
Patients' characteristics and follow-up
| Patient . | Age (years) . | Sex . | Diagnosis . | CBU TNC (×107/kg)* . | HLA match PD-CBU vs† . | Status per May 2016 . | aGVHD . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Recipient class I/II . | NE-CBU class I/II . | Follow-up . | Relapse . | ||||||||
| CBU1 . | CBU2 . | Months . | Alive . | ||||||||
| 1 | 56 | Male | AML | 3.6 | 2.1 | 4/5 | 3/5 | 90+ | Yes | No | Yes |
| 2 | 66 | Female | foll NHL | 3.2 | 4.3 | 3/3 | 6/3 | 43 | No | No | No |
| 3 | 31 | Female | AML | 1.8 | 1.6 | 2/2 | 6/5 | 83+ | Yes | No | No |
| 4 | 42 | Female | CLL | 3.9 | 2.8 | 4/2 | 5/3 | 2 | No | No | Yes |
| 5 | 65 | Male | B-ALL | 3.2 | 3.2 | 3/4 | 2/2 | 27+ | Yes | No | Yes |
| 6 | 53 | Female | T-ALL | 1.7 | 2.8 | 3/5 | 3/4 | 11 | No | No | No |
| 7 | 35 | Male | CML | 2.4 | 2.3 | 4/4 | 6/5 | 60+ | Yes | No | Yes |
| 8 | 39 | Female | AA | 3.4 | 1.9 | 4/1 | 3/2 | 7 | No | No | Yes |
| 9 | 54 | Female | AML | 2.6 | 5.1 | 6/2 | 6/0 | 11 | No | No | Yes |
| 10 | 46 | Male | AML | 2.9 | 2.3 | 5/3 | 5/5 | 54+ | Yes | No | Yes |
| 11 | 62 | Male | AML | 2.0 | 2.8 | 4/5 | 2/5 | 8 | No | Yes | Yes |
| Median | 53 | 2.9 | 2.8 | 4/3 | 5/4 | 43 | |||||
| Patient . | Age (years) . | Sex . | Diagnosis . | CBU TNC (×107/kg)* . | HLA match PD-CBU vs† . | Status per May 2016 . | aGVHD . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Recipient class I/II . | NE-CBU class I/II . | Follow-up . | Relapse . | ||||||||
| CBU1 . | CBU2 . | Months . | Alive . | ||||||||
| 1 | 56 | Male | AML | 3.6 | 2.1 | 4/5 | 3/5 | 90+ | Yes | No | Yes |
| 2 | 66 | Female | foll NHL | 3.2 | 4.3 | 3/3 | 6/3 | 43 | No | No | No |
| 3 | 31 | Female | AML | 1.8 | 1.6 | 2/2 | 6/5 | 83+ | Yes | No | No |
| 4 | 42 | Female | CLL | 3.9 | 2.8 | 4/2 | 5/3 | 2 | No | No | Yes |
| 5 | 65 | Male | B-ALL | 3.2 | 3.2 | 3/4 | 2/2 | 27+ | Yes | No | Yes |
| 6 | 53 | Female | T-ALL | 1.7 | 2.8 | 3/5 | 3/4 | 11 | No | No | No |
| 7 | 35 | Male | CML | 2.4 | 2.3 | 4/4 | 6/5 | 60+ | Yes | No | Yes |
| 8 | 39 | Female | AA | 3.4 | 1.9 | 4/1 | 3/2 | 7 | No | No | Yes |
| 9 | 54 | Female | AML | 2.6 | 5.1 | 6/2 | 6/0 | 11 | No | No | Yes |
| 10 | 46 | Male | AML | 2.9 | 2.3 | 5/3 | 5/5 | 54+ | Yes | No | Yes |
| 11 | 62 | Male | AML | 2.0 | 2.8 | 4/5 | 2/5 | 8 | No | Yes | Yes |
| Median | 53 | 2.9 | 2.8 | 4/3 | 5/4 | 43 | |||||
Patients (except patients 1 and 6) were treated in the Hovon 106 study,7 with a preconditioning regimen of Flu/Cy/TBI 2x2Gy and all received graft-versus-host disease prophylaxe (cyclosporin A and mycophenolate mofetil). AA, aplastic anemia; AML, acute myeloid leukemia; aGVHD, acute graft-versus-host disease; B-ALL, acute B lymphoblastic leukemia; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; Cy, cyclophosphamide; Flu, fludarabine; foll NHL, follicular non-Hodgkin lymphoma; T-ALL, acute T lymphoblastic leukemia; TBI, total body irradiation; TNC, total nucleated cell number.
Total nucleated cell count per kilogram recipient weight resulting from CBU1 and CBU2.
HLA match grade at HLA class I (HLA-A,-B,-C) allele level, maximum 6 (=6/6) and HLA class II (HLA-DRB1,-DQB1,-DPB1) allele level, maximum 6 (=6/6) between the PD-CBU and the recipient and between the PD-CBU and the NE-CBU.
Characteristics of post-dUCBT patient PBMC samples
| Patient no. . | Months post-dUCBT . | Donor T-cell chimerism* . | CD3+ cells (109/L) . | CD3+CD4+ cells (% of T cells) . |
|---|---|---|---|---|
| 1 | 2.1 | >95% D2 | 0.226 | 88.9 |
| 1 | 2.9 | >95% D2 | 0.166 | 96.4 |
| 2 | 2.1 | >95% D1 | 0.487 | 80.1 |
| 2 | 3.4 | >95% D1 | 0.699 | 94.1 |
| 3 | 2.0 | >95% D2 | 0.093 | 69.9 |
| 3 | 3.9 | ND† | 0.282 | 67.4 |
| 4 | 1.1 | >95% D1 | 0.546 | 58.4 |
| 5 | 1.1 | >95% D2 | 0.139 | 93.5 |
| 6 | 2.0 | ND† | 0.072 | 73.6 |
| 6 | 3.0 | >95% D2 | 0.442 | 58.8 |
| 6 | 6.0 | >95% D2 | 0.290 | 40.7 |
| 7 | 3.5 | ND†, 92% R | 0.034 | 88.2 |
| 7 | 6.7 | 2% D1, <5% D2† | 0.030 | 86.7 |
| 8 | 5.3 | >95% D1 | 0.155 | 91.0 |
| 9 | 2.1 | >95% D1 | 0.207 | 68.1 |
| 10 | 1.1 | >95% D2 | 0.059 | 72.9 |
| 10 | 2.1 | >95% D2 | 0.211 | 74.4 |
| 11 | 1.1 | >95% D1 | 0.102 | 66.6 |
| 11 | 2.8 | >95% D1 | 0.452 | 41.8 |
| Patient no. . | Months post-dUCBT . | Donor T-cell chimerism* . | CD3+ cells (109/L) . | CD3+CD4+ cells (% of T cells) . |
|---|---|---|---|---|
| 1 | 2.1 | >95% D2 | 0.226 | 88.9 |
| 1 | 2.9 | >95% D2 | 0.166 | 96.4 |
| 2 | 2.1 | >95% D1 | 0.487 | 80.1 |
| 2 | 3.4 | >95% D1 | 0.699 | 94.1 |
| 3 | 2.0 | >95% D2 | 0.093 | 69.9 |
| 3 | 3.9 | ND† | 0.282 | 67.4 |
| 4 | 1.1 | >95% D1 | 0.546 | 58.4 |
| 5 | 1.1 | >95% D2 | 0.139 | 93.5 |
| 6 | 2.0 | ND† | 0.072 | 73.6 |
| 6 | 3.0 | >95% D2 | 0.442 | 58.8 |
| 6 | 6.0 | >95% D2 | 0.290 | 40.7 |
| 7 | 3.5 | ND†, 92% R | 0.034 | 88.2 |
| 7 | 6.7 | 2% D1, <5% D2† | 0.030 | 86.7 |
| 8 | 5.3 | >95% D1 | 0.155 | 91.0 |
| 9 | 2.1 | >95% D1 | 0.207 | 68.1 |
| 10 | 1.1 | >95% D2 | 0.059 | 72.9 |
| 10 | 2.1 | >95% D2 | 0.211 | 74.4 |
| 11 | 1.1 | >95% D1 | 0.102 | 66.6 |
| 11 | 2.8 | >95% D1 | 0.452 | 41.8 |
Predominant donor 1 (D1) or D2 CBU; method STR PCR analysis on magnetic-activated cell sorting–sorted peripheral blood CD3+ T cells.
No data (ND) T-cell chimerism, but PB/BM >95% D2 (patient 3, patient 6); PB/BM >95% D1 (pat 7).
Characteristics of post-dUCBT patient PBMC samples before and after HLA class II allele-specific T-cell propagation. (A) Absolute numbers of CD3+ T cells in PB samples analyzed for HLA class II allele immune recognition. Graphs represent individual and median (line) values. Dotted line, lower limit of normal values of CD3+ T cells in blood of healthy individuals. (B) Percentage of CD4+ T cells in PB (before) and in T-cell cultures after HLA class II–specific amplification, with either HeLa cells transduced with HLA class II mismatched alleles (after, MM) or with HLA class II third-party alleles (after, 3P). Depending on the number of mismatched alleles in the dUCBT combination, from 1 to 7 T-cell amplification cultures per PB sample were performed. Graphs represent individual and median (line) observations. Gray area, normal range of CD3+CD4+ values in blood of healthy individuals; significance in nonpaired t tests is shown. (C) Increase in T-cell numbers in post-dUCBT PBMC samples following processing in HLA class II–specific amplification cultures, expressed as fold increase relative to the T-cell number at start of culture (see B).
Characteristics of post-dUCBT patient PBMC samples before and after HLA class II allele-specific T-cell propagation. (A) Absolute numbers of CD3+ T cells in PB samples analyzed for HLA class II allele immune recognition. Graphs represent individual and median (line) values. Dotted line, lower limit of normal values of CD3+ T cells in blood of healthy individuals. (B) Percentage of CD4+ T cells in PB (before) and in T-cell cultures after HLA class II–specific amplification, with either HeLa cells transduced with HLA class II mismatched alleles (after, MM) or with HLA class II third-party alleles (after, 3P). Depending on the number of mismatched alleles in the dUCBT combination, from 1 to 7 T-cell amplification cultures per PB sample were performed. Graphs represent individual and median (line) observations. Gray area, normal range of CD3+CD4+ values in blood of healthy individuals; significance in nonpaired t tests is shown. (C) Increase in T-cell numbers in post-dUCBT PBMC samples following processing in HLA class II–specific amplification cultures, expressed as fold increase relative to the T-cell number at start of culture (see B).
High-resolution HLA class I and II typing results are presented in Table 3. HLA class II allele mismatches between the PD-CBU and both NE-CBU and recipient are depicted in Table 4. The median number of HLA class II mismatch alleles between the 2 units was 3 (range, 1-7). In total, 33 of the 34 HLA class II mismatch alleles were analysed for T-cell response including 16 mismatched DR alleles, of which 7 were paralog alleles (9 of 16 were unique), 7 were mismatched DQ alleles (6 unique), and 10 were mismatched DP alleles (7 unique). The total number of unique alleles that was tested was 22. Nineteen post-dUCBT PBMC samples were analyzed for HLA-class II allele-specific T-cell alloreactivity.
High-resolution HLA class II typing of recipient and CBUs
| Patient no. . | . | HLA-A . | HLA-B . | HLA-C . | HLA-DRB1* . | HLA-DQ . | HLA-DP . |
|---|---|---|---|---|---|---|---|
| 1 | Recipient | 03:01, 32:01 | 27:05, 40:01 | 02:02, 03:04 | 08:01, 13:01 | 04:02, 06:03 | 04:01, 02:02 |
| 1 | PD-CBU | 03:01, 02:01 | –, 40:01 | –, 03:04 | 08:01, 13:01 | 04:02, 0603 | 04:01, 14:01 |
| 1 | NE-CBU | 03:01, 23:01 | 40:02, 40:01, | 02:02, 03:04 | 08:01, 13:01 | 04:02, 06xx | 04:01, 04:02 |
| 2 | Recipient | 02:05, 68:02 | 14:02, 50:01 | 06:02, 08:02 | 04:01, 04:08 | 03:01, 03:02 | 04:01, 04:02 |
| 2 | PD-CBU | 02:01, 68:02 | 14:02, 44:02 | 05:01, 08:02 | 04:01, 13:03 | 03:01,– | 04:01,– |
| 2 | NE-CBU | 02:01, 68:02 | 14:02, 44:xx | 05:01, 08:02 | 04:01, 01:02 | 03:01, 05:01 | 04:01, 13:01 |
| 3 | Recipient | 23:01, 24:03 | 38:01, 57:03 | 07:01, 12:03 | 03:02, 13:01 | 02:03, 06:03 | 02:01, 01:01 |
| 3 | PD-CBU | 24:02, 26:01 | 38:01, 57:01 | 06:02, 12:03 | 07:01, 13:01 | 03:03, 06;03 | –, 04:01 |
| 3 | NE-CBU | 24;02, 26:01 | 38:01, 57:01 | 06:02, 12:03 | 07:01, 13:01 | 03:03, 06:03 | 02:01, 04:01 |
| 4 | Recipient | 02:01, 32:01 | 15:01, 37:01 | 03:04, 06:02 | 03:01, 10:01 | 02:01, 05:01 | 01:01, 0201 |
| 4 | PD-CBU | 02:01, 25:01 | 15:01, 37:01 | 03:03, 06:02 | 15:01, 10:01 | 06:02, 05:01 | 04:01,– |
| 4 | NE-CBU | 01:01, 02:01 | 15:01, 37:01 | 03:03, 06:02 | 08:01, 10:01 | 04:02, 05:01 | 04:01, 02:01 |
| 5 | Recipient | 01:01, 24:02 | 15:01, 44:02 | 01:02,– | 01:01, 16:01 | 05:01, 05:02 | 02:01, 04:02 |
| 5 | PD-CBU | 01:01, 24:02 | 35:01, 44:05 | 02:02, 04:01 | 01:01, 16:01 | 05:01, 05:02 | 01:01, 16:01 |
| 5 | NE-CBU | 01:01, 24:02 | 15:01, 44:03 | 03:03, 07:01 | 01:01, 07:01 | 05:01, 02:02 | 10:01, 15:01 |
| 6 | Recipient | 25:01, 31:01 | 49:01, 57:01 | 06:02, 07:01 | 01:01, 08:01 | 04:02, 05:04 | 04:01, 16:01 |
| 6 | PD-CBU | 11:01, 31:01 | 39:06, 49:01 | 07:01, 07:02 | 01:01, 08:01 | 04:02, 05:04 | 03:01, 04:01 |
| 6 | NE-CBU | 26:01, 31:01 | 49:01, 15:01 | 03:03, 07:01 | 01:01, 08:01 | 04:02, 05:04 | 02:01, 13:01 |
| 7 | Recipient | 01:01, 31:01 | 44:02, 57:01 | 02:02, 06:02 | 04:04, 08:06 | 03:02, 05:01 | ND |
| 7 | PD-CBU | 01:01, 31:01 | 40:01, 57:01 | 03:04, 06:02 | 04:04, 07:01 | 03:02, 03:03 | 04:01,– |
| 7 | NE-CBU | 01:01, 31:01 | 40:01, 57:01 | 03:04, 06:02 | 04:04, 07:01 | 03:02, 03:03 | 04:01, 06:01 |
| 8 | Recipient | 03:01, 68:01 | 53:01, 57:03 | 06:02, 07:01 | 09:01, 14:54 | 02:02, 0:503 | 17:01,– |
| 8 | PD-CBU | 03:01, 68:02 | 53:01, 57:03 | 04:01, 07:18 | 13:02, 15:03 | 06:09, 06:02 | 02:01,– |
| 8 | NE-CBU | 03:02, 68:02 | 53:01, 57:01 | 04:01, 06:02 | 07:01, 15:01 | 03:03, 06:02 | 04:01,– |
| 9 | Recipient | 01:01, 31:01 | 37:01, 40:01 | 03:04, 06:02 | 01:01, 13:03 | 03:01, 05:01 | 04:01,– |
| 9 | PD-CBU | 01:01, 31:01 | 37:01, 40:01 | 03:04, 06:02 | 01:01, 10:01 | 05:01, 05:01 | 02:01, 04:02 |
| 9 | NE-CBU | 01:01, 31:01 | 37:01, 40:01 | 03:04, 06:02 | 04:04, 15:01 | 03:02, 06:02 | 04:01,– |
| 10 | Recipient | 31:01, 33:01 | 07:02, 14:02 | 07:02, 08:02 | 04:03, 15:01 | 03:02, 06:02 | 03:01, 04:01 |
| 10 | PD-CBU | 03:01, 33:01 | 07:xx, 14:02 | 07:02, 08:02 | 01:02, 15:01 | 05:01, 06:02 | 02:01, 04:01 |
| 10 | NE-CBU | 31:01, 33:01 | 07:02, 14:02 | 07:02, 08:02 | 11:04, 15:01 | 03:01, 06:02 | 02:01, 04:01 |
| 11 | Recipient | 01:01, 11:01 | 07:02, 44:02 | 07:02, 07:04 | 04:04, 11:01 | 03:01, 03:02 | 03:01,– |
| 11 | PD-CBU | 01:01, 01:xx | 07:02, 44:02 | 07:02, 05:01 | 04:04, 11:01 | 03:01, 03:02 | 04:01,– |
| 11 | NE-CBU | 01:01, 29:02 | 07:02, 44:03 | 04:01, 07:01 | 15:01, 11:01 | 03:01, 06:02 | 04:01,– |
| Patient no. . | . | HLA-A . | HLA-B . | HLA-C . | HLA-DRB1* . | HLA-DQ . | HLA-DP . |
|---|---|---|---|---|---|---|---|
| 1 | Recipient | 03:01, 32:01 | 27:05, 40:01 | 02:02, 03:04 | 08:01, 13:01 | 04:02, 06:03 | 04:01, 02:02 |
| 1 | PD-CBU | 03:01, 02:01 | –, 40:01 | –, 03:04 | 08:01, 13:01 | 04:02, 0603 | 04:01, 14:01 |
| 1 | NE-CBU | 03:01, 23:01 | 40:02, 40:01, | 02:02, 03:04 | 08:01, 13:01 | 04:02, 06xx | 04:01, 04:02 |
| 2 | Recipient | 02:05, 68:02 | 14:02, 50:01 | 06:02, 08:02 | 04:01, 04:08 | 03:01, 03:02 | 04:01, 04:02 |
| 2 | PD-CBU | 02:01, 68:02 | 14:02, 44:02 | 05:01, 08:02 | 04:01, 13:03 | 03:01,– | 04:01,– |
| 2 | NE-CBU | 02:01, 68:02 | 14:02, 44:xx | 05:01, 08:02 | 04:01, 01:02 | 03:01, 05:01 | 04:01, 13:01 |
| 3 | Recipient | 23:01, 24:03 | 38:01, 57:03 | 07:01, 12:03 | 03:02, 13:01 | 02:03, 06:03 | 02:01, 01:01 |
| 3 | PD-CBU | 24:02, 26:01 | 38:01, 57:01 | 06:02, 12:03 | 07:01, 13:01 | 03:03, 06;03 | –, 04:01 |
| 3 | NE-CBU | 24;02, 26:01 | 38:01, 57:01 | 06:02, 12:03 | 07:01, 13:01 | 03:03, 06:03 | 02:01, 04:01 |
| 4 | Recipient | 02:01, 32:01 | 15:01, 37:01 | 03:04, 06:02 | 03:01, 10:01 | 02:01, 05:01 | 01:01, 0201 |
| 4 | PD-CBU | 02:01, 25:01 | 15:01, 37:01 | 03:03, 06:02 | 15:01, 10:01 | 06:02, 05:01 | 04:01,– |
| 4 | NE-CBU | 01:01, 02:01 | 15:01, 37:01 | 03:03, 06:02 | 08:01, 10:01 | 04:02, 05:01 | 04:01, 02:01 |
| 5 | Recipient | 01:01, 24:02 | 15:01, 44:02 | 01:02,– | 01:01, 16:01 | 05:01, 05:02 | 02:01, 04:02 |
| 5 | PD-CBU | 01:01, 24:02 | 35:01, 44:05 | 02:02, 04:01 | 01:01, 16:01 | 05:01, 05:02 | 01:01, 16:01 |
| 5 | NE-CBU | 01:01, 24:02 | 15:01, 44:03 | 03:03, 07:01 | 01:01, 07:01 | 05:01, 02:02 | 10:01, 15:01 |
| 6 | Recipient | 25:01, 31:01 | 49:01, 57:01 | 06:02, 07:01 | 01:01, 08:01 | 04:02, 05:04 | 04:01, 16:01 |
| 6 | PD-CBU | 11:01, 31:01 | 39:06, 49:01 | 07:01, 07:02 | 01:01, 08:01 | 04:02, 05:04 | 03:01, 04:01 |
| 6 | NE-CBU | 26:01, 31:01 | 49:01, 15:01 | 03:03, 07:01 | 01:01, 08:01 | 04:02, 05:04 | 02:01, 13:01 |
| 7 | Recipient | 01:01, 31:01 | 44:02, 57:01 | 02:02, 06:02 | 04:04, 08:06 | 03:02, 05:01 | ND |
| 7 | PD-CBU | 01:01, 31:01 | 40:01, 57:01 | 03:04, 06:02 | 04:04, 07:01 | 03:02, 03:03 | 04:01,– |
| 7 | NE-CBU | 01:01, 31:01 | 40:01, 57:01 | 03:04, 06:02 | 04:04, 07:01 | 03:02, 03:03 | 04:01, 06:01 |
| 8 | Recipient | 03:01, 68:01 | 53:01, 57:03 | 06:02, 07:01 | 09:01, 14:54 | 02:02, 0:503 | 17:01,– |
| 8 | PD-CBU | 03:01, 68:02 | 53:01, 57:03 | 04:01, 07:18 | 13:02, 15:03 | 06:09, 06:02 | 02:01,– |
| 8 | NE-CBU | 03:02, 68:02 | 53:01, 57:01 | 04:01, 06:02 | 07:01, 15:01 | 03:03, 06:02 | 04:01,– |
| 9 | Recipient | 01:01, 31:01 | 37:01, 40:01 | 03:04, 06:02 | 01:01, 13:03 | 03:01, 05:01 | 04:01,– |
| 9 | PD-CBU | 01:01, 31:01 | 37:01, 40:01 | 03:04, 06:02 | 01:01, 10:01 | 05:01, 05:01 | 02:01, 04:02 |
| 9 | NE-CBU | 01:01, 31:01 | 37:01, 40:01 | 03:04, 06:02 | 04:04, 15:01 | 03:02, 06:02 | 04:01,– |
| 10 | Recipient | 31:01, 33:01 | 07:02, 14:02 | 07:02, 08:02 | 04:03, 15:01 | 03:02, 06:02 | 03:01, 04:01 |
| 10 | PD-CBU | 03:01, 33:01 | 07:xx, 14:02 | 07:02, 08:02 | 01:02, 15:01 | 05:01, 06:02 | 02:01, 04:01 |
| 10 | NE-CBU | 31:01, 33:01 | 07:02, 14:02 | 07:02, 08:02 | 11:04, 15:01 | 03:01, 06:02 | 02:01, 04:01 |
| 11 | Recipient | 01:01, 11:01 | 07:02, 44:02 | 07:02, 07:04 | 04:04, 11:01 | 03:01, 03:02 | 03:01,– |
| 11 | PD-CBU | 01:01, 01:xx | 07:02, 44:02 | 07:02, 05:01 | 04:04, 11:01 | 03:01, 03:02 | 04:01,– |
| 11 | NE-CBU | 01:01, 29:02 | 07:02, 44:03 | 04:01, 07:01 | 15:01, 11:01 | 03:01, 06:02 | 04:01,– |
HLA mismatches between both recipient and NE-CBU vs PD-CBU are depicted in bold.
ND, no data available; –, no second allele identified; xx, allele digits not specified.
In the case of a specific mismatched DRB1 allele (between UCBs), we also tested the mismatching paralogue allele (selection based on frequency prediction); see Table 4.
HLA class II alleles evaluated for immune recognition by post-dUCBT patient PBMCs: mismatched alleles
| Patient no. . | HLA-DR . | B3† . | B4 . | B5 . | HLA-DQ . | HLA-DP . | |||
|---|---|---|---|---|---|---|---|---|---|
| B1 . | B1 . | B1 . | B1 . | B1 . | B1 . | ||||
| 1 | 04:02 | ||||||||
| 2 | 01:02‡(01:01) | 01:01 | 05:01 | 13:01 | |||||
| 3 | 02:01 | ||||||||
| 4 | 08:01 | 04:02 | 02:01 | ||||||
| 5 | 07:01 | 01:01‡(01:03) | 02:02§ | 10:01 | 15:01 | ||||
| 6 | 13:01 | ||||||||
| 7 | 06:01 | ||||||||
| 8 | 07:01 | 15:01 | 01:01 | 03:03 | 04:01 | ||||
| 9 | 04:04‡(04:01) | 15:01 | 01:01‡(01:03) | 01:01 | 03:02 | 06:02 | 04:01 | ||
| 10 | 11:04‡(11:01) | 02:02 | 03:01 | ||||||
| 11 | 15:01 | 01:01 | 06:02 | ||||||
| Patient no. . | HLA-DR . | B3† . | B4 . | B5 . | HLA-DQ . | HLA-DP . | |||
|---|---|---|---|---|---|---|---|---|---|
| B1 . | B1 . | B1 . | B1 . | B1 . | B1 . | ||||
| 1 | 04:02 | ||||||||
| 2 | 01:02‡(01:01) | 01:01 | 05:01 | 13:01 | |||||
| 3 | 02:01 | ||||||||
| 4 | 08:01 | 04:02 | 02:01 | ||||||
| 5 | 07:01 | 01:01‡(01:03) | 02:02§ | 10:01 | 15:01 | ||||
| 6 | 13:01 | ||||||||
| 7 | 06:01 | ||||||||
| 8 | 07:01 | 15:01 | 01:01 | 03:03 | 04:01 | ||||
| 9 | 04:04‡(04:01) | 15:01 | 01:01‡(01:03) | 01:01 | 03:02 | 06:02 | 04:01 | ||
| 10 | 11:04‡(11:01) | 02:02 | 03:01 | ||||||
| 11 | 15:01 | 01:01 | 06:02 | ||||||
Mismatched alleles between predominant CBU and nonengrafting UCB. Allelic variants of DRB1 are linked with either none or one of the paralogue genes DRB3, DRB4, and DRB5. There are related pseudogenes, DRB2, DRB6, DRB7, DRB8, and DRB9, that are not expressed. DRB1*01, DRB1*08, and DRB1*10 have no paralogue genes; DRB1*03 and DRB1*11/12/13/14 have paralogue gene DRB3; DRB1*04 and DRB1*07/09 have paralogue gene DRB4; and DRB1*15 and DRB1*16 have paralogue gene DRB5.
Although DRB1 is expressed at a level 5 times higher than its paralogs, we included in the analysis of a specific DRB1 mismatch allele the most probable DRB1 paralog as well.
Lack of relevant DNA sequences prohibited generation of allele-specific HeLa; most related available allele was used instead (in parentheses).
Generation of HLA-DQB1*02:02 expressing HeLa cell not successful; this allele was not tested.
Post-dUCBT blood T cells show consistent and robust alloreactivity toward mismatched HLA class II alleles of the nonengrafting CBU
First, we defined the optimal method to assess HLA class II alloreactivity in post-dUCBT patient PBMCs (supplemental Materials). In this method, patient post-dUCBT PBMCs were first propagated by HLA class II allele-specific stimulation in coculture with irradiated HeLa cells transduced with a single HLA class II (mismatched or third party neither expressed by the PD-NE nor recipient) molecule (14-21 days), followed by analysis of the T-cell culture for alloreactivity. To analyze T-cell alloreactivity, the propagated T cells were cocultured for 2 to 20 hours with HeLa cells transduced with the selected mismatched, matched, or third party HLA class II alleles followed by flow cytometry assessment of measures of specific immune reactivity in propagated T cells, typically being upregulation of the activation marker CD137 (supplemental Figure 3A).
The HLA class II allele-specific propagation of the post-dUCBT PBMC samples increased T-cell numbers 29-fold (median; range, 2-787) and further skewed post-dUBCT blood T cells toward CD4+ (Figure 1B-C). Results of the alloreactivity assays of the 19 post-dUCBT PBMC samples taken from the 11 patients are summarized in Figures 2 and 3. In all 11 patients, alloreactive CD4+ T-cells toward 1 or more HLA class II mismatched alleles of the NE-CBU were detectable (Figure 2A). In total, CD4+ T-cell alloreactivity toward 29 of 33 (88%) mismatched alleles was detectable, including 15 of 16 (94%) for DR, 7 of 7 (100%) for DQ, and 7 of 10 (70%) for DP alleles. All HLA class II mismatched alleles but 1 (DPB1*04:01) elicited a CD4+ T-cell response. CD4+ T-cell reactivities toward DR and DQ alleles in comparison with DP alleles were higher, with a median fold increase of CD137 upregulation on CD4+ T cells of 10.1-fold (range, 1.1- to 84.6-fold) and 9.8-fold (range, 4.3- to 71.5-fold), respectively, vs 4.3-fold (range, 1.4- to 48.0-fold) for DP alleles, which may be explained by its lower expression.
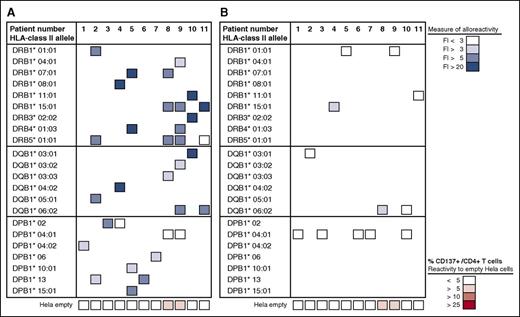
Anti-HLA class II allele-specific immune reactivity in early post-dUCBT PB samples. Nineteen PBMC samples obtained from 11 patients at 1 to 6 months after dUCBT were propagated toward mismatched HLA class II alleles of the nonengrafting CBU and subsequently tested for alloreactivity toward mismatched and matched HLA class II alleles. The response was quantified by CD137 upregulation on CD4+ T cells and presented as fold increase (FI; %CD137+/CD4+) of reactivity toward HeLa cells transduced with (mis)matched HLA class II alleles relative to reactivity toward the nontransduced empty HeLa cell (control) (supplemental Figure 3A). In case >1 PBMC sample per patient was tested, the best result (ie, highest FI) was considered, generally obtained with the PBMC sample with the shortest follow-up after dUBCT. (A) Patient PBMCs were propagated toward mismatched alleles in a 14- to 21-day coculture and subsequently assayed toward the same mismatched alleles (as specified in the left column). The level of response is presented in a 4-color grading scale, as depicted and was as follows: DR alleles, FI = 11.6 (median; range, 2-84; n = 16); DQ alleles, FI = 8.9 (median; range, 4-87; n = 7); DP alleles, FI = 3.6 (median; range, 1.0-12; n = 10). The level of reactivity toward the HLA class II negative (empty) HeLa cell is shown at the bottom row of the figure, is presented as the mean % CD137+/CD4+ of 1 to 7 observations per patient (median %CD137+/CD4+, 2.2; range, 0.5-8.7; n = 11), and is depicted in a 4-color-grading scale. (B) Patient PBMCs were propagated toward 1 to 7 mismatched alleles per patient (as shown in A) and subsequently assayed toward a single matched allele of the engrafting CBU (as specified in the left column). The mean value of response of 1 to 7 observations per patient is presented. The level of response was as follows: DR alleles, FI = 1.6 (median; range, 1.2-3.6; n = 4); DQ alleles, FI = 2.3 (median; range, 1.3-4.3; n = 3); DP alleles, FI = 1.1 (median; range, 0.6-1.6; n = 5; see A).
Anti-HLA class II allele-specific immune reactivity in early post-dUCBT PB samples. Nineteen PBMC samples obtained from 11 patients at 1 to 6 months after dUCBT were propagated toward mismatched HLA class II alleles of the nonengrafting CBU and subsequently tested for alloreactivity toward mismatched and matched HLA class II alleles. The response was quantified by CD137 upregulation on CD4+ T cells and presented as fold increase (FI; %CD137+/CD4+) of reactivity toward HeLa cells transduced with (mis)matched HLA class II alleles relative to reactivity toward the nontransduced empty HeLa cell (control) (supplemental Figure 3A). In case >1 PBMC sample per patient was tested, the best result (ie, highest FI) was considered, generally obtained with the PBMC sample with the shortest follow-up after dUBCT. (A) Patient PBMCs were propagated toward mismatched alleles in a 14- to 21-day coculture and subsequently assayed toward the same mismatched alleles (as specified in the left column). The level of response is presented in a 4-color grading scale, as depicted and was as follows: DR alleles, FI = 11.6 (median; range, 2-84; n = 16); DQ alleles, FI = 8.9 (median; range, 4-87; n = 7); DP alleles, FI = 3.6 (median; range, 1.0-12; n = 10). The level of reactivity toward the HLA class II negative (empty) HeLa cell is shown at the bottom row of the figure, is presented as the mean % CD137+/CD4+ of 1 to 7 observations per patient (median %CD137+/CD4+, 2.2; range, 0.5-8.7; n = 11), and is depicted in a 4-color-grading scale. (B) Patient PBMCs were propagated toward 1 to 7 mismatched alleles per patient (as shown in A) and subsequently assayed toward a single matched allele of the engrafting CBU (as specified in the left column). The mean value of response of 1 to 7 observations per patient is presented. The level of response was as follows: DR alleles, FI = 1.6 (median; range, 1.2-3.6; n = 4); DQ alleles, FI = 2.3 (median; range, 1.3-4.3; n = 3); DP alleles, FI = 1.1 (median; range, 0.6-1.6; n = 5; see A).
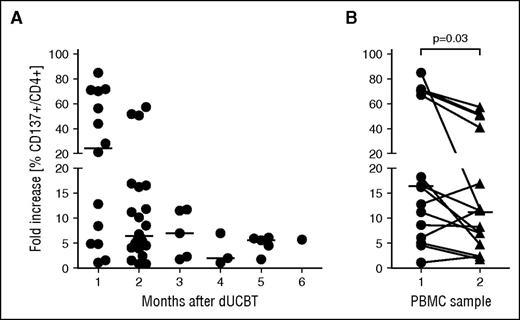
Kinetics of anti-HLA class II allele-specific immune reactivity in early post-dUCBT PB samples. Patient PBMCs were propagated and assayed as described in Figure 2. (A) Immune reactivity toward individual alleles (n = 33) tested in 19 PBMC samples as a function of PBMC sampling time after dUCBT; individual and median (line) values are shown. (B) Immune reactivity toward individual alleles in paired PBMC samples of 5 patients: sample 1, 1 month (median; range, 1-2 months) after dUCBT; sample 2, 2 months (median; range, 2-4 months) after dUCBT. Individual and median (line) values are shown; P value, paired Student t test.
Kinetics of anti-HLA class II allele-specific immune reactivity in early post-dUCBT PB samples. Patient PBMCs were propagated and assayed as described in Figure 2. (A) Immune reactivity toward individual alleles (n = 33) tested in 19 PBMC samples as a function of PBMC sampling time after dUCBT; individual and median (line) values are shown. (B) Immune reactivity toward individual alleles in paired PBMC samples of 5 patients: sample 1, 1 month (median; range, 1-2 months) after dUCBT; sample 2, 2 months (median; range, 2-4 months) after dUCBT. Individual and median (line) values are shown; P value, paired Student t test.
CD8+ T-cell alloreactivity toward transduced HeLa cells was present as well, albeit less prominent than CD4+ T-cell alloreactivity, largely due to the low numbers of CD4− (≈CD8+) T cells. In addition, assessment of CD8+ T-cell alloreactivity by CD137 upregulation was hampered by high background reactivity toward the control (empty) HeLa cells (supplemental Figure 3B).
Notably, the highest CD4+ T-cell alloreactive responses were observed in samples taken at 1 month after UCBT (Figure 3A). In 5 patients, PBMC samples collected at multiple time points after dUCBT were analyzed for CD4+ T-cell alloreactivity toward a total of 15 HLA class II alleles and showed significantly higher alloreactive responses in the sample with the shortest follow-up after dUCBT (t test, P = 0.03; Figure 3B).
Analysis for reactivity toward matched alleles of the PD-CBU showed positive results for 2 of 12 alleles (17%; Table 5; Figure 2B). These propagated T-cell cultures were also analyzed for reactivity toward irrelevant third party alleles (not present in recipient, PD-CBU, or NE-CBU) of the same HLA class (DR, DQ, or DP) and showed positive results for 3 of 17 alleles (18%), of which 2 were DR alleles and 1 was a DQ allele (Table 6; Figure 4A).
HLA class II alleles evaluated for immune recognition by post-dUCBT patient PBMCs: matched alleles
| Patient no. . | HLA-DR (B1) . | HLA-DQ (B1) . | HLA-DP (B1) . |
|---|---|---|---|
| 1 | 04:01 | ||
| 2 | 03:01 | ||
| 3 | 04:01 | ||
| 4 | 15:01 | ||
| 5 | 01:01 | ||
| 6 | 04:01 | ||
| 7 | 04:01 | ||
| 8 | 06:02 | ||
| 9 | 01:01 | ||
| 10 | 06:02 | 04:01 | |
| 11 | 11:01 |
| Patient no. . | HLA-DR (B1) . | HLA-DQ (B1) . | HLA-DP (B1) . |
|---|---|---|---|
| 1 | 04:01 | ||
| 2 | 03:01 | ||
| 3 | 04:01 | ||
| 4 | 15:01 | ||
| 5 | 01:01 | ||
| 6 | 04:01 | ||
| 7 | 04:01 | ||
| 8 | 06:02 | ||
| 9 | 01:01 | ||
| 10 | 06:02 | 04:01 | |
| 11 | 11:01 |
Matched alleles between predominant CBU and nonengrafting UCB.
HLA class II alleles evaluated for immune recognition by post-dUCBT patient PBMCs: third party alleles
| Patient no. . | HLA-DR . | HLA-DQ (B1) . | HLA-DP (B1) . | |
|---|---|---|---|---|
| B1 . | B4 . | |||
| 1 | 01:01 | 06:01 13:01 | ||
| 2 | ||||
| 3 | ||||
| 4 | 07:01 | 03:03 | 13:01 | |
| 5 | 15:01 | 06:02 | 06:01 | |
| 6 | ||||
| 7 | ||||
| 8 | 01:03 | 04:02 | 06:01 | |
| 9 | 08:01 | 04:02 | 13:01 | |
| 10 | 01:01 | |||
| 11 | ||||
| Patient no. . | HLA-DR . | HLA-DQ (B1) . | HLA-DP (B1) . | |
|---|---|---|---|---|
| B1 . | B4 . | |||
| 1 | 01:01 | 06:01 13:01 | ||
| 2 | ||||
| 3 | ||||
| 4 | 07:01 | 03:03 | 13:01 | |
| 5 | 15:01 | 06:02 | 06:01 | |
| 6 | ||||
| 7 | ||||
| 8 | 01:03 | 04:02 | 06:01 | |
| 9 | 08:01 | 04:02 | 13:01 | |
| 10 | 01:01 | |||
| 11 | ||||
Third party alleles, not present in recipient, predominant UCB, or nonengrafting UCB.
Alloreactivity of post-dUCBT blood T cells toward third party HLA class II molecules. (A) PBMCs from 6 patients (patients 1, 4, 5, 8, 9, and 10) were propagated toward 1 to 7 mismatched alleles per patient (as specified in Figure 2A) and subsequently assayed toward a third party allele of the same family (as specified in the left column). The mean value of response of 1 to 4 observations per allele per patient is presented. The level of response was as follows: DR alleles, FI = 2.2 (median; range, 1.4-5.1; n = 6); DQ alleles, FI = 2.3 (median; range, 0.2-4.4; n = 4); DP alleles, FI = 1.5 (median; range, 0.2-2.8; n = 6). (B) PBMCs from 6 patients were propagated toward third party alleles (as specified in the left column) and assayed toward the same third party allele (Figure 2A). The level of response was as follows: DR alleles, FI = 3.3 (median; range, 1.3-21.2; n = 6); DQ alleles, FI = 3.9 (median; range, 1.4-8.8; n = 4); DP alleles, FI = 3.0 (median; range, 1.3-18.6; n = 6). The level of reactivity toward (empty) HeLa cell: median %CD137+/CD4+, 6.8 (range, 2.0-15.0; n = 11).
Alloreactivity of post-dUCBT blood T cells toward third party HLA class II molecules. (A) PBMCs from 6 patients (patients 1, 4, 5, 8, 9, and 10) were propagated toward 1 to 7 mismatched alleles per patient (as specified in Figure 2A) and subsequently assayed toward a third party allele of the same family (as specified in the left column). The mean value of response of 1 to 4 observations per allele per patient is presented. The level of response was as follows: DR alleles, FI = 2.2 (median; range, 1.4-5.1; n = 6); DQ alleles, FI = 2.3 (median; range, 0.2-4.4; n = 4); DP alleles, FI = 1.5 (median; range, 0.2-2.8; n = 6). (B) PBMCs from 6 patients were propagated toward third party alleles (as specified in the left column) and assayed toward the same third party allele (Figure 2A). The level of response was as follows: DR alleles, FI = 3.3 (median; range, 1.3-21.2; n = 6); DQ alleles, FI = 3.9 (median; range, 1.4-8.8; n = 4); DP alleles, FI = 3.0 (median; range, 1.3-18.6; n = 6). The level of reactivity toward (empty) HeLa cell: median %CD137+/CD4+, 6.8 (range, 2.0-15.0; n = 11).
In additional control experiments, post-dUCBT PBMC samples were propagated by third party alleles and showed moderate T-cell expansion in these cultures. That is, cell numbers only increased 12-fold (median; range, 2-98; Figure 1C). CD4+ T-cell reactivity of these cultures toward the matched alleles of the PD-CBU was negligible, whereas CD4+ T-cell alloreactivity toward corresponding third party alleles was shown for 5 of 11 (45%) tested alleles (1 DR, 2 DQ, and 2 DP), and 4 of 9 (44%) unique alleles (1 DR, 2 DQ, and 1 DP; Figure 4B; supplemental Figure 3A,C).
The magnitude of the CD4+ T-cell alloreactive response was significantly higher toward the mismatched alleles, ie, median FI (%CD137+ of CD4+) 7.4 (range, 1.1- to 84.6-fold; n = 33), than toward matched alleles, ie, median FI 1.4 (range, 0.3-12.5; n = 12; t test, P = .04) or third party alleles, ie, median FI 2.9 (range, 1.3-42.5; n = 11; not significant).
We also questioned whether allo-reactive CD4+ T cells would be able to exert additional effector T-cell functions, besides upregulated expression of CD137. Therefore, we included multiple T-cell activation and effector markers in the analysis and showed that allo-reactive CD4+ T cells upregulate multiple T-cell activation (CD134 [data not shown] and PD1), as well as effector markers (IFNγ and CD107; supplemental Figure 3A).
Post-dUCBT blood T cells show alloreactivity toward mismatched HLA class II alleles of primary leukemic cells
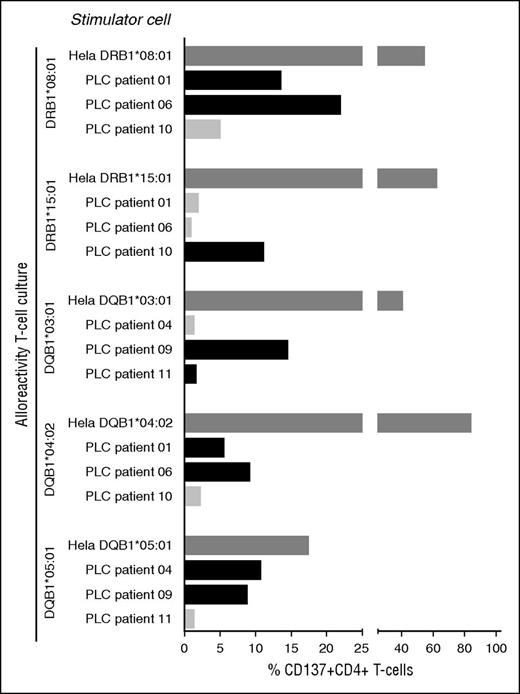
Next, we addressed whether PD-CBU–derived T cells with alloreactivity toward mismatched HLA class II alleles of the NE-CBU would recognize PLCs via mismatched HLA class II alleles shared between the NE-CBU and leukemic cells. As our cohort lacked patients with shared mismatched HLA class II alleles between the NE-CBU and recipient PLCs, we assayed allogeneic combinations. To that end we selected 5 T-cell cultures with reactivity toward DRB1*08:01 (derived from patient 4), DRB1*15:01 (patient 11), DQB1*03:01 (patient 10), DQB1*04:02 (patient 4), and DQB1*05:01 (patient 2). These T-cell cultures were assayed for alloreactivity toward PLCs obtained from 6 patients at study entry and expressing 1 to 2 of the corresponding alleles (Figure 5). Complete HLA class II typing of patients 1, 4, 6, 9, 10, and 11 is presented in Table 3. PLCs of 5 of 6 patients were recognized by the HLA class II alloreactive CD4+ T cells at 1 or 2 alleles, whereas irrelevant combinations were negative (Figure 5). Unexpectedly, the T-cell culture reactive toward DQB1*03:01 did not recognize PLCs of DQB1*03:01-positive patient 11. Cocultures with the corresponding HLA class II expressing HeLa cells confirmed specificity of the alloreactive T cells (positive control). Thus, alloreactive post-dUCBT blood T cells that were amplified by HLA class II HeLa stimulation recognized naturally expressed HLA class II molecules on leukemic cells.
Anti-HLA class II allele-specific immune reactivity in early post-dUCBT PB samples contributes to antileukemic cell reactivity. Post-dUCBT patient PBMC samples were propagated on HeLa cells transduced with mismatched HLA class II alleles of the nonengrafting CBU as specified in Figure 2A. T-cell cultures (n = 5) with proven HLA class II alloreactivity toward DRB1*08:01 (derived from patient 4), DRB1*15:01 (patient 11), DQB1*03:01 (patient 10), DQB1*04:02 (patient 4), and DQB1*05:01 (patient 2) were tested for reactivity toward the corresponding HLA class II molecule expressing HeLa and toward PLCs obtained from 6 patients, ie, patient 1 (DRB1*08:01, DQB1*04:02), patient 4 (DQB1*05:01), patient 6 (DRB1*08:01, DQB1*04:02), patient 9 (DQB1*03:01, DQB1*05:01), patient 10 (DRB1*15:01), and patient 11 (DRB1*11:01, DQB1*03:01). Complete HLA class II typing of the patients is presented in Table 3. x-axis: net % CD137 upregulation, ie, %CD137 upregulation on alloreactive CD4+ T cells following coculture with HLA class II–expressing HeLa cells or PLCs minus reactivity toward the empty HeLa cell (positive response: net CD137 upregulation >5%); y-axis: stimulator cells (Hela or PLC) per alloreactivity of the tested T-cell culture. Dark gray bar: positive control, HLA class II alloreactive T-cell culture with the corresponding HLA class II allele transduced HeLa cell; black bar: HLA class II alloreactive T-cell culture with PLCs expressing the corresponding HLA class II allele; light gray bar: HLA class II alloreactive T-cell culture with PLCs not expressing the corresponding HLA class II allele.
Anti-HLA class II allele-specific immune reactivity in early post-dUCBT PB samples contributes to antileukemic cell reactivity. Post-dUCBT patient PBMC samples were propagated on HeLa cells transduced with mismatched HLA class II alleles of the nonengrafting CBU as specified in Figure 2A. T-cell cultures (n = 5) with proven HLA class II alloreactivity toward DRB1*08:01 (derived from patient 4), DRB1*15:01 (patient 11), DQB1*03:01 (patient 10), DQB1*04:02 (patient 4), and DQB1*05:01 (patient 2) were tested for reactivity toward the corresponding HLA class II molecule expressing HeLa and toward PLCs obtained from 6 patients, ie, patient 1 (DRB1*08:01, DQB1*04:02), patient 4 (DQB1*05:01), patient 6 (DRB1*08:01, DQB1*04:02), patient 9 (DQB1*03:01, DQB1*05:01), patient 10 (DRB1*15:01), and patient 11 (DRB1*11:01, DQB1*03:01). Complete HLA class II typing of the patients is presented in Table 3. x-axis: net % CD137 upregulation, ie, %CD137 upregulation on alloreactive CD4+ T cells following coculture with HLA class II–expressing HeLa cells or PLCs minus reactivity toward the empty HeLa cell (positive response: net CD137 upregulation >5%); y-axis: stimulator cells (Hela or PLC) per alloreactivity of the tested T-cell culture. Dark gray bar: positive control, HLA class II alloreactive T-cell culture with the corresponding HLA class II allele transduced HeLa cell; black bar: HLA class II alloreactive T-cell culture with PLCs expressing the corresponding HLA class II allele; light gray bar: HLA class II alloreactive T-cell culture with PLCs not expressing the corresponding HLA class II allele.
Discussion
Given the observations that following dUCBT, (1) CD4+ T cells show an early expansion, (2) CD4+ T-cell chimerism may predict graft predominance, and (3) many HLA class II mismatches are generally present between the 2 CBUs,7,16 we hypothesized that HLA class II–specific CD4+ T cells of the winning unit (PD-CBU) might exert an immediate and targeted alloreactive immune response toward mismatched HLA class II alleles of the rejected unit (NE-CBU). Here, we generated support for that hypothesis by showing the presence of a CD4+ T-cell alloreactive immune response by post-dUCBT blood T cells toward the majority (88%) of mismatched HLA class II alleles of the NE-CBU shortly after transplantation. Although alloreactivity was detected toward HLA-DR, -DQ, and -DP alleles, the strongest reactivity was observed toward HLA-DR and -DQ alleles. Except for the dose of CD3+ T cells and HPCs in the PD-CBU and TNC of the NE-CBU, to date, no graft characteristics or indications of dominant mismatches have been associated with graft predominance.13,14,21
We applied HeLa stimulator cells, transduced with a single HLA class II molecule,22 to in vitro amplify T cells present in early post-dUCBT PBMCs with alloreactivity toward mismatched HLA class II alleles of the NE-CBU. Following this amplification step, these cultures were characterized by outgrowth of predominantly CD4+ T cells that displayed significant levels of alloreactivity toward the majority of mismatched HLA class II alleles of the NE-CBU, except for HLA-DP*04:01, one of the most frequent HLA-DP molecules in the human population worldwide.23 In these cultures, alloreactivity toward matched and third party alleles was low, but not negligible. In third party allele-specific amplified cultures, the CD4+ T-cell alloreactivity was significant, but was accompanied by high background reactivity toward the control (empty) HeLa cells. This observation may be expected considering the broad naïve T-cell repertoire in UCB, which significantly contributes to the alloreactive T-cell pool.24,25 Our in vitro expansion approach may imply a bias toward a preferred detection of allele-specific CD4+ T cells. Although this urges for cautious interpretation, both the time frame and high percentages observed provide support for a dominant role of alloreactive effector CD4+ T cells. In fact, we documented CD4+ T-cell alloreactivity toward 88% of mismatch alleles of the NE-CBU in all patients. The levels of alloreactivity were highest at 1 month after dUCBT and rapidly declined thereafter. Collectively, these observations suggest an early in vivo expansion of the mismatch-specific alloreactive CD4+ T-cell pool and a rapid decline after the elimination of the nonengrafting unit. Of note, the use of steroids, as was needed for acute graft-versus-host disease treatment in 8 of 11 patients, might possibly have contributed to the latter observation.
Our investigations were dominated by CD4+ T-cell responses, yet CD8+ T cells, present in relatively low numbers, did show alloreactivity, albeit accompanied by relatively high background reactivity toward the control (empty) HeLa cell. The alloreactive CD4+ T cells were armored with T-cell effector mechanisms (degranulation and IFNγ production) and thus are expected to elicit an immediate and targeted (cytotoxic) immune response toward mismatched HLA class II molecules expressed by the HSCs of the NE-CBU. In contrast to Gutman et al,15 who identified alloreactive IFNγ-secreting CD8+ T cells in 14- to 28-day post-dUCBT PBMCs, our findings suggest CD4+ T cells to be a major player in the phenomenon of graft-versus-graft alloreactivity, although we have not directly compared the CD4+ and CD8+ T-cell responses. The presence of a T cell–mediated graft-versus-graft alloreactive immune reaction originally comes from clinical observations26,27 and experimental models.28,29
Clinically, single UCBT is associated with similar relapse rates but reduced incidence and severity of graft-versus-host disease compared with T cell–depleted unrelated donor HSCT, whereas the incidence of relapse may be lower when compared with haploidentical HSCT.1,3,30-34 Interestingly, in adult patients with hematologic malignancies, dUCBT may be associated with a lower incidence of relapse,35-39, possibly due to stronger T-cell alloreactivity. Here, we show that PD-CBU–derived T cells with alloreactivity toward mismatched alleles of the NE-CBU also recognize leukemic cells, when the mismatched HLA class II alleles were shared between NE-CBU and the leukemic cells. The weaker reactivity toward fresh leukemic cells vs HLA class II mismatched HeLa cells might be due to variable expression by leukemic cells of HLA class II molecules and ligands for costimulatory and coinhibitory molecules.40-42 Thus, our data suggest that a CD4+ T cell-mediated graft-versus-graft alloreactivity potentially adds to GVL in the dUCBT setting. A possible contribution of CD4+ T cells in GVL was earlier suggested by Stevanovic et al, showing that alloreactive CD4+ T cells directed against mismatched HLA class II molecules were capable of directly eliminating leukemic cells in an experimental setting.43
To exploit this phenomenon in dUCBT, the 2 CBUs might be selected for HLA class II mismatches between the 2 units that also mismatch with the recipient. In this setting, the generated graft-versus-graft HLA class II alloreactivity may reduce relapse incidence and improve clinical outcome by specific targeting of HLA class II molecules expressed by leukemic cells. It implies also that pursuing HLA class II matching between the 2 units can be abandoned, a goal for which no evidence is available.
Taken together, our results suggest that immediate cytotoxicity exerted by CD4+ T cells from the PD-CBU represents a novel mechanism of rapid rejection of the NE-CBU after dUCBT following a non–antithymocyte globulin conditioning regimen. These observations might possibly also provide an explanation for a lower incidence of graft failure after dUCBT in adult patients compared with single unit UCBT with relatively low (<2.7 TNC/kg) cell counts. It could at least in part result from an immunologic, graft-potentiating effect evoked by mismatched HLA class II alleles expressed by the rejected graft. Furthermore, our findings suggest that graft-versus-graft alloreactivity might potentially facilitate GVL after dUCBT. The latter result argues for reconsidering allele matching for HLA class II between the 2 units. In fact, HLA class II mismatches between the CBUs might even be aimed for, which is to be confirmed in a prospective study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Corrien Groot-van Ruyven and Simone Luxemburg-Heijs for technical assistance.
This work was supported financially by Erasmus Medical Center Translational Research program 2012 and the Dutch Cancer Society (UL 2008-4263).
Authorship
Contribution: C.H.J.L., R.W., C.A.M.v.B., J.A.E.S., E.B., J.W.G., R.D., J.H.F.F., and J.J.C. designed research; R.W., C.A.M.v.B., and C.H.J.L. performed experiments; and C.H.J.L. and R.W. analyzed the data and performed statistical analysis; and C.H.J.L. and J.J.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cor H. J. Lamers, Laboratory of Tumor Immunology, Department of Medical Oncology, Erasmus MC Cancer Institute, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: c.lamers@erasmusmc.nl.