Key Points
Vegfb, Vegfd, and Pgfb can sustain vascular development in the absence of Vegfaa.
Engineering and in vivo validation of dominant negative Vegfaa molecules that block vascular development.
Abstract
The mechanisms that allow cells to bypass anti–vascular endothelial growth factor A (VEGFA) therapy remain poorly understood. Here we use zebrafish to investigate this question and first show that vegfaa mutants display a severe vascular phenotype that can surprisingly be rescued to viability by vegfaa messenger RNA injections at the 1-cell stage. Using vegfaa mutants as an in vivo test tube, we found that zebrafish Vegfbb, Vegfd, and Pgfb can also rescue these animals to viability. Taking advantage of a new vegfr1 tyrosine kinase–deficient mutant, we determined that Pgfb rescues vegfaa mutants via Vegfr1. Altogether, these data reveal potential resistance routes against current anti-VEGFA therapies. In order to circumvent this resistance, we engineered and validated new dominant negative Vegfa molecules that by trapping Vegf family members can block vascular development. Thus, our results show that Vegfbb, Vegfd, and Pgfb can sustain vascular development in the absence of VegfA, and our newly engineered Vegf molecules expand the toolbox for basic research and antiangiogenic therapy.
Introduction
Blood vessels are tubular structures lined by endothelial cells that carry blood and nutrients throughout the body. Abnormal vascular formation is a hallmark of pathology often seen in cancer and in the neovascular form of age-related vascular degeneration. The latter is the leading cause of blindness in humans.1 Vascular endothelial growth factors (VEGFs) and their receptors tightly control processes such as vasculogenesis and angiogenesis during blood vessel development. In mammals, the VEGF family consists of 5 members, VEGFA, B, C, and D and placenta growth factor (PGF). The biological effects of VEGFs are mediated by receptor tyrosine kinases, VEGF receptor 1, 2, and 3 (VEGFR1–3) also known as Fms-related tyrosine kinase 1 (FLT1), KDR, and FLT4, respectively, as well as coreceptors, including neuropilins (NRPs). Biochemical and cell culture studies have shown that VEGFA and B and PGF bind to VEGFR1, that VEGFA binds to VEGFR2, and that VEGFC and D bind to VEGFR3 and, after proteolytic processing, also to VEGFR2.2 Several proangiogenic factors, including VEGFA and PGF, are consistently upregulated during diverse forms of pathological angiogenesis.3
VEGFA blockers have been extensively used in the past few years to treat different types of cancers and hypervascularization conditions including age-related vascular degeneration. However, it is now widely accepted that cells can survive and proliferate even in the absence of VEGFA function.4 Recent studies have implicated additional VEGF family members, notably PGF and VEGFB, in the pathology of ocular vascular diseases and in some cancers.5 Interestingly, PGF-deficient and VEGFB-deficient mice are viable, fertile, and do not exhibit any dramatic vascular phenotype.6 How these factors sustain vascular growth in the absence of VEGFA function remains poorly understood as our current knowledge is mainly derived from in vitro studies.2
Here we used the zebrafish model whose conserved developmental pathways recapitulate human vascular physiology and disease pathology to investigate this question.
Methods
Fish stocks
All zebrafish husbandry was performed under standard conditions in accordance with institutional and national ethical and animal welfare guidelines. The following transgenic line was used for this work: TgBAC(etv2:EGFP)ci1.7
Genome editing
TAL effector nucleases (TALENs) were designed targeting vegfaa (Figure 1A) using TALEN targeter (https://tale-nt.cac.cornell.edu/) and constructed using Golden Gate assembly.8 Zebrafish embryos were injected into the cell at the 1-cell stage with 100 pg total TALEN RNA. Genome engineering was performed as previously described. The vegfaa TALEN recognition sites are GCTGGTAGACATCATC and GAGATCGAGCACACGT. Guide RNA and CAS9 plasmids were purchased from Addgene.9 Clustered regularly interspaced short palindromic repeat (CRISPR) guide RNA targeting vegfab exon 2 was designed using CRISPR design (http://crispr.mit.edu/).
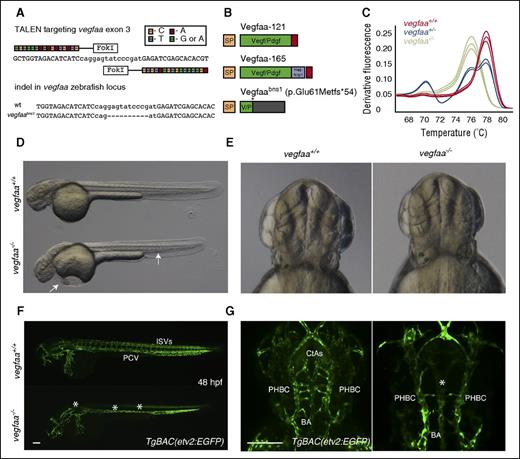
The vegfaabns1 mutants exhibit severe vascular defects. (A) A TALEN was designed to target exon 3 of vegfaa; sequence alignment of the TALEN generated allele. vegfaabns1 has a 10 nucleotide deletion leading to a premature stop codon. (B) Cartoon representation of wild-type (WT) Vegfaa isoforms ∼121 and -165 and Vegfaabns1. Vegfaa-121 consists of a signal peptide and a knot motif, whereas Vegfaa-165 has an additional heparin and Nrp1 binding domain. vegfaabns1 encodes a truncated polypeptide (p.Glu61Metfs*54). (C) Genotyping example of single embryos sampled from a population of vegfaa wt (red), bns1/+ (blue), and bns1/bns1 (green) using high resolution melt analysis (HRMA). (D-E) Brightfield micrographs of 48 hours postfertilization (hpf) WT sibling and vegfaabns1 mutant embryos in lateral and dorsal views. Arrows point to pericardial edema and blood pooling in the tail. (F-G) Confocal micrographs of 48 hpf TgBAC(etv2:EGFP) WT sibling and vegfaabns1 mutant embryos in lateral (F) and dorsal (G) views. Asterisks denote lack of intersegmental vessels (ISVs), axial vessel (F), and reduced central artery (CtA) sprouting (F-G). Bars represent 100 µm.
The vegfaabns1 mutants exhibit severe vascular defects. (A) A TALEN was designed to target exon 3 of vegfaa; sequence alignment of the TALEN generated allele. vegfaabns1 has a 10 nucleotide deletion leading to a premature stop codon. (B) Cartoon representation of wild-type (WT) Vegfaa isoforms ∼121 and -165 and Vegfaabns1. Vegfaa-121 consists of a signal peptide and a knot motif, whereas Vegfaa-165 has an additional heparin and Nrp1 binding domain. vegfaabns1 encodes a truncated polypeptide (p.Glu61Metfs*54). (C) Genotyping example of single embryos sampled from a population of vegfaa wt (red), bns1/+ (blue), and bns1/bns1 (green) using high resolution melt analysis (HRMA). (D-E) Brightfield micrographs of 48 hours postfertilization (hpf) WT sibling and vegfaabns1 mutant embryos in lateral and dorsal views. Arrows point to pericardial edema and blood pooling in the tail. (F-G) Confocal micrographs of 48 hpf TgBAC(etv2:EGFP) WT sibling and vegfaabns1 mutant embryos in lateral (F) and dorsal (G) views. Asterisks denote lack of intersegmental vessels (ISVs), axial vessel (F), and reduced central artery (CtA) sprouting (F-G). Bars represent 100 µm.
Confocal microscopy
An LSM 700 (Zeiss, Germany) was used for live imaging. Embryos and larvae were anesthetized with a low dose of tricaine, placed in a glass-bottom petri dish (MatTek) with a layer of 1.2% low-melt agarose, and imaged using a Plan-Apochromat 10x/0.3 and LCI Plan-Neofluar 25x/0.8.
Plasmids
Total RNA extraction was performed using TRIZOL (Life Technologies, Carlsbad, CA) and used for complementary DNA (cDNA) synthesis using SuperScript second strand (Life Technologies). cDNAs encoding the Vegfaa-121, Vegfaa-165, Vegfab-171, Vegfab-209, Vegfba, Vegfbb, Vegfc, Vegfd, and Pgfb sequences were PCR-amplified using the primers listed in supplemental Table 1 (available on the Blood Web site) from whole embryo cDNA as template. PCR fragments were ligated into the mammalian expression vector pcDNA3.1 myc-HIS tag between HindIII and EcoRI.
Mutagenesis
Site-directed mutagenesis was performed using PfuUltra Fusion HS (Agilent). The polymerase chain reaction (PCR) protocol was 95°C for 1 minute; then 18 cycles of 95°C for 50 seconds, 60°C for 50 seconds, and 58°C to 68°C for 1 minute per kilobase of plasmid length; then 68°C for 7 minutes and 4°C hold. Dpn1 (1 µL) was added to the PCR reaction and incubated at 37°C for 1 hour to digest parental DNA and transform into competent cells. Mutations were introduced using the primers listed in supplemental Table 1.
Rescue experiments
Zebrafish embryos were injected into the cell at the 1-cell stage with 50 pg of vegfaa-121 or 200 pg of vegfaa-165 of RNA,10 and 200 pg was used for the other Vegf family members. Messenger RNA (mRNA) encoding full-length zebrafish Vegfc or human VEGFC lacking the N- and C-terminal domains were injected at different concentrations including 50, 100, 200, and 300 pg. The rescue potential was measured as a percentage of embryos showing blood flow circulation and lack of pericardial edema (supplemental Video 1). Control injections were performed using eGFP mRNA. For additional details, see supplemental Methods.
Results
Generation and characterization of vegfaa and vegfab mutant zebrafish
The zebrafish genome contains 2 Vegfa paralogs, vegfaa and vegfab; vegfaa encodes the 2 main isoforms Vegfaa-121 and -165, whereas vegfab encodes Vegfab-171 and -210.11 To investigate the role of Vegf signaling during zebrafish development, we generated vegfaa and vegfab mutants using TALENs and CRISPRs. We targeted the third exon of vegfaa (Figure 1A) and second exon of vegfab (supplemental Figure 2). Two frameshift mutations were identified and recovered (Figure 1A; supplemental Figure 2). The vegfaa mutant allele (vegfaabns1) harbors a 10-bp deletion, and the vegfab mutant allele (vegfabbns92) a 7-bp insertion (Figure 1A; supplemental Figure 2). vegfaabns1 (p.Glu61Metfs*54) is predicted to encode a truncated polypeptide containing a stretch of 54 incorrect amino acids past the lesion point at position 61 (Figure 1B). vegfabbns92 (p.Met88Trpfs*18) is predicted to encode a truncated polypeptide containing a stretch of 18 incorrect amino acids past the lesion point at position 88 (supplemental Figure 2). In both mutants, the cysteines that characterize the knot motif are lost, thus affecting Vegf dimerization. Therefore, vegfaabns1 and vegfabbns92 are likely to be severe or null alleles. HRMA was used to genotype vegfaabns1 mutants (Figure 1C), whereas allele-specific PCR was used for vegfabbns92. To analyze Vegfaa and Vegfab function during development, vegfaabns1/+ and vegfabbns92/+ fish were crossed into the TgBAC(etv2:EGFP) background.7
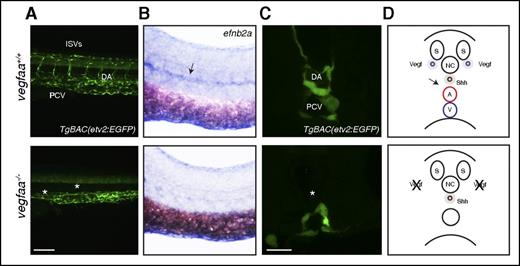
Brightfield microscopy analysis revealed severe pericardial edema, blood pooling, small eyes, and lack of circulation in all vegfaabns1 mutants (Figure 1D). Interestingly, no differences in gross morphology were evident between vegfabbns92 WT and mutant siblings (supplemental Figure 3). Defective formation of an axial vessel in the region of the dorsal aorta (DA) and lack of arterial ISVs were observed in vegfaabns1 mutants at 48 hpf using confocal microscopy (Figure 1D,F), as previously observed in vegfaa morphants.12 The vegfabbns92 mutants exhibit mild angiogenesis defects at 48 hpf (supplemental Figure 3). The VEGF signaling pathway also regulates hindbrain blood vessel formation.10 The vegfaabns1 and vegfabbns92 mutants appear to be unaffected in terms of head morphology and primordial hindbrain channel (PHBC) formation (Figure 1E,G; supplemental Figure 3). However, they are defective in forming the basilar artery (BA) and CtAs (Figure 1G; supplemental Figure 3). Reduced Vegf signaling has been associated with loss of arterial endothelial cell differentiation.13 Hence, we investigated arterial endothelial cell differentiation by examining the vascular pattern of efnb2a mRNA expression and observed a profound reduction in 26 hpf vegfaabns1 mutants compared with their WT siblings (Figure 2B). Expression of the venous endothelial marker flt4 appeared comparable between vegfaabns1 mutants and their WT siblings at 26 hpf (supplemental Figure 1B). Moreover, only a single vessel is present in the trunk of vegfaabns1 mutant embryos at 26 hpf (Figure 2C-D).
vegfaa is required for arterial differentiation. (A) Confocal micrographs of 48 hpf TgBAC(etv2:EGFP) WT sibling and vegfaabns1 mutant embryos in lateral views. Bar represents 100 µm. Asterisks denote lack of ISVs, axial vessel, and reduced CtA sprouting. (B) Brightfield whole-mount in situ hybridization of efnb2a expression in 26 hpf embryos in lateral views. Arrow points to a reduction of efnb2a expression in the DA region. (C) Confocal micrographs of 26 hpf TgBAC(etv2:EGFP) WT sibling and vegfaabns1 mutant embryos in transverse views. Asterisk denotes lack of vessel in DA region. Bar represents 30 µm. (D) Cartoon representations of a cross section through the zebrafish trunk. A, dorsal aorta; NC, notochord; S, somite; V, posterior cardinal vein.
vegfaa is required for arterial differentiation. (A) Confocal micrographs of 48 hpf TgBAC(etv2:EGFP) WT sibling and vegfaabns1 mutant embryos in lateral views. Bar represents 100 µm. Asterisks denote lack of ISVs, axial vessel, and reduced CtA sprouting. (B) Brightfield whole-mount in situ hybridization of efnb2a expression in 26 hpf embryos in lateral views. Arrow points to a reduction of efnb2a expression in the DA region. (C) Confocal micrographs of 26 hpf TgBAC(etv2:EGFP) WT sibling and vegfaabns1 mutant embryos in transverse views. Asterisk denotes lack of vessel in DA region. Bar represents 30 µm. (D) Cartoon representations of a cross section through the zebrafish trunk. A, dorsal aorta; NC, notochord; S, somite; V, posterior cardinal vein.
vegfaa mRNA injections can rescue vegfaa mutants to adult viability
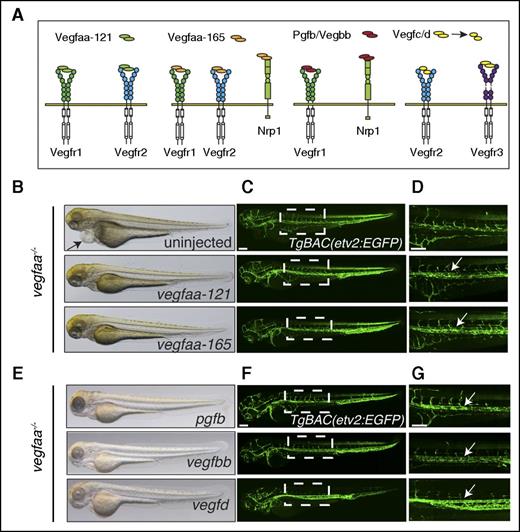
Our data show that the inactivation of Vegfaa leads to dramatic vascular defects, incompatible with life. Because the vegfabbns92 mutant animals only show mild vascular defects during embryogenesis, we decided to focus on Vegfaa as its inactivation recapitulates the dramatic vascular phenotypes of its mammalian ortholog.2 We next reasoned that the vegfaabns1 mutant phenotype could potentially be rescued by mRNA injections based on 2 considerations. First, Vegfa is strongly expressed during early vascular development where it triggers a wide range of responses in endothelial cells, including proliferation, migration, and survival.2 Second, previous work had shown that vegfaa mRNA injections could rescue the arterial vascular phenotype of shh mutants.13 However, Vegfa might need to be supplied in a gradient fashion for the effective control of arterial cell differentiation.14 Embryos derived from an in-cross of vegfaabns1/+ adults were injected with mRNA encoding either Vegfaa-121 or -165 (Figure 3A). Strikingly, a majority of the injected animals lacked pericardial edema and developed a functional circulatory loop and normally sized eyes (Figure 3B). Confocal micrographs of TgBAC(etv2:EGFP) mutant larvae show that upon injection of vegfaa-121 or vegfaa-165 mRNA, the endothelial cells of the DA region formed a tubular structure (Figure 3C-D). However, ISVs formation was impaired, and the length of the circulatory loop was variable in these embryos (Figure 3D). Nevertheless, vegfaa-injected vegfaabns1 mutant larvae survived to become fertile adults. Together, these data indicate that Vegfaa is dispensable for viability after zebrafish embryogenesis.
vegfaa mRNA injections can rescue vegfaa mutants to adult viability. (A) Schematic representation of Vegf molecules binding to their receptors. (B,E) Brightfield micrographs of 72-hpf vegfaabns1 mutants injected with indicated mRNA in lateral views. Arrow points to pericardial edema. (C,D,F,G) Confocal micrographs of 72-hpf TgBAC(etv2:EGFP);vegfaabns1 mutants injected with indicated mRNA in lateral views. Arrows point to lumenized vessel. Bars represent 100 µm.
vegfaa mRNA injections can rescue vegfaa mutants to adult viability. (A) Schematic representation of Vegf molecules binding to their receptors. (B,E) Brightfield micrographs of 72-hpf vegfaabns1 mutants injected with indicated mRNA in lateral views. Arrow points to pericardial edema. (C,D,F,G) Confocal micrographs of 72-hpf TgBAC(etv2:EGFP);vegfaabns1 mutants injected with indicated mRNA in lateral views. Arrows point to lumenized vessel. Bars represent 100 µm.
Vascular growth can be sustained by multiple Vegf family members
Fertile vegfaabns1 mutant fish represent a unique and powerful tool for in vivo endothelial cell studies. We exploited this simple and straightforward in vivo rescue assay to systematically test the biological activity of different Vegf molecules, as well as >20 synthetic variants, in the absence of Vegfaa. Although the in vitro endothelial cell tube formation assay is a widely used model to study angiogenic activity, it suffers from a number of drawbacks. Frequently cited limitations include the lack of neighboring cells, a poor representation of extracellular matrix components, a loss of cell identity, and the reported ability of other cultured cells such as fibroblasts and astrocytes to form tubelike structures in response to Matrigel.15 In contrast, our in vivo assay system provides a physiological environment.
vegfab, vegfba, vegfbb, vegfc, vegfd, and pgfb (zebrafish orthologs of human VEGFA, VEGFB, VEGFC, VEGFD, and PGF, respectively) were cloned, in vitro transcribed, and injected into vegfaabns1 mutants at the 1-cell stage. Their rescue potential was measured as a percentage of embryos showing blood flow and lack of pericardial edema at 3 days postfertilization (Table 1). Brightfield images show absence of pericardial edema in pgfb, vegfbb, and vegfd (Figure 3E) injected larvae, and confocal analyses (Figure 3F) of TgBAC(etv2:EGFP) expression show lumenized vessels in the DA region of these injected animals (Figure 3G). Vegfc and Vegfd are synthesized and secreted as large precursors that are proteolytically processed into mature forms. Unprocessed peptides preferentially signal through Vegfr3, whereas only the mature forms efficiently trigger Vegfr2 signaling (Figure 3A).6 Thus, Vegfc failure to rescue vegfaabns1 mutants could be because of poor cleavage during early embryonic development or low affinity toward Vegfr2. To distinguish between these 2 possibilities, we injected a short isoform of human VEGFC lacking the N- and C- terminal domains but retaining the signal peptide. The proteolytic processing sites of zebrafish Vegfc have not yet been identified, whereas human VEGFC has been examined in detail and is functional in zebrafish embryos.16,17 Similar to full-length zebrafish vegfc, human VEGFC injections failed to rescue vegfaabns1 mutants (Table 1), indicating that VEGFC poorly activates Vegfr2. However, Vegfd did rescue vegfaabns1 mutants suggesting that Vegfr3 or Vegfr2 can signal and promote vascular growth in the absence of Vegfaa.
Vegf molecules injected into vegfaa mutant animals and rescue percentages
| Ligand or amino acid . | Rescue % . |
|---|---|
| Vegf | |
| Vegfaa-121 | 57.7 ± 5.2 |
| Vegfaa-165 | 76.8 ± 4.1 |
| Vegfbb | 84.4 ± 3.5 |
| Vegfd | 89.3 ± 4.9 |
| Plgfb | 97.9 ± 1.2 |
| Vegfab-171 | 3 ± 1.3 |
| Vegfab-210 | 0 |
| Vegfba | 0 |
| Vegfc | 1 ± 1 |
| ∆C∆NVEGFC | 0 |
| Vegfab-171* | 81 ± 5 |
| Vegfaa-121 Vegfr2 determinants | |
| F17A | 0 |
| M18A | 0 |
| I43A | 0 |
| T46A | 0 |
| Y47A | 0 |
| E64A | 0 |
| V83A | 0 |
| K84A | 0 |
| Vegfaa-165 Vegfr1 determinant | |
| D63A/E64A/E67A | 52.5 |
| Vegfaa-165 Vegfr2 determinants | |
| F17A | 88.9 ± 7.8 |
| K84A | 89 ± 5.4 |
| Y47A/K84A | 0 |
| E64A/K84A | 0 |
| Vegfaa-165 Nrp1 determinants | |
| E153A | 77.5 ± 3.7 |
| R165A | 81.1 ± 3.9 |
| Pgfb Nrp1 determinants | |
| R158A | 89.8 ± 5.9 |
| Pgfb Vegfr2 determinants | |
| F17A | 93.2 ± 6.1 |
| E64A | 91 ± 4.5 |
| K84A | 95 ± 3.1 |
| F17A/K84A | 94.7 ± 2.1 |
| E64A/K84A | 89.9 ± 7.5 |
| F17A/E64A/K84A | 88 ± 5.9 |
| Pgfb Vegfr1 determinant | |
| D63A/E64A/E67A | 43 |
| Ligand or amino acid . | Rescue % . |
|---|---|
| Vegf | |
| Vegfaa-121 | 57.7 ± 5.2 |
| Vegfaa-165 | 76.8 ± 4.1 |
| Vegfbb | 84.4 ± 3.5 |
| Vegfd | 89.3 ± 4.9 |
| Plgfb | 97.9 ± 1.2 |
| Vegfab-171 | 3 ± 1.3 |
| Vegfab-210 | 0 |
| Vegfba | 0 |
| Vegfc | 1 ± 1 |
| ∆C∆NVEGFC | 0 |
| Vegfab-171* | 81 ± 5 |
| Vegfaa-121 Vegfr2 determinants | |
| F17A | 0 |
| M18A | 0 |
| I43A | 0 |
| T46A | 0 |
| Y47A | 0 |
| E64A | 0 |
| V83A | 0 |
| K84A | 0 |
| Vegfaa-165 Vegfr1 determinant | |
| D63A/E64A/E67A | 52.5 |
| Vegfaa-165 Vegfr2 determinants | |
| F17A | 88.9 ± 7.8 |
| K84A | 89 ± 5.4 |
| Y47A/K84A | 0 |
| E64A/K84A | 0 |
| Vegfaa-165 Nrp1 determinants | |
| E153A | 77.5 ± 3.7 |
| R165A | 81.1 ± 3.9 |
| Pgfb Nrp1 determinants | |
| R158A | 89.8 ± 5.9 |
| Pgfb Vegfr2 determinants | |
| F17A | 93.2 ± 6.1 |
| E64A | 91 ± 4.5 |
| K84A | 95 ± 3.1 |
| F17A/K84A | 94.7 ± 2.1 |
| E64A/K84A | 89.9 ± 7.5 |
| F17A/E64A/K84A | 88 ± 5.9 |
| Pgfb Vegfr1 determinant | |
| D63A/E64A/E67A | 43 |
Values represent the average percentage of rescue from 3 to 10 biological replicates ± standard error. Vegfab-171* refers to a chimera where the Vegfab signal peptide was replaced with that of Vegfaa.
Pgfb and Vegfbb bind only to Vegfr1 and Nrp1 (Figure 3A), and in contrast to Vegfaa, neither Pgfb nor Vegfbb are essential for vascular development or physiological angiogenesis in mouse.6 Nevertheless, they are both implicated in pathological vascular remodeling in the context of ocular vascular diseases as well as in some cancers.3
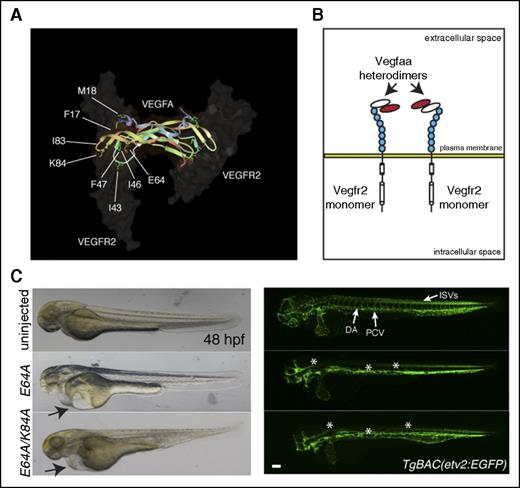
In order to understand the contribution of the different receptors allowing the rescue of vegfaabns1 mutants, Vegfaa and Pgfb isoforms were engineered to selectively bind specific receptors. Vegfaa-121 is freely diffusible and binds to Vegfr1 and Vegfr2, whereas Vegfaa-165 is associated with the extracellular matrix and binds Nrp1 (Figure 3A).2 Thus, we aimed to investigate the structural determinants of Vegfaa binding to Vegfr2 and Nrp1 in vivo. We examined crystal structures, biochemical data from previous in vitro experiments, and amino acid conservation to identify key residues to mutate.18,19 The codons for several amino acids shown in Figure 4A were mutagenized to code for alanine, and the resulting vegfaa variants were injected into vegfaabns1 mutants. We found that the Vegfaa-121 F17, M18, I43, T46, Y47, E64, V83, and K84 residues are crucial for interaction with Vegfr2 in vivo as substitution of each of these amino acids in Vegfaa-121 abrogated its rescue ability (Table 1). Interestingly, these mutations did not significantly affect Vegfaa-165–mediated rescue of vegfaabns1 mutants (Table 1). In order to examine the binding of these molecules to Vegfr2, we performed cell culture experiments. Cos-1 cells were transiently transfected with Vegfr2 and either WT or K84A myc-tagged vegfaa-121 or -165 followed by immunofluorescence analysis. Epifluorescence micrographs (supplemental Figure 4A) show plasma membrane staining for both WT Vegfaa isoforms indicating Vegfr2 binding. When the same experiment was performed with K84A myc-tagged variants, Vegfaa-121 but not Vegfaa-165 staining was lost (supplemental Figure 4A), suggesting important binding differences between the 2 isoforms.
Vegfaa-engineered molecules block vascular development. (A) VEGFA-VEGFR2 complex highlighting VEGFA amino acids responsible for VEGFR2 interactions. (B) Schematic representation of heterodimers of dominant negative Vegfaa (red) and WT Vegfaa (white), which block Vegfr2 dimerization. (C) Brightfield micrographs of 48 hpf embryos injected with indicated mRNA in lateral views (left). Arrows point to pericardial edema. Confocal micrographs of 48 hpf TgBAC(etv2:EGFP) embryos injected with indicated mRNA in lateral views (right). Asterisks denote lack of ISVs and absent or abnormal axial vessel and brain vasculature. Bars represent 100 µm.
Vegfaa-engineered molecules block vascular development. (A) VEGFA-VEGFR2 complex highlighting VEGFA amino acids responsible for VEGFR2 interactions. (B) Schematic representation of heterodimers of dominant negative Vegfaa (red) and WT Vegfaa (white), which block Vegfr2 dimerization. (C) Brightfield micrographs of 48 hpf embryos injected with indicated mRNA in lateral views (left). Arrows point to pericardial edema. Confocal micrographs of 48 hpf TgBAC(etv2:EGFP) embryos injected with indicated mRNA in lateral views (right). Asterisks denote lack of ISVs and absent or abnormal axial vessel and brain vasculature. Bars represent 100 µm.
The heparin or Nrp1 binding domain present in Vegfaa-165 but not Vegfaa-121 might account for these differences. We speculated that the heparin binding domain might stabilize Vegfaa-Vegfr2 interactions while Nrp1 might enhance Vegfaa-165 activity. Therefore, we introduced multiple mutations in Vegfaa-165 and found that whenever 2 of the mutations listed previously were combined, Vegfaa-165 also failed to rescue vegfaabns1 mutants (supplemental Figure 4B; Table 1). We also investigated Vegfa-Nrp1 signaling by mutagenizing the last arginine 165 (R165) in Vegfaa-165, which is responsible for Nrp1 binding.20 Interestingly, the point mutation R165A did not significantly reduce Vegfaa-165’s ability to rescue vegfaabns1 mutants, indicating that in this context Nrp1 contribution is mild (supplemental Figure 4C; Table 1).
These data suggest that Vegfr2 signaling likely drives the rescue of the vegfaabns1 mutants, whereas Nrp1 function, a controversial topic in Vegf biology, does not play a role in this process.21 Furthermore, Vegfaa-165 rescued vegfaabns1 mutants more efficiently than Vegfaa-121 did (Table 1). The difference in activity between the 2 isoforms has been reported in other species as well.6 Our data indicate that Vegfaa-165 binding to Vegfr2 is more stable than Vegfaa-121 binding to Vegfr2 because it can tolerate an amino acid substitution in the domains responsible for interaction with Vegfr2, whereas the short isoform cannot do so. It is therefore likely that the heparin binding domain of Vegfaa-165 helps it form a complex with Vegfr2 thereby enhancing Vegfaa-165–dependent signaling activity.
pgfb mRNA injections rescue vegfaa mutants via Vegfr1
Pgfb and Vegfbb both rescue the viability of vegfaabns1 mutants; however, Vegfbb is poorly annotated in the zebrafish genome. Thus, we decided to focus on Pgfb. Site-directed mutagenesis was performed in order to understand how Pgfb mediates its biological activity in vegfaabns1 mutants. To test the possibility that zebrafish Pgfb retains the ability to bind Vegfr2, we mutagenized those conserved amino acids that might sustain this interaction. The codons for several amino acids shown in Table 1 were mutagenized. However, none of these mutations significantly reduced Pgfb’s ability to rescue vegfaabns1 mutants, indicating that Pgfb does not signal directly through Vegfr2 (Table 1). We therefore investigated the role of Pgfb-Nrp1 binding by mutagenizing arginine 158 (R158) of Pgfb, which is responsible for this interaction.20 Interestingly, Pgfb R158A mRNA injections also rescued vegfaabns1 mutants (Table 1), indicating that Vegfr1 is likely responsible for the ability of Pgfb, and probably also of Vegfbb, to rescue vegfaabns1 mutants, either directly or through Vegfr2 transphosphorylation.6
In order to investigate the contribution of Vegfr1 (aka Flt1) signaling in the rescue of vegfaa mutants, we used an N-ethyl-N-nitrosourea–induced flt1 mutant (flt1fh390) that is predicted to lack both transmembrane and tyrosine kinase domains without affecting the soluble form (supplemental Figure 5A).22 Interestingly, flt1fh390 mutants do not display any obvious vascular phenotype compared with their WT siblings (data not shown). We thus raised double heterozygous vegfaa;flt1 animals and first observed that 25% of their progeny exhibited a vegfaa mutant phenotype (32/129) (supplemental Figure 5B,D). In comparison, when injected with pgfb mRNA, only 12.9% of these embryos (from vegfaabns1/+;flt1fh390/+ in-crosses) (supplemental Figure 5C) exhibited severe vascular defects (24/185) (supplemental Figure 5D), suggesting a 50% rescue. Genotyping of the affected embryos (n = 24) revealed that pgfb failed to rescue vegfaa;flt1 double mutants (11/185 were double mutants, corresponding to the expected 1/16 Mendelian ratio), as well as some of the vegfaabns1/bns1;flt1fh390/+ embryos (n = 10) (supplemental Figure 5E), supporting a model where pgfb modulates vasculogenesis in a Vegfr1-dependent process. Similarly, coinjection of a vegfaa morpholino12 and 200 pg of pgfb mRNA in flt1fh390/+ in-crosses had little effect on flt1+/+ embryos but caused severe vascular defects in their flt1fh390/+ or flt1fh390/fh390 siblings at 48 hpf (8/24 and 12/24, respectively) (data not shown). In addition, we mutagenized Vegfaa-165 and Pgfb at 3 conserved amino acids (D63, E64, and E67) that are reported to modulate Vegfr1 binding (supplemental Figure 6A) and compared their embryos derived from a vegfaabns1/bns1 in-cross.20 These mutations moderately decreased Vegfaa-165’s rescue efficiency to ∼73%, whereas Pgfb’s rescue efficiency decreased to ∼43% (both compared with WT mRNA) (supplemental Figure 6D; Table 1). Altogether, these experiments indicate that Pgfb mediates its rescue effects through Vegfr1.
Vegfaa dominant negative molecules block vascular development
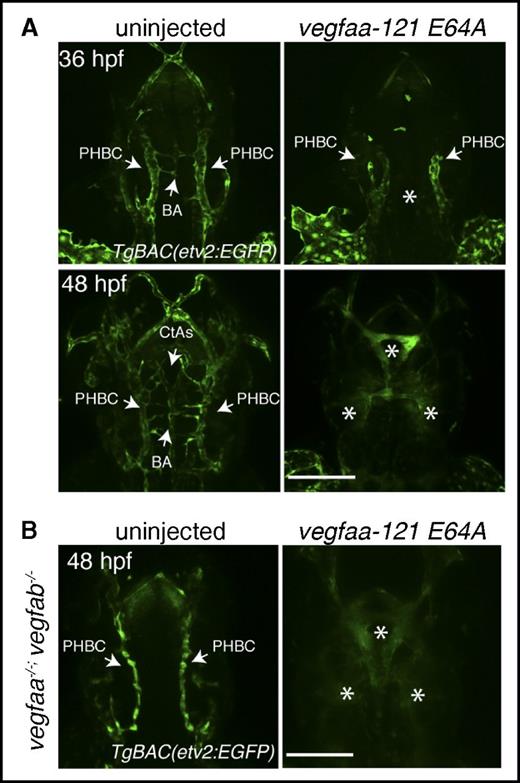
Our data indicate that other Vegf molecules can sustain vascular development independently of Vegfa. We thus reasoned that Vegfaa molecules that cannot bind to Vegfr2 but retain the ability to dimerize with other Vegf family members should behave in a dominant negative (dn) fashion and thereby block vascular development (Figure 4A-B). WT embryos were injected with mRNAs encoding dn isoforms of Vegfaa at the 1-cell stage. Brightfield microscopy analysis revealed pericardial edema, blood pooling, small eyes, and lack of circulation in the injected embryos at 48 hpf (Figure 4C). Moreover, TgBAC(etv2:EGFP) confocal micrographs show impaired trunk vessel formation and lack of ISVs in the dn-vegfaa injected embryos at 24 and 48 hpf (Figure 4C, supplemental Figures 7 and 8). Slight differences in activity were noticed between dn forms of Vegfaa-121 and Vegfaa-165, with dn forms of the shorter isoform causing greater vascular inhibition (Figure 4C). Interestingly, TgBAC(etv2:EGFP) confocal micrographs show that the PHBCs, BA, and CtAs did not develop properly in the dn-vegfaa injected embryos at 36 hpf (Figure 5A). Moreover, lack of vasculature and diffuse enhanced green fluorescent protein fluorescence in the head of the injected embryos at 48 hpf suggest endothelial cell death (Figure 5A). Next, we aimed to understand whether the dn-Vegfaa molecules could interfere with other Vegf molecules. In order to answer this question, embryos derived from an in-cross of vegfaabns1/+;vegfabbns92/+ adults were injected with mRNA encoding the dn-Vegfaa-121 E64A isoform. Confocal micrographs of vegfaabns1;vegfabbns92 double mutant embryos show a brain vasculature pattern similar to that observed in kdrlhu5088 mutants with unaffected PHBCs but lack of BA and CtAs (Figure 5B).23 However, the entire brain vasculature was lost in the dn-vegfaa injected embryos, and only diffused enhanced green fluorescent protein fluorescence was visible (Figure 5B). Reduced ISV formation was also evident in these embryos (supplemental Figure 9). Furthermore, confocal microscopy analyses showed that WT larvae injected with dn-Vegf molecules lacked brain vasculature, the subintestinal vein and interconnecting vessels at 72 hpf (supplemental Figure 10). Because PHBC formation has been suggested to be regulated by Vegfc/Vegfr3,24 and subintestinal vein and interconnecting vessel formation by Vegfaa/Vegfab,25 these observations indicate that the dn-Vegf molecules can interfere with other Vegf family members.
Vegfaa-engineered molecules block brain angiogenesis. (A) Confocal micrographs of 36- or 48-hpf uninjected (left) or vegfaa-121 E64A mRNA injected (right) embryos. Arrows point to PHBC, BA, and CtAs in the uninjected embryos and to reduced PHBC formation in the injected embryos. Asterisks denote lack of brain vasculature in the injected embryos. (B) Confocal micrographs of 48-hpf TgBAC(etv2:EGFP) vegfaabns1;vegfabbns92 double mutants uninjected (left) or vegfaa-121 E64 mRNA injected (right) embryos in dorsal views. Arrows point to PHBC in the uninjected embryos. Asterisks denote lack of brain vasculature in the injected embryos. Bars represent 100 µm.
Vegfaa-engineered molecules block brain angiogenesis. (A) Confocal micrographs of 36- or 48-hpf uninjected (left) or vegfaa-121 E64A mRNA injected (right) embryos. Arrows point to PHBC, BA, and CtAs in the uninjected embryos and to reduced PHBC formation in the injected embryos. Asterisks denote lack of brain vasculature in the injected embryos. (B) Confocal micrographs of 48-hpf TgBAC(etv2:EGFP) vegfaabns1;vegfabbns92 double mutants uninjected (left) or vegfaa-121 E64 mRNA injected (right) embryos in dorsal views. Arrows point to PHBC in the uninjected embryos. Asterisks denote lack of brain vasculature in the injected embryos. Bars represent 100 µm.
Discussion
Recognition of the role of VEGF as an important regulator of pathological angiogenesis to promote tumor growth in several types of cancer has provided a rationale for the development of VEGF inhibitors, with the goal of blocking angiogenesis and restrict tumor growth and progression.2-6 Despite the encouraging beneficial effects of anti-VEGF therapy, several patients develop resistance that is often associated with poor prognosis. Proangiogenic escape has been proposed as a mechanism to bypass anti-VEGF therapy, whereby tumors upregulate alternate proangiogenic signaling pathways to reestablish neovascularization.3,4,26,27
Our experiments show that vegfaa mutants, which display a dramatic vascular phenotype, can be rescued to viability through mRNA injections using different Vegf family members. These molecules can sustain vascular development in the absence of Vegfaa, potentially explaining the resistance to current anti-VEGFA therapies.
More specifically, our studies suggest that targeting PGF, VEGFB, and VEGFD, in addition to VEGFA, might lead to additional benefits in treating angiogenic disorders.
Interestingly, in our system both full-length zebrafish Vegfc and human VEGFC lacking the N- and C-terminal domains but retaining the signal peptide failed to rescue vegfaa mutants. It has been previously shown that VEGFC signaling through VEGFR2 works synergistically with and not independently of VEGFA.28 Furthermore, in mice, an excess of VEGFC signaling through VEGFR2 induced the disturbance of vasculogenesis and hematopoiesis during embryogenesis.28
The various commercially available VEGF blockers differ in their affinity for VEGFA, relative antiangiogenic activity, and/or the ability to bind different VEGF isoforms and family members. However, none of these molecules inhibits Vegfr2 dimerization and/or signaling by interacting with other molecules such as Gremlin29 and VEGFD.
Furthermore, it has been recently shown that Vegfr2 can form dimers, which are phosphorylated, in the absence of ligands suggesting that pathological angiogenesis might occur even if Vegf family members are inhibited.30 Thus, blocking both Vegf family members and Vegfr2 signaling might lead to improved antiangiogenic activity.
Working toward this goal, we engineered and validated Vegf molecules that can interfere with Vegfr2 dimerization by trapping Vegfa as well as other Vegf family members. Our data show for the first time that dn-Vegf molecules can block vascular development in vivo, thereby offering an alternative approach to the existing VEGFA blockers.
In addition, we anticipate that vegfaa mutants and dn-Vegf molecules will represent powerful tools to generate and study organ specific avascular models, providing insights into vascularization during development and revascularization during organ regeneration. Furthermore, these avascular models can serve as platforms for small-molecule screens to identify novel proangiogenic compounds to treat ischemic diseases among others.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Zacharias Kontarakis, Christian Helker, Saulius Sumanas, Claudia Carlantoni, Radiance Lim, and Rubén Marín-Juez for discussions, comments on the manuscript, and/or reagents, and Stefan Schulte-Merker and Chenghua Gu for sharing reagents.
This work was supported by funds from the Max Planck Society.
Authorship
Contribution: All authors were involved in experimental design and data analysis; experiments were performed by all authors except D.Y.R.S., who supervised the project; and A.R., S.G., and D.Y.R.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrea Rossi, Department of Developmental Genetics, Max Planck Institute for Heart and Lung Research, Ludwigstr 43, 61231 Bad Nauheim, Germany; e-mail: andrea.rossi@mpi-bn.mpg.de; and Didier Y. R. Stainier, Department of Developmental Genetics, Max Planck Institute for Heart and Lung Research, Ludwigstr 43, 61231 Bad Nauheim, Germany; e-mail: didier.stainier@mpi-bn.mpg.de.
References
Author notes
A.R. and S.G. contributed equally to this study.