Key Points
MICA-129 matching improves survival in uHSCT.
MICA-129 mismatches were observed in 6.7% of all transplant patients.
Major histocompatibility complex class I polypeptide-related sequence A (MICA) is a highly polymorphic ligand of the activating NKG2D receptor on natural killer (NK) cells, γδ-T cells, and NKT cells. MICA incompatibilities have been associated with an increased graft-versus-host disease (GVHD) incidence, and the MICA-129 (met/val) dimorphism has been shown to influence NKG2D signaling in unrelated hematopoietic stem cell transplantation (uHSCT). We investigated the effect of MICA matching on survival after uHSCT. We sequenced 2172 patients and their respective donors for MICA. All patients and donors were high-resolution HLA-typed and matched for 10/10 (n = 1379), 9/10 (n = 636), or 8/10 (n = 157) HLA alleles. Within each HLA match group, cases matched and mismatched for MICA and MICA-129 were analyzed for the end points overall survival (OS), disease-free survival (DFS), nonrelapse mortality (NRM), relapse-incidence (RI), and GVHD. Mismatches at the MICA locus as well as MICA-129 increased with the number of HLA mismatches (MICA mismatched 10/10, 9.2% [n = 127]; 9/10, 22.3% [n = 142]; 8/10, 38.2% [n = 60]; MICA-129 mismatched 10/10, 3.9% [n = 54]; 9/10, 10.2% [n = 65]; 8/10, 17.2% [n = 27]). Adverse OS was observed in the 10/10 match group if MICA-129 was mismatched (10/10, hazard ratio [HR], 1.77; confidence interval [CI], 1.22-2.57; P = .003). MICA-129 mismatches correlated with a significantly worse outcome for DFS in the 10/10 HLA match group (HR, 1.77; CI, 1.26-2.50; P = .001). Higher rates of aGVHD were seen in MICA-129 mismatched cases. Our results indicate that MICA-129 matching is relevant in uHSCT. Prospective typing of patients and donors in unrelated donor search may identify mismatches for MICA-129, and compatible donor selection may improve outcome for this small but high-risk subgroup.
Introduction
The major histocompatibility complex class I related chain A gene (MICA) is a highly polymorphic gene closely linked to the HLA-B locus.1,2 It encodes a cell stress–inducible glycoprotein, which mediates an activatory signal toward the NKG2D receptor expressed on natural killer (NK) cells, γδ-T cells, and NKT cells.3 At this time, there are 106 known MICA alleles encoding for 82 different proteins (World Health Organization HLA database 3.26.0, 2016-10).4 MICA molecules show sequence homology and similarities in the tertiary structure to classical HLA alleles, but they do not bind peptides and do not associate with β2-microglobulin.5,6
MICA transcripts have been found in almost all tissues except the brain, and MICA has been shown to be expressed on the cell surface of the gastrointestinal epithelium, as well as endothelial cells, fibroblasts, and monocytes, but not on quiescent CD4+, CD8+, and CD19+ lymphocytes.7,8
MICA alloimmunization influences outcome of solid organ transplantation, and the presence of MICA alloantibodies correlates with increased risk for kidney allograft loss.9 In the context of hematopoietic stem cell transplantation (HSCT), MICA polymorphisms are of interest because they interact with cells, which are known to be relevant for relapse control. In addition, costimulatory effects of such an interaction might trigger T-cell alloreactivity, leading to an increased risk for transplant-associated morbidity, particularly the incidence of graft-versus-host disease (GVHD).10 In addition to direct receptor–ligand interactions, presentation of polymorphic MICA peptide fragments on classical HLA molecules might give rise to alloreactive T-cell clones because of mismatched MICA in the setting of allogeneic stem cell transplantation.11,-13 Furthermore, the MICA-129 (met/val) dimorphism has been shown to influence NKG2D signaling.14 Therefore, we hypothesized that mismatches at the MICA locus are associated with adverse survival outcomes in unrelated HSCT (uHSCT). We tested this hypothesis in a large cohort of adult German patients transplanted for malignant diseases of the lymphohematopoietic system. As secondary end points, we defined nonrelapse mortality (NRM) and relapse incidence (RI), where, according to our hypothesis, higher incidences of adverse events would be expected in patients transplanted with MICA-mismatched grafts.
Methods
Study design
The search unit for unrelated stem cell donors in Ulm, Germany, currently conducts donor search for 25 German transplant centers totaling more than 1100 searches per year. We conducted a collaborative retrospective study to analyze the effect of MICA matching on outcome of uHSCT. Clinical data were provided by the German Registry for Stem Cell Transplantation in pseudonymized form. These data were collected by the transplant centers at day 100 after transplantation, and yearly thereafter, according to the European Society for Blood and Marrow Transplantation (EBMT) guidelines (Med-AB forms manual). The data were recorded in the EBMT ProMISe database, of which the German Registry for Stem Cell Transplantation represents the subset of German patients. Consent for registration, clinical data collection, and scientific analysis was obtained on registration in the EBMT database. Consent for histocompatibility typing was obtained on initiation of donor search. The project was approved by the ethics committee of the University of Ulm (no. 263/09).
Patients and donors
We included 2172 adult patients with malignant diseases of the lymphohematopoietic system transplanted for the first time with an allogeneic-unrelated donor between 2000 and 2011. Graft sources were T-cell-replete bone marrow or peripheral blood stem cell. All patients and donors were 10/10, 9/10, or 8/10 HLA matched on allelic level for the loci HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1. DPB1 typing, carried out by an in-house next generation sequencing protocol, was available for 64.1% of the patients and donors at the time of analysis. All searches were conducted by the search unit Ulm. The disease entities were restricted to the 5 largest groups: acute myeloid leukemia, myelodysplastic syndrome, non-Hodgkin lymphoma, acute lymphoblastic leukemia, and acute leukemia not specified as acute myeloid leukemia or acute lymphoblastic leukemia (undifferentiated, biphenotypic, or secondary acute leukemia). Patient characteristics are summarized in Table 1.
Patient characteristics
| Category . | Total . | MICA-129 matched . | MICA-129 mismatched . | P value . |
|---|---|---|---|---|
| Age | .258 | |||
| Median | 54 | 54 | 53.5 | |
| Range | 18-76 | 18-76 | 18-75 | |
| Diagnosis | .777 | |||
| AML | 808 (37.2) | 757 (37.4) | 51 (34.9) | |
| MDS | 552 (25.4) | 511 (25.2) | 41 (28.1) | |
| NHL | 339 (15.6) | 314 (15.5) | 25 (17.1) | |
| ALL | 309 (14.2) | 288 (14.2) | 21 (14.4) | |
| Acute leukemia | 164 (7.6) | 156 (7.7) | 8 (5.5) | |
| Disease stage | .513 | |||
| Early | 958 (44.1) | 895 (44.2) | 63 (43.2) | |
| Intermediate | 630 (29.0) | 582 (28.7) | 48 (32.9) | |
| Advanced | 584 (26.9) | 549 (27.1) | 35 (24.0) | |
| Year of transplantation | .105 | |||
| 2000-2004 | 205 (9.4) | 184 (9.1) | 21 (14.4) | |
| 2005-2008 | 926 (42.6) | 868 (42.8) | 58 (39.7) | |
| 2009-2011 | 1041 (47.9) | 974 (48.1) | 67 (45.9) | |
| HLA matching | <.001 | |||
| 10/10 | 1379 (63.5) | 1325 (65.4) | 54 (37.0) | |
| 9/10 | 636 (29.3) | 571 (28.2) | 65 (44.5) | |
| 8/10 | 157 (7.2) | 130 (6.4) | 27 (18.5) | |
| Conditioning regimen | .618 | |||
| RIC | 838 (38.6) | 785 (38.7) | 53 (36.3) | |
| MAC | 1334 (61.4) | 1241 (61.3) | 93 (63.7) | |
| Graft source | 1.000 | |||
| BM | 123 (5.7) | 115 (5.7) | 8 (5.5) | |
| Peripheral blood stem cells | 2049 (94.3) | 1911 (94.3) | 138 (94.5) | |
| ATG treatment | .254 | |||
| Yes | 1194 (55.0) | 1123 (55.4) | 71 (48.6) | |
| No | 609 (28.0) | 564 (27.8) | 45 (30.8) | |
| Data missing | 369 (17.0) | 339 (16.7) | 30 (20.5) | |
| KPS | .016 | |||
| 80-100 | 1501 (69.1) | 1412 (69.7) | 89 (61.0) | |
| <80 | 102 (4.7) | 89 (4.4) | 13 (8.9) | |
| Data missing | 569 (26.2) | 525 (25.9) | 44 (30.1) | |
| Patient–donor CMV status combination | .097 | |||
| neg neg | 582 (26.8) | 550 (27.1) | 32 (21.9) | |
| neg pos | 184 (8.5) | 166 (8.2) | 18 (12.3) | |
| pos neg | 496 (22.8) | 460 (22.7) | 36 (24.7) | |
| pos | 559 (25.7) | 529 (26.1) | 30 (20.5) | |
| Data missing | 351 (16.2) | 321 (15.8) | 30 (20.5) | |
| Sex match | .494 | |||
| Other | 1906 (87.8) | 1781 (87.9) | 125 (85.6) | |
| Male patient-female donor | 266 (12.2) | 245 (12.1) | 21 (14.4) |
| Category . | Total . | MICA-129 matched . | MICA-129 mismatched . | P value . |
|---|---|---|---|---|
| Age | .258 | |||
| Median | 54 | 54 | 53.5 | |
| Range | 18-76 | 18-76 | 18-75 | |
| Diagnosis | .777 | |||
| AML | 808 (37.2) | 757 (37.4) | 51 (34.9) | |
| MDS | 552 (25.4) | 511 (25.2) | 41 (28.1) | |
| NHL | 339 (15.6) | 314 (15.5) | 25 (17.1) | |
| ALL | 309 (14.2) | 288 (14.2) | 21 (14.4) | |
| Acute leukemia | 164 (7.6) | 156 (7.7) | 8 (5.5) | |
| Disease stage | .513 | |||
| Early | 958 (44.1) | 895 (44.2) | 63 (43.2) | |
| Intermediate | 630 (29.0) | 582 (28.7) | 48 (32.9) | |
| Advanced | 584 (26.9) | 549 (27.1) | 35 (24.0) | |
| Year of transplantation | .105 | |||
| 2000-2004 | 205 (9.4) | 184 (9.1) | 21 (14.4) | |
| 2005-2008 | 926 (42.6) | 868 (42.8) | 58 (39.7) | |
| 2009-2011 | 1041 (47.9) | 974 (48.1) | 67 (45.9) | |
| HLA matching | <.001 | |||
| 10/10 | 1379 (63.5) | 1325 (65.4) | 54 (37.0) | |
| 9/10 | 636 (29.3) | 571 (28.2) | 65 (44.5) | |
| 8/10 | 157 (7.2) | 130 (6.4) | 27 (18.5) | |
| Conditioning regimen | .618 | |||
| RIC | 838 (38.6) | 785 (38.7) | 53 (36.3) | |
| MAC | 1334 (61.4) | 1241 (61.3) | 93 (63.7) | |
| Graft source | 1.000 | |||
| BM | 123 (5.7) | 115 (5.7) | 8 (5.5) | |
| Peripheral blood stem cells | 2049 (94.3) | 1911 (94.3) | 138 (94.5) | |
| ATG treatment | .254 | |||
| Yes | 1194 (55.0) | 1123 (55.4) | 71 (48.6) | |
| No | 609 (28.0) | 564 (27.8) | 45 (30.8) | |
| Data missing | 369 (17.0) | 339 (16.7) | 30 (20.5) | |
| KPS | .016 | |||
| 80-100 | 1501 (69.1) | 1412 (69.7) | 89 (61.0) | |
| <80 | 102 (4.7) | 89 (4.4) | 13 (8.9) | |
| Data missing | 569 (26.2) | 525 (25.9) | 44 (30.1) | |
| Patient–donor CMV status combination | .097 | |||
| neg neg | 582 (26.8) | 550 (27.1) | 32 (21.9) | |
| neg pos | 184 (8.5) | 166 (8.2) | 18 (12.3) | |
| pos neg | 496 (22.8) | 460 (22.7) | 36 (24.7) | |
| pos | 559 (25.7) | 529 (26.1) | 30 (20.5) | |
| Data missing | 351 (16.2) | 321 (15.8) | 30 (20.5) | |
| Sex match | .494 | |||
| Other | 1906 (87.8) | 1781 (87.9) | 125 (85.6) | |
| Male patient-female donor | 266 (12.2) | 245 (12.1) | 21 (14.4) |
Acute leukemia, acute leukemia not specified as AML or ALL (undifferentiated, biphenotypic, or secondary acute); ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ATG, antithymocyte globulin; CMV, cytomegalovirus; KPS, Karnofsky performance score; MAC, myeloablative conditioning; MDS, myelodysplastic syndrome; neg, negative; NHL, non-Hodgkin lymphoma; pos, positive; RIC, reduced intensity conditioning.
MICA genotyping and matching
MICA genotyping was performed by sequence-based typing, as reported previously15 : Exons 2 to 5 were sequenced in both directions, using the Sanger technique. Exon 5 contains a short tandem repeat with variable length. The exact allelic sequence of exon 5 was resolved by allele separation, using group-specific polymerase chain reaction, or by comparison of heterozygous sequences with a database of all known possible combinations.
Genetic variations leading to a different protein within the extracellular (exons 2-4) and the transmembrane domains (exon 5) were considered for MICA matching purposes, as exon 1 encodes a leader sequence and exon 6 the intracytoplasmic region.
Differences between patient and donor were considered as mismatches, irrespective of the vector of the combination (bidirectional, unidirectional GVH, unidirectional HVG).16
The MICA-129 (met/val) phenotype in patients and donors was inferred from the genomic sequence.
Statistical analysis
Univariate analyses of the effect of MICA mismatches on OS and DFS were performed using Kaplan-Meier analysis and logrank testing. Multivariate analyses were carried out by Cox regression modeling. NRM and RI were analyzed using competing risks analysis.17 Stratification was used to account for heterogeneity of diagnosis. A center effect was adjusted using a γ-frailty term.18 MICA shows genetic linkage with HLA-B. To account for possible confounding of MICA mismatches by concomitant presence of HLA mismatches, subgroup analysis was performed according to the degree of HLA compatibility. Statistical models evaluated the following clinical predictors: patient age as continuous variable, disease stage, HLA compatibility, donor–recipient sex combination, donor–recipient CMV combination, KPS lower than 80 vs 80 to 100, year of transplantation (2000-2004, 2005-2008, 2009-2011), conditioning regimen intensity, stem cell source (bone marrow vs peripheral blood stem cells), and treatment with antithymocyte globulin. Early disease stage was defined as transplantation in first complete remission for acute leukemia as well as untreated or in first complete remission for myelodysplastic syndrome and non-Hodgkin lymphoma. Intermediate disease stage was defined as transplantation in second complete remission for acute leukemia, in second complete remission or in partial remission for myelodysplastic syndrome, and in second complete remission, in partial remission, or stable disease for non-Hodgkin lymphoma. More advanced disease status was classified as advanced disease stage. KPS, CMV status, and treatment with antithymocyte globulin values were missing in 569 (26.2%), 351 (16.2%), and 369 (17.0%) of the entire transplant cases, respectively. Models were validated by inclusion of missing cases as a separate group, as well as by omission of cases with missing values.19,20 During validation, no bias was found. Acute GVHD (aGVHD) prevalences are reported descriptively, and chronic GVHD (cGVHD) incidences were analyzed using competing risks analysis. All models were checked for proportional hazards assumption, and no violations were found. Statistical significance was set to P = .05. All statistical procedures were performed with R, version 3.2.2.
Results
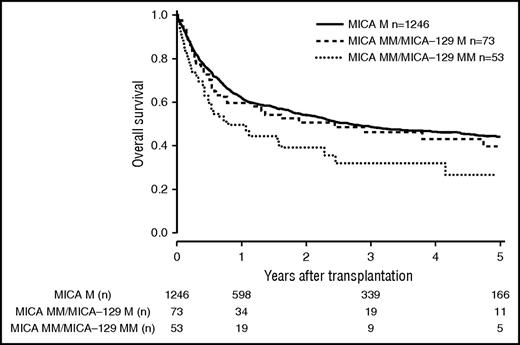
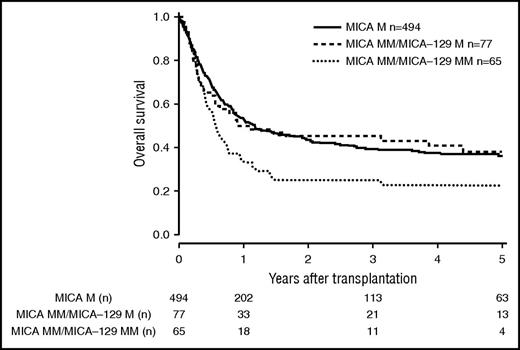
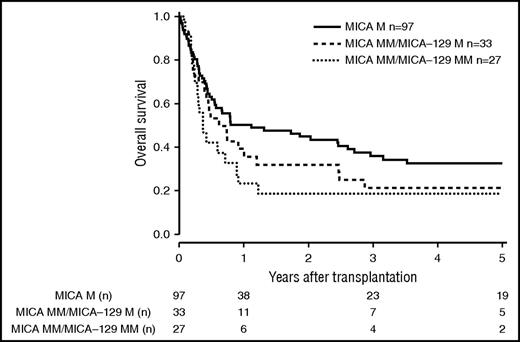
Patients and donor characteristics are summarized in Table 1. Median follow-up time was 43 months. Mismatches at the MICA locus were observed in 9.2% (n = 127) of the 10/10 HLA-matched, 22.3% (n = 142) of the 9/10 HLA-matched, and 38.2% (n = 60) of the 8/10 HLA-matched group. MICA-129 mismatches were observed in 3.9% (n = 54, 10/10), 10.2% (n = 65, 9/10), and 17.2% (n = 27, 8/10) of the transplant pairs. Of 329 cases mismatched for MICA, 37 (11.3%) were mismatched unidirectional in HVG direction, 263 (79.9%) were mismatched bidirectional, and 29 (8.8%) were mismatched in GVH direction. For MICA-129 matching, 13 (8.9%) were mismatched unidirectional in HVG direction, 123 (84.2%) were mismatched bidirectional, and 10 (6.9%) were mismatched in GVH direction. HLA-B mismatches were frequently associated with MICA mismatches (n = 87; 58.8%), Almost all MICA mismatched cases were single mismatches. Double MICA mismatches were observed only in 11 cases (10/10 matched group, n = 1 [0.1%]; 9/10 matched group, n = 5 [0.8%]; 8/10 matched group, n = 5 [3.2%]). The distribution of clinical predictors was balanced within MICA matched and mismatched groups with the exception of KPS and HLA match grade, with the latter indicating confounding of HLA mismatches with MICA mismatches. Therefore, subgroup analyses were performed according to the number of HLA mismatches. In univariate analysis, the presence of mismatched MICA-129 was associated with adverse OS in the 10/10 and the 9/10 HLA match groups (5-year OS 10/10, 0.44 vs 0.27 [P = .013], and 5-year OS 9/10, 0.37 vs 0.23 [P = .006]; Figures 1 and 2; Table 2). No significantly different adverse 5-year OS was found for the 8/10 HLA match group (5-year OS, 0.30 vs 0.19 [P = .082]; Figure 3; Table 2). For DFS, outcome was significantly worse if mismatched MICA-129 was present in all HLA match groups (10/10, 5-year DFS, 0.36 vs 0.20 [P = .010]; 9/10, 5-year DFS, 0.30 vs 0.14 [P = .018]; 8/10, 5-year DFS, 0.25 vs 0.09 [P = .019]).
OS of patients with 10/10 compatible donors transplanted with a MICA matched donor (solid black line), a MICA mismatched/MICA-129 matched donor (dashed line), and a MICA mismatched/MICA-129 mismatched donor (dotted line) (P = .037). M, match; MM, mismatch.
OS of patients with 10/10 compatible donors transplanted with a MICA matched donor (solid black line), a MICA mismatched/MICA-129 matched donor (dashed line), and a MICA mismatched/MICA-129 mismatched donor (dotted line) (P = .037). M, match; MM, mismatch.
OS of patients with 9/10 compatible donors transplanted with a MICA matched donor (solid black line), a MICA mismatched/MICA-129 matched donor (dashed line), and a MICA mismatched/MICA-129 mismatched donor (dotted line) (P = .022).
OS of patients with 9/10 compatible donors transplanted with a MICA matched donor (solid black line), a MICA mismatched/MICA-129 matched donor (dashed line), and a MICA mismatched/MICA-129 mismatched donor (dotted line) (P = .022).
Univariate analysis
| End points . | 10/10 HLA match group . | P value . | 9/10 HLA match group . | P value . | 8/10 HLA match group . | P value . | |||
|---|---|---|---|---|---|---|---|---|---|
| MICA-129 Matched . | MICA-129 Mismatched . | MICA-129 Matched . | MICA-129 Mismatched . | MICA-129 Matched . | MICA-129 Mismatched . | ||||
| OS | .013 | .006 | .082 | ||||||
| 1 y | 0.62 (0.59-0.65) | 0.50 (0.37-0.67) | 0.52 (0.48-0.57) | 0.33 (0.23-0.48) | 0.47 (0.39-0.57) | 0.23 (0.11-0.50) | |||
| 3 y | 0.49 (0.46-0.52) | 0.32 (0.20-0.51) | 0.40 (0.36-0.45) | 0.25 (0.16-0.40) | 0.32 (0.24-0.42) | 0.19 (0.08-0.45) | |||
| 5 y | 0.44 (0.41-0.47) | 0.27 (0.15-0.48) | 0.37 (0.32-0.42) | 0.23 (0.14-0.37) | 0.30 (0.22-0.40) | 0.19 (0.08-0.45) | |||
| DFS | .010 | .018 | .019 | ||||||
| 1 y | 0.55 (0.53-0.58) | 0.37 (0.26-0.55) | 0.46 (0.42-0.51) | 0.26 (0.16-0.40) | 0.41 (0.33-0.51) | 0.09 (0.03-0.35) | |||
| 3 y | 0.43 (0.40-0.46) | 0.24 (0.14-0.41) | 0.35 (0.31-0.40) | 0.23 (0.14-0.38) | 0.30 (0.22-0.40) | 0.09 (0.03-0.35) | |||
| 5 y | 0.36 (0.33-0.39) | 0.20 (0.10-0.38) | 0.30 (0.26-0.34) | 0.14 (0.06-0.31) | 0.25 (0.18-0.35) | 0.09 (0.03-0.35) | |||
| NRM | .035 | .901 | .105 | ||||||
| 1 y | 0.26 (0.23-0.28) | 0.37 (0.23-0.51) | 0.31 (0.27-0.35) | 0.33 (0.22-0.46) | 0.38 (0.29-0.47) | 0.63 (0.39-0.80) | |||
| 3 y | 0.33 (0.30-0.36) | 0.46 (0.30-0.60) | 0.38 (0.34-0.42) | 0.35 (0.23-0.48) | 0.48 (0.38-0.57) | 0.63 (0.39-0.80) | |||
| 5 y | 0.36 (0.33-0.39) | 0.51 (0.33-0.67) | 0.40 (0.36-0.45) | 0.38 (0.25-0.50) | 0.49 (0.39-0.58) | 0.63 (0.39-0.80) | |||
| RI | .411 | .033 | .592 | ||||||
| 1 y | 0.22 (0.19-0.25) | 0.30 (0.17-0.43) | 0.28 (0.24-0.32) | 0.46 (0.32-0.58) | 0.28 (0.20-0.36) | 0.42 (0.21-0.62) | |||
| 3 y | 0.29 (0.26-0.32) | 0.38 (0.23-0.54) | 0.33 (0.29-0.38) | 0.46 (0.32-0.58) | 0.35 (0.26-0.44) | 0.42 (0.21-0.62) | |||
| 5 y | 0.33 (0.30-0.36) | 0.38 (0.23-0.54) | 0.37 (0.32-0.42) | 0.56 (0.39-0.70) | 0.38 (0.29-0.47) | 0.42 (0.21-0.62) | |||
| cGVHD incidence | .988 | .324 | .146 | ||||||
| 1 y | 0.32 (0.28-0.35) | 0.31 (0.15-0.48) | 0.38 (0.32-0.43) | 0.30 (0.15-0.47) | 0.37 (0.25-0.50) | 0.67 (0.30-0.88) | |||
| 3 y | 0.40 (0.36-0.43) | 0.38 (0.20-0.56) | 0.43 (0.37-0.48) | 0.30 (0.15-0.47) | 0.47 (0.34-0.59) | 0.67 (0.30-0.88) | |||
| 5 y | 0.40 (0.36-0.43) | 0.38 (0.20-0.56) | 0.45 (0.39-0.50) | 0.38 (0.17-0.60) | 0.47 (0.34-0.59) | NA | |||
| End points . | 10/10 HLA match group . | P value . | 9/10 HLA match group . | P value . | 8/10 HLA match group . | P value . | |||
|---|---|---|---|---|---|---|---|---|---|
| MICA-129 Matched . | MICA-129 Mismatched . | MICA-129 Matched . | MICA-129 Mismatched . | MICA-129 Matched . | MICA-129 Mismatched . | ||||
| OS | .013 | .006 | .082 | ||||||
| 1 y | 0.62 (0.59-0.65) | 0.50 (0.37-0.67) | 0.52 (0.48-0.57) | 0.33 (0.23-0.48) | 0.47 (0.39-0.57) | 0.23 (0.11-0.50) | |||
| 3 y | 0.49 (0.46-0.52) | 0.32 (0.20-0.51) | 0.40 (0.36-0.45) | 0.25 (0.16-0.40) | 0.32 (0.24-0.42) | 0.19 (0.08-0.45) | |||
| 5 y | 0.44 (0.41-0.47) | 0.27 (0.15-0.48) | 0.37 (0.32-0.42) | 0.23 (0.14-0.37) | 0.30 (0.22-0.40) | 0.19 (0.08-0.45) | |||
| DFS | .010 | .018 | .019 | ||||||
| 1 y | 0.55 (0.53-0.58) | 0.37 (0.26-0.55) | 0.46 (0.42-0.51) | 0.26 (0.16-0.40) | 0.41 (0.33-0.51) | 0.09 (0.03-0.35) | |||
| 3 y | 0.43 (0.40-0.46) | 0.24 (0.14-0.41) | 0.35 (0.31-0.40) | 0.23 (0.14-0.38) | 0.30 (0.22-0.40) | 0.09 (0.03-0.35) | |||
| 5 y | 0.36 (0.33-0.39) | 0.20 (0.10-0.38) | 0.30 (0.26-0.34) | 0.14 (0.06-0.31) | 0.25 (0.18-0.35) | 0.09 (0.03-0.35) | |||
| NRM | .035 | .901 | .105 | ||||||
| 1 y | 0.26 (0.23-0.28) | 0.37 (0.23-0.51) | 0.31 (0.27-0.35) | 0.33 (0.22-0.46) | 0.38 (0.29-0.47) | 0.63 (0.39-0.80) | |||
| 3 y | 0.33 (0.30-0.36) | 0.46 (0.30-0.60) | 0.38 (0.34-0.42) | 0.35 (0.23-0.48) | 0.48 (0.38-0.57) | 0.63 (0.39-0.80) | |||
| 5 y | 0.36 (0.33-0.39) | 0.51 (0.33-0.67) | 0.40 (0.36-0.45) | 0.38 (0.25-0.50) | 0.49 (0.39-0.58) | 0.63 (0.39-0.80) | |||
| RI | .411 | .033 | .592 | ||||||
| 1 y | 0.22 (0.19-0.25) | 0.30 (0.17-0.43) | 0.28 (0.24-0.32) | 0.46 (0.32-0.58) | 0.28 (0.20-0.36) | 0.42 (0.21-0.62) | |||
| 3 y | 0.29 (0.26-0.32) | 0.38 (0.23-0.54) | 0.33 (0.29-0.38) | 0.46 (0.32-0.58) | 0.35 (0.26-0.44) | 0.42 (0.21-0.62) | |||
| 5 y | 0.33 (0.30-0.36) | 0.38 (0.23-0.54) | 0.37 (0.32-0.42) | 0.56 (0.39-0.70) | 0.38 (0.29-0.47) | 0.42 (0.21-0.62) | |||
| cGVHD incidence | .988 | .324 | .146 | ||||||
| 1 y | 0.32 (0.28-0.35) | 0.31 (0.15-0.48) | 0.38 (0.32-0.43) | 0.30 (0.15-0.47) | 0.37 (0.25-0.50) | 0.67 (0.30-0.88) | |||
| 3 y | 0.40 (0.36-0.43) | 0.38 (0.20-0.56) | 0.43 (0.37-0.48) | 0.30 (0.15-0.47) | 0.47 (0.34-0.59) | 0.67 (0.30-0.88) | |||
| 5 y | 0.40 (0.36-0.43) | 0.38 (0.20-0.56) | 0.45 (0.39-0.50) | 0.38 (0.17-0.60) | 0.47 (0.34-0.59) | NA | |||
NA, no cases remaining in follow-up.
OS of patients with 8/10 compatible donors transplanted with a MICA matched donor (solid black line), a MICA mismatched/MICA-129 matched donor (dashed line), and a MICA mismatched/MICA-129 mismatched donor (dotted line) (P = .096).
OS of patients with 8/10 compatible donors transplanted with a MICA matched donor (solid black line), a MICA mismatched/MICA-129 matched donor (dashed line), and a MICA mismatched/MICA-129 mismatched donor (dotted line) (P = .096).
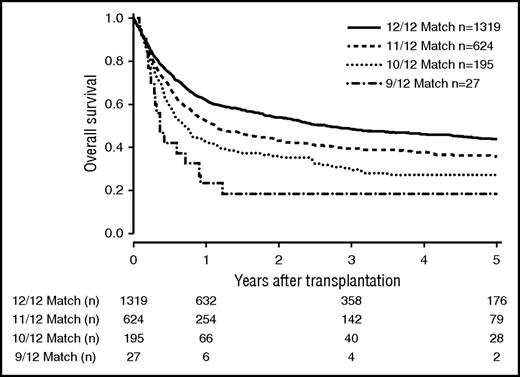
The number of mismatches on 6 loci level (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, MICA-129) strongly correlated with outcome (Figure 4). OS at 5 years was 0.44 (CI, 0.41-0.47) in the 12/12 group, 0.36 (CI, 0.32-0.41) in the 11/12 group, 0.27 (CI, 0.21-0.35) in the 10/12 group, and 0.19 (CI, 0.08-0.45) in the 9/12 group (P < .001).
OS of patients matched for 6 loci (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-MICA-129). Kaplan-Meier estimators are depicted depending on the match grade (P < .001).
OS of patients matched for 6 loci (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-MICA-129). Kaplan-Meier estimators are depicted depending on the match grade (P < .001).
A significantly higher NRM was seen for transplant pairs mismatched for MICA-129 in the 10/10 HLA match group (5-year NRM, 0.51 vs 0.36; P = .035), and a significantly higher RI was observed in the 9/10 HLA match group if MICA-129 mismatches were present (5-year RI, 0.56 vs 0.37; P = .033).
In multivariate analysis of OS, MICA-129 mismatches significantly affected outcome in the 10/10 HLA matched group (HR, 1.77; CI, 1.22-2.57; P = .003). For the end point DFS, MICA-129 mismatches were associated with increased risk for adverse events in the 10/10 HLA matched group (HR, 1.77; CI, 1.26-2.50; P = .001). Other relevant predictors in multivariate analysis were recipient age, donor age, disease stage, HLA mismatches, CMV− recipient − CMV+ donor, and KPS lower than 80 (Table 3).
Multivariate analysis
| Variables . | HR . | 95% CI . | P value . |
|---|---|---|---|
| OS | |||
| Patient age | 1.01 | 1.01-1.02 | <.001 |
| Donor age | 1.01 | 1.00-1.02 | .002 |
| Intermediate disease stage | 1.18 | 1.00-1.40 | .049 |
| Advanced disease stage | 1.79 | 1.53-2.10 | <.001 |
| HLA 9/10 | 1.21 | 1.04-1.39 | .011 |
| HLA 8/10 | 1.47 | 1.13-1.90 | .004 |
| Recipient CMV− − donor CMV+ | 1.31 | 1.06-1.63 | .013 |
| KPS < 80 | 1.75 | 1.35-2.27 | <.001 |
| 10/10 HLA, MICA-129 mismatched | 1.77 | 1.22-2.57 | .003 |
| 9/10 HLA, MICA-129 mismatched | 1.19 | 0.84-1.68 | .327 |
| 8/10 HLA, MICA-129 mismatched | 1.37 | 0.76-2.44 | .295 |
| DFS | |||
| Patient age | 1.01 | 1.01-1.02 | <.001 |
| Donor age | 1.01 | 1.00-1.01 | .013 |
| Intermediate disease stage | 1.30 | 1.11-1.51 | <.001 |
| Advanced disease stage | 1.87 | 1.61-2.17 | <.001 |
| HLA 9/10 | 1.18 | 1.03-1.35 | .015 |
| HLA 8/10 | 1.34 | 1.04-1.71 | .022 |
| Recipient CMV− − donor CMV+ | 1.29 | 1.06-1.58 | .013 |
| KPS < 80 | 1.79 | 1.40-2.30 | <.001 |
| 10/10 HLA, MICA-129 mismatched | 1.77 | 1.26-2.50 | .001 |
| 9/10 HLA, MICA-129 mismatched | 1.08 | 0.77-1.50 | .657 |
| 8/10 HLA, MICA-129 mismatched | 1.64 | 0.96-2.80 | .068 |
| Variables . | HR . | 95% CI . | P value . |
|---|---|---|---|
| OS | |||
| Patient age | 1.01 | 1.01-1.02 | <.001 |
| Donor age | 1.01 | 1.00-1.02 | .002 |
| Intermediate disease stage | 1.18 | 1.00-1.40 | .049 |
| Advanced disease stage | 1.79 | 1.53-2.10 | <.001 |
| HLA 9/10 | 1.21 | 1.04-1.39 | .011 |
| HLA 8/10 | 1.47 | 1.13-1.90 | .004 |
| Recipient CMV− − donor CMV+ | 1.31 | 1.06-1.63 | .013 |
| KPS < 80 | 1.75 | 1.35-2.27 | <.001 |
| 10/10 HLA, MICA-129 mismatched | 1.77 | 1.22-2.57 | .003 |
| 9/10 HLA, MICA-129 mismatched | 1.19 | 0.84-1.68 | .327 |
| 8/10 HLA, MICA-129 mismatched | 1.37 | 0.76-2.44 | .295 |
| DFS | |||
| Patient age | 1.01 | 1.01-1.02 | <.001 |
| Donor age | 1.01 | 1.00-1.01 | .013 |
| Intermediate disease stage | 1.30 | 1.11-1.51 | <.001 |
| Advanced disease stage | 1.87 | 1.61-2.17 | <.001 |
| HLA 9/10 | 1.18 | 1.03-1.35 | .015 |
| HLA 8/10 | 1.34 | 1.04-1.71 | .022 |
| Recipient CMV− − donor CMV+ | 1.29 | 1.06-1.58 | .013 |
| KPS < 80 | 1.79 | 1.40-2.30 | <.001 |
| 10/10 HLA, MICA-129 mismatched | 1.77 | 1.26-2.50 | .001 |
| 9/10 HLA, MICA-129 mismatched | 1.08 | 0.77-1.50 | .657 |
| 8/10 HLA, MICA-129 mismatched | 1.64 | 0.96-2.80 | .068 |
Age was modeled as continuous variable.
The prevalence of grade II to IV aGVHD was slightly higher in the MICA-129 mismatched groups for 10/10 and 8/10 matched transplantations (10/10, 29.6% vs 25.4%; 8/10, 40.7% vs 29.2%), but not for 9/10 matched cases (9/10, 32.3 vs 35.0%). The prevalence of grade III to IV aGVHD for MICA-129 mismatched cases was higher in all HLA match categories (10/10, 16.3% vs 11.0%; 9/10, 17.7% vs 16.6%; and 8/10, 25.0% vs 17.4%). In competing risks analysis for cGVHD, no significant differences were observed between MICA-129 matched and mismatched transplantations.
Discussion
Our analysis indicated that MICA-129 mismatches significantly influence survival outcome in unrelated HSCT. Because of the strong linkage within extended HLA haplotypes, and with HLA-B in particular, MICA mismatches, and consequentially also MICA-129 mismatches, are observed with increasing frequency when the number of HLA mismatches rises.21 As HLA matching must be considered as a confounder, we designed our analysis to obtain estimates for each HLA match group. Within each of these groups, OS rates were lower if a MICA-129 mismatch was present, whereas transplant pairs mismatched for MICA and matched for MICA-129 had similar survival rates as patients transplanted with MICA-matched donors. The differences were statistically significant for the 10/10 HLA as well as for the 9/10 HLA match groups (Figures 1 and 2; Table 2). The number of mismatches on 6 loci allele level, including MICA-129 to HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 matching, strongly correlated with OS. In previous studies, MICA mismatches have been associated with adverse outcome in uHSCT.10,22 We also saw survival differences between MICA matched and mismatched cases, which could be explained by MICA-129 matching. We conclude from our results that MICA-129 mismatching should be considered an independent risk factor in uHSCT. Because of strong linkage with HLA-B, we observed MICA-129 mismatches in 3.9% of the otherwise 10/10 HLA-matched cases. The effect on survival (11% to 17% 5-year OS difference) is in a magnitude, which is comparable to that of any other mismatch for “classical” HLA alleles.23,24
MICA is a highly polymorphic ligand of the activating CD94/NKG2D receptor.3 In a transplantation setting, MICA polymorphisms may influence alloreactivity of immune effector cells bearing the cognate receptor, leading to differences in relapse control.25,-27 In particular, the MICA-129 dimorphism (met/val) has been associated with differential binding affinity toward the NKG2D receptor.28 It has been shown that this, in turn, leads to differences in downstream phosphorylation of SRC kinases.14,28 Furthermore, MICA polymorphisms might lead to differences in the antigenic repertoires between donor and recipient in mismatched MICA allotypes.27,29,30 Mismatched MICA-129 might therefore induce direct T-cell alloreactivity.31,32 The immunogenic potential of MICA polymorphisms has been also demonstrated in solid organ transplantation, in which processing of MICA antigens in host antigen-presenting cells may lead to alloantibodies, causing graft rejection.9 This dual mechanism (CD94/NKG2D signaling and direct immunological recognition of MICA allotypes) may explain the findings in our study, as we found a tendency for higher relapse rates as a result of impaired relapse control in the 9/10 HLA-matched group and for higher NRM in the 10/10 HLA-matched group if MICA-129 were mismatched. Both effects add up and drive the significant differences on survival end points we observed. The CD94/NKD2D-mediated pathway is well characterized, and relevance for uHSCT has already been demonstrated.14
MICA mismatches have been associated with increased GVHD incidences.10 Although Anderson et al have questioned this relationship, the recent report of Carapito et al conclusively confirmed this association.33,34 We also saw higher rates of aGVHD in patients mismatched for MICA-129. However, Carapito et al also described a higher risk for cGVHD and a lower relapse rate in MICA mismatched cases, which we could not confirm for MICA-129 mismatched cases.34 We therefore speculate that an impaired NK-alloreactivity in the subgroup of MICA + MICA-129 mismatched cases, as opposed to MICA-mismatched + MICA-129 matched patients, may account for this difference. Carapito et al also analyzed matching for MICA-129 and saw no differences for all end points.34 Given the low frequency of MICA-129 mismatched cases, the absence of a difference in this subanalysis may be a result of low case numbers or random sampling. In the 10/10 and 9/10 HLA-matched groups, Kaplan-Meier analysis indicated an increase in mortality around 6 months after transplant, which may correlate to late-onset aGVHD or progressive onset of cGVHD in MICA-129 mismatched cases, possibly requiring prolonged intensive immunosuppression treatment in these patients. In a previous report, carriers of a “MICA-129-met” allele have shown better survival outcomes, and in another study, Boukouaci et al showed that patients with a “MICA-129-val/val” phenotype associated with higher cGVHD incidence.14,25 Interpretation of these data are limited by sample size, and the data of Isernhagen et al are additionally difficult to interpret because of the presence of HLA mismatches.14 In our analysis, the MICA-129 phenotype of patients or donors did not correlate with any survival outcome within the different HLA match groups. MICA-129 mismatches are quite rare, and therefore high numbers of transplants need to be analyzed to achieve sufficient statistical power, which may be the reason why the effect we describe has remained unnoticed in previous studies. Although MICA-129 mismatches are rare, according to our data, they are associated with a high-risk situation, which is of clinical relevance. Our results underscore that the MICA-129 dimorphism is an important regulator of NKG2D receptor-bearing cells and suggest that the MICA-129 dimorphism also plays a role in maturation or licensing of these immune effector cells (NK cells, γδ-T cells, or NKT cells).
Multivariate analysis was performed for OS and DFS. As in univariate analysis, a significantly increased risk was found for 10/10 HLA-matched patients if mismatched MICA-129 was present. The estimates obtained confirmed the independent effect of MICA mismatches on survival end points for 10/10 matched transplantations. We also confirm a significant effect of donor age on survival after uHSCT (Table 3).35 Conditioning regimen intensity was not a significant predictor in multivariate analysis, and further subanalyses for differences of MICA-129 matching in the different categories of conditioning intensity was not possible because of power considerations. Similarly, subanalysis according to the vector of mismatches for MICA/MICA-129 was not performed because of the small resulting subgroups.
A limitation is that HLA-DPB1 typing was available only for 64.1% of the transplantations. Nonpermissive HLA-DPB1 mismatches have been shown to influence outcome of allogeneic stem cell transplantation adversely.36 No significant difference in the distribution of HLA-DPB1 T-cell epitope group (TCE) mismatches and MICA mismatches was found (P = .788). Also, no significant difference between HLA-DBP1 TCE mismatches and MICA-129 matching was found (P = .444). Inclusion of DPB1 TCE matching for cases with available HLA-DPB1 data in multivariate analysis did not significantly improve our models, and was therefore not included in the final analysis. In particular, an interaction between MICA-129 mismatches and HLA-DPB1 matching was not found in this subset; this, however, should be reevaluated in a confirmatory prospective trial.37
The MICA-129 dimorphisms were inferred from genomic sequencing. According to our data, assignment of the MICA-129 dimorphism would be sufficient for clinical purposes. This could be done quickly and inexpensively, using sequence-specific oligonucleotides or TaqMan assays.25,38 Our cohort contains a relevant proportion of older patients (ie, patients aged 65 years or older, n = 325; 14.9%), and almost half of the transplantations were performed after 2008 (n = 1041; 47.9%). Therefore, our cohort reflects the current transplantation situation in Germany.39 To limit disease-associated heterogeneity, we included the largest 5 disease entities currently coordinated by our search unit.
To overcome the HLA match confounding in the MICA compatibility analysis in different HLA match subgroups, some statistical calculations with a reduced power had to be accepted. In particular, for competing risks’ end points, more cases are necessary to show potential effects more clearly. Our interpretation regarding aGVHD should be viewed with caution, as no competing risks analysis was possible because of a missing aGVHD onset date in the ProMISe database.
In summary, we show that MICA-129 mismatches are an independent risk factor for unrelated HSCT. Transplantations with mismatched MICA-129 define a small but high-risk subset. If our results can be confirmed in a prospective trial including HLA-DPB1 typing, MICA-129 typing and matching may be recommended for donor search in unrelated HSCT. Inclusion of MICA typing in registry typing protocols may be beneficial for identification of MICA compatible donors for patients with rare or unusual HLA-B/MICA combinations.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Deutsche José Carreras Leukämie-Stiftung e.V. (Grant DJCLS 11/10) and the German Red Cross Blood Transfusion Service, Baden-Württemberg/Hessen.
Authorship
Contribution: D.F., J.M., H.S., and C.M. are principal investigators. They designed the study, performed data analysis/interpretation, and wrote the manuscript. C.N., D.M., and C.T. contributed in MICA typing and in editing of the manuscript. D.N., D.B., M.G., E.W., G.W., B.G., M.P., H.E., R.A., G.S., K.S.-E., S.F., J.C., M.K., M.W., B.H., S.K., and M.R. contributed patients, reviewed the data, and coedited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joannis Mytilineos, Institute of Clinical Transfusion Medicine and Immunogenetics Ulm, Department of Transplantation Immunology, Helmholtzstrasse 10, 89081 Ulm, Germany; e-mail: j.mytilineos@blutspende.de.