Abstract
Introduction
2016 NCCN guideline recommended that induction therapy used conventional chemotherapy such as proteasome inhibitors and immunomodulatory drugs for newly diagnosis Multiple Myeloma (MM) patients. Now, HDAC inhibitor, CS-1 Antibody agents, and novel proteasome inhibitor were available for the treatment of MM. Thereby, the outcome of MM was significantly improved regarding response rates and overall survival. However, relapse is inevitable in almost all patients and the cure of myeloma is difficult even now. Recurrence of myeloma is typically more aggressive with each relapse, leading to the development of treatment-refractory disease. For each treatment, we have to choose appropriate new agents. However, we do not know how to choose these new agents Therefore, we try to use next generation sequencer as a tool of drug choosing system. The aim of this study is to identify genetic alterations in MM cells by next generation sequencer for determining the optimal drug and predicting drug resistance in the future.
Patients and Methods:
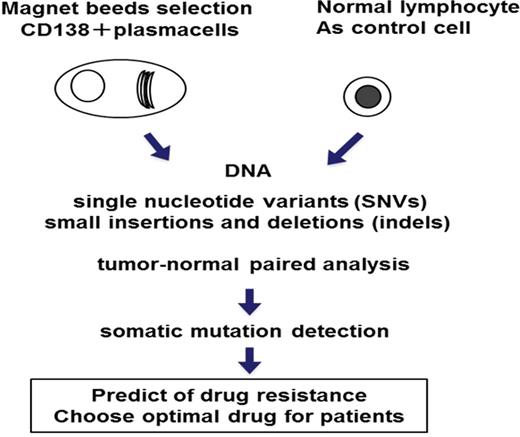
At first, we reviewed newly diagnosed 11 patients with MM (Male: Female 6:5) The median age was 62.27 years (range, 37-78 ). Patients received novel agents including bortezomib in Sapporo Medical University Hospital. Patient plasma cell DNA was extracted from magnetic bead-enriched bone marrow CD138-positive fraction.CD138-negative cells and peripheral lymphocyte were used as matched non-tumor cells. Next, forty nanograms of DNA were used for multiplex PCR amplification with an Ion Ampliseq Comprehensive Cancer Panel. This cancer panel offers targeted coverage of all exons in 409 tumor suppressor genes and oncogenes frequently cited and frequently mutated in human cancers. (covered regions: 95.4% of total). We sequenced 15,992 regions which obtained more than 1.5 megabases of target sequence.
Result:
Each sample underwent on average 8.3 million sequencing reads after quality filtering. The mean read depths were 539x, and >95% of targeted bases were represented by at least 20 reads. The average number of synonymous mutations detected per patient was 5.8 (range 0-11).
We also detected copy number variations which segments of genome can be deleted from sequencing data. We found genetic alterations that associated with poor prognosis and refractory to chemotherapy of MM patients.
We detected mutationin several patients such as EGFR,IKBKB,ERBB3,MYH11,CYLD,TP53,CDH2. These genomes are mainly involved in several important pathways, including cell cycle regulation, RTK-MAPK-PI3K and NF-kB. And then, we can detect main mutation pathway of cancer cells and choose the pathway blockable agents.
Conclusion:
We performed targeted next-generation sequencing gene analysis of malignant plasma cells from patients with MM. Next generation sequencing analysis of Myeloma cells can detect mutation and copy number variations. These data predict of drug resistance and facilitate improvements in the treatment of MM patients. We can use targeted next-generation sequencing as tool of drug choosing system. This method is useful for determining the optimal drug for patients with MM in the future.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal