Key Points
A platform for the generation of clinical-grade CD19-CAR–modified TSCM.
CD19-CAR–modified TSCM mediate superior antitumor responses compared with CD19-CAR T cells currently used in clinical trials.
Abstract
Long-lived, self-renewing, multipotent T memory stem cells (TSCM) can trigger profound and sustained tumor regression but their rareness poses a major hurdle to their clinical application. Presently, clinically compliant procedures to generate relevant numbers of this T-cell population are undefined. Here, we provide a strategy for deriving large numbers of clinical-grade tumor-redirected TSCM starting from naive precursors. CD8+CD62L+CD45RA+ naive T cells enriched by streptamer-based serial-positive selection were activated by CD3/CD28 engagement in the presence of interleukin-7 (IL-7), IL-21, and the glycogen synthase-3β inhibitor TWS119, and genetically engineered to express a CD19-specific chimeric antigen receptor (CD19-CAR). These conditions enabled the generation of CD19-CAR–modified CD8+ TSCM that were phenotypically, functionally, and transcriptomically equivalent to their naturally occurring counterpart. Compared with CD8+ T cells generated with clinical protocols currently under investigation, CD19-CAR–modified CD8+ TSCM exhibited enhanced metabolic fitness and mediated robust, long-lasting antitumor responses against systemic acute lymphoblastic leukemia xenografts. This clinical-grade platform provides the basis for a phase 1 trial evaluating the activity of CD19-CAR–modified CD8+ TSCM in patients with B-cell malignancies refractory to prior allogeneic hematopoietic stem cell transplantation.
Introduction
Adoptive transfer of tumor-specific T lymphocytes is an effective treatment of patients with advanced cancer.1,2 Advances in gene transfer technology permit the conveyance of de novo cancer reactivity to any type of T cell through genetic engineering of tumor-reactive T-cell receptors (TCRs) or chimeric antigen receptors (CARs).1,2 Akin to other tissues, T cells exist in a continuum of differentiation states characterized by the gradual acquisition or loss of phenotypic traits, functional properties, and gene expression patterns.3,4 At the extremes of the differentiation spectrum are antigen-inexperienced naive T cells (TN) and terminally differentiated effectors (TTE).3,4 Memory T cells represent cells at an intermediate state of differentiation that can be further divided into memory stem cells (TSCM), central memory cells (TCM), and effector memory cells (TEM) along a progressive developmental path.3,4 Which T-cell subsets should be used for adoptive immunotherapy has been debated for many years,5 but cumulating evidence in mice indicates that the infusion of less-differentiated T cells results in greater cell expansion, persistence, and tumor destruction.6-11 In particular, TSCM have been shown to eradicate large tumors even when limited numbers of cells were transferred, a condition in which other memory and effector subsets had little impact.9,10
Despite overwhelming preclinical data indicating the benefit of tumor-redirecting less-differentiated T-cell subsets, clinical trials have largely used TCR or CAR-engineered T cells derived from unfractionated peripheral blood mononuclear cells (PBMCs). This strategy not only simplifies the manufacturing process, but it also generates inconsistent cell products because the PBMC composition can significantly vary between individuals as a consequence of age,12,13 pathogen exposure,14 and prior systemic treatments.15 Moreover, unselected populations, especially those skewed toward TEM and TTE, often fail to generate viable clinical products due to poor in vitro cell expansion.16 Recently, several clinical trials in which tumor-redirected T cells were derived from TCM have been reported.17,18 However, clinical exploitation of TSCM has so far been hampered by their relative paucity in the circulation and the lack of robust, clinical-grade protocols capable of isolating and maintaining this cell type in vitro.19
Activation of TN in the presence of interleukin-7 (IL-7) and IL-15 has been reported to promote the generation of TSCM-like cells.20-22 However, cells generated under these conditions have some discrepancies with the phenotype of TSCM as they express CD45RO,20,21 which is absent from the surface of naturally occurring TSCM.9,23 Here, we report that clinical-grade tumor-redirected TSCM can be efficiently induced by activating naive precursors in the presence of IL-7, IL-21, and the glycogen synthase-3β (GSK-3β) inhibitor TWS119. These cells display the phenotype, functions, and a gene expression profile equivalent to their naturally occurring counterpart. More importantly, tumor-redirected CD8+ TSCM mediated superior and more durable antitumor responses than CD8+ T cells generated with protocols currently used in clinical trials.
Materials
Manufacturing of CD19-CAR–modified T cells
PBMCs were obtained from healthy donors (Transfusion Medicine Department, Clinical Center, National Institutes of Health [NIH]) or patients enrolled in clinical trials approved by the National Cancer Institute (NCI) Institutional Review Board. PBMCs were either frozen (“standard” cell product) or further enriched for naive CD8+CD62L+CD45RA+cells by serial-positive magnetic bead enrichment using clinical-grade (Stage Cell Therapeutics GmbH) or research-grade Fab streptamers (IBA GmbH) before freezing (TSCM-enriched product), as described in the supplemental Methods (available on the Blood Web site). CD19-CAR–modified standard cells were generated from thawed PBMCs as previously described.24 To generate CD19-CAR–modified TSCM-enriched cells, naive CD8+ T cells were thawed and activated with anti-CD3/CD28 beads (1:1 bead-to-cell ratio) (Dynabeads Human T-Expander CD3/CD28; Thermo Fisher Scientific) in AIM-V (Thermo Fisher Scientific) 5% human AB serum (Valley Biomedical) supplemented with 2 mM glutamax (Thermo Fisher Scientific) in the presence of 5 ng/mL IL-7, 30 ng/mL IL-21 (Cellgenix), and 5 μM TWS119 (Cayman Chemical, revialed by the NIH Pharmaceutical Development Section under current Good Manufacturing Practice aseptic conditions). Cells were transduced on days 2 and 3 with the CD19-CAR (FMC63-28-ζ) retrovirus25 and expanded for 5 more days in media containing IL-7, IL-21, and TWS119 after removal of the activating beads on day 4.
Cell lines
SUDHL4 (diffuse large B-cell lymphoma line), CCRF-CEM (acute lymphoblastic T-cell leukemia line), and NALM6-GL (acute lymphoblastic leukemia line, stably transfected with green fluorescent protein and luciferase) were cultured in RPMI 1640 media supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM glutamax, and 1 mM sodium pyruvate (Thermo Fisher Scientific).
Flow cytometry and intracellular cytokine staining
A detailed list of the antibodies used in flow cytometry experiments can be found in the supplemental Methods. Intracellular cytokine staining of CD19-CAR–modified T-cell products was performed as previously described26 after a 6-hour coculture in the presence of SUDHL4 or CCRF-CEM. A LSRII (BD Biosciences) was used for flow cytometry acquisition. Samples were analyzed with FlowJo software (TreeStar).
Cytokine release
CD19-CAR–modified T-cell products were seeded at the density of 150 000 cells per well in 96-well round-bottom plates (Corning) in complete medium without cytokines. T cells were cultured alone or cocultured with SUDHL4 B-cells or CCRF-CEM (1:1 ratio). After 16 hours of culture, supernatants were collected and assayed using the Bio-PlexPro Human Cytokine 8-plex assay (Bio-Rad). Data were acquired with a Bio-Plex MAGPIX Multiplex reader and the results were analyzed with Bio-Plex Manager MP Software and Bio-Plex Data Pro Software (Bio-Rad).
Bioenergetic analyses
Glycolyis and mitochondrial stress test were performed using the XFe-24 metabolic extracellular flux analyzer (Seahorse Bioscience). Assay conditions and reagents are described in detail in the supplemental Methods.
Animal studies
Animal studies were carried out under protocols approved by the NCI Bethesda Animal Care and Use Committee. NOD scid γ, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice were purchased from The Jackson Laboratory. Two million NALM6-GL were injected IV, followed 3 days later by 2.5 × 105 CD19-CAR+CD8+ T cells enriched using the CD8+ T-cell isolation kit (Miltenyi Biotec). Recombinant human IL-15 (NCI) was injected intraperitoneally every other day(1 μg per mouse). Tumor burden was measured using the Xenogen IVIS Lumina (Caliper Life Sciences) as previously described.27
Microarray analysis
Microarray analysis of standard and TSCM-enriched products was performed as previously described9 using the Whole Transcript Human Gene 1.0 ST arrays (Affymetrix). A detailed description of the analyses is found in the supplemental Methods.
Quantitative reverse transcription PCR
Total RNA was reverse-transcribed using the SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) or the Transcriptor High Fidelity cDNA Synthesis kit (Roche) followed by polymerase chain reaction (PCR) on an Applied Biosystems ViiA 7 or a CFX Connect Real-Time PCR Detection System. Primer sequences and probes are provided in the supplemental Methods.
Statistical analyses
A 2-tailed Wilcoxon matched-pairs signed rank test was used for data comparison. A log-rank (MantelCox) test was used for comparison of survival curves. Pearson correlation was used to correlate messenger RNA levels determined by quantitative reverse transcription PCR (qRT-PCR) with log2-transformed microarray intensity values.
Results
Efficient enrichment of naive CD8+ T cells by Fab-streptamer technology
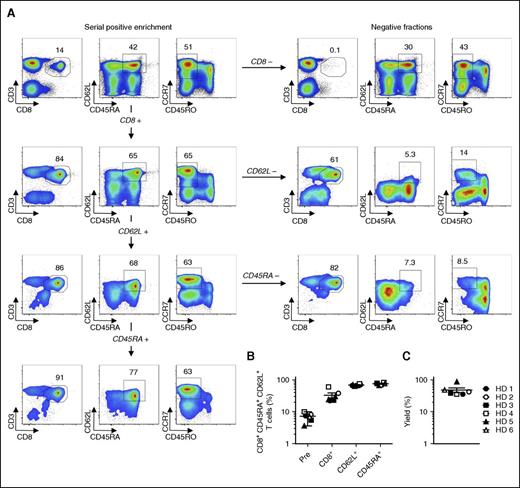
Because T-cell differentiation is tightly linked to cell division,28-30 we sought to avoid excessively expanding T cells by isolating large numbers of naive precursors that could be efficiently programmed to differentiate into TSCM under culture conditions that restrain proliferation.9,10 Although the purification of naive cells adds a layer of complexity to the manufacturing procedure, it is a necessary step because the presence of memory T cells during stimulation of TN has been shown to accelerate their differentiation.31 We took advantage of a reversible multimer technology based on clinical-grade Fab streptamers32 to enrich naive CD8+ T lymphocytes by sequentially purifying CD8+, CD62L+, and CD45RA+ cells with paramagnetic beads. A representative example of the entire cell separation procedure, including all positive and negative fractions, is shown in Figure 1A. The enrichment protocol was highly reproducible across 6 independent healthy donors, resulting in final cell products with a median purity of 76.8% ± 5.9% (Figure 1A-B). The cell yields were also consistent across all donors, with a median value of about 40% (Figure 1C). From leukapheresis products ranging from 3.73 to 7.95 × 109 cells, we purified a median of 1.53 × 108 ± 0.6 × 108 naive CD8+CD62L+CD45RA+ T cells (supplemental Table 1). These data demonstrate that reversible Fab-streptamer technology can be effectively used to enrich large numbers of TN using clinically compliant reagents.
Enrichment of naive CD8+ T cells by Fab-streptamer technology. (A) Flow cytometry analyses of fresh human PBMCs from a healthy donor (HD) prior to and after sequential enrichment of CD8+, CD62L+, and CD45RA+ cells with Fab multimers conjugated with Strep-Tactin–functionalized magnetic beads. Living lymphocytes in the respective positive and negative fractions of each selection step are shown. Data are shown after gating on live cells. Numbers indicate the percentage of cells in each gate. (B) Percentage of CD8+CD62L+CD45RA+ T cells prior to and after each selection step from 6 HDs; mean value ± standard error of the mean (SEM) is indicated. (C) Percentage yields of the target CD8+CD62L+CD45RA+ T cells from 6 HD; mean value ± SEM is indicated.
Enrichment of naive CD8+ T cells by Fab-streptamer technology. (A) Flow cytometry analyses of fresh human PBMCs from a healthy donor (HD) prior to and after sequential enrichment of CD8+, CD62L+, and CD45RA+ cells with Fab multimers conjugated with Strep-Tactin–functionalized magnetic beads. Living lymphocytes in the respective positive and negative fractions of each selection step are shown. Data are shown after gating on live cells. Numbers indicate the percentage of cells in each gate. (B) Percentage of CD8+CD62L+CD45RA+ T cells prior to and after each selection step from 6 HDs; mean value ± standard error of the mean (SEM) is indicated. (C) Percentage yields of the target CD8+CD62L+CD45RA+ T cells from 6 HD; mean value ± SEM is indicated.
Generation of CD19-CAR–modified TSCM from naive precursors
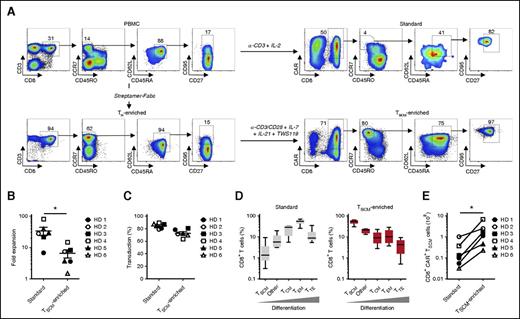
TSCM can be generated and maintained in vitro by stimulating TN in the presence of small molecules activating the WNT/β-catenin pathway (ie, GSK-3β inhibitors).9,10 To maximize the induction of TSCM, we developed a new protocol that involves the use of the GSK-3β inhibitor TWS119 and a cocktail of cytokines implicated in the generation and maintenance of less-differentiated T-cell subsets. Specifically, we used IL-7, which provides key instructive signals for the formation of TSCM,20 and IL-21, which has profound inhibitory effects on effector T-cell differentiation33 and maintains high-level expression of CD28 compared with IL-1534 (supplemental Figure 1). Although IL-21 has been shown to preserve the expression of the WNT/β-catenin transcription factors,33 blockade of GSK-3β by TWS119 was necessary for β-catenin stabilization and maximal expression of TCF7 and LEF1 (supplemental Figure 2). Fab-streptamer–enriched CD8+ TN were activated by anti-CD3/CD28 beads in the presence of IL-7, IL-21, and TWS119 and transduced with a γ-retroviral vector encoding the CD19-CAR (FMC63-28-ζ) currently used in clinical trials.24,35-39 As controls, we generated standard CD19-CAR T cells from the PBMCs of the same donors using culture conditions (ie, soluble anti-CD3 antibody and IL-2) presently adopted to produce TCR- or CAR-engineered T cells for clinical trials at our institution.24,35,36,39-41 A scheme and representative phenotypes of T cells at different steps of the manufacturing process are provided in Figure 2A.
CD8+ T-cell subset composition of CD19-CAR–modified standard and TSCM-enriched products. (A) Flow cytometry analyses of cells from a representative healthy donor (HD) at different steps of the 9-day manufacturing processes for the generation of CD19-CAR–modified standard and TSCM-enriched products. Data are shown after gating on live cells (left panels) or live CD3+ T cells (right panels). Numbers indicate the percentage of cells in each gate. (B) Fold expansion of CD19-CAR–modified T cells generated under standard and TSCM-enriched culture conditions. Data represent results from 6 HD; mean value ± SEM is indicated (*P < .05; Wilcoxon matched-pairs signed rank test). (C) Percentage of CD19-CAR–transduced T cells in standard and TSCM-enriched products. Data represent results from 6 HDs; mean value ± SEM is indicated. (D) Percentage of CD8+ T-cell subsets in the CD19-CAR–modified standard and TSCM-enriched products. TSCM were defined as CD3+CD8+CD45RO−CCR7+CD45RA+CD62L+CD27+CD95+ cells; TCM as CD3+CD8+CCR7+CD45RO+; TEM as CD3+CD8+CCR7−CD45RO+; TTE as CD3+CD8+CCR7−CD45RO−; other cells as CD3+CD8+CCR7+CD45RO− not displaying the full TSCM phenotype. Data representing results from 6 HDs are shown as box-and-whisker plots extending to minimum and maximum values. Bands inside the box represent median values. (E) Theoretical number of CD19-CAR+CD8+ TSCM obtainable from stimulation of 1 × 108 PBMCs (standard products) or Fab-streptamer–enriched CD8+ TN (TSCM-enriched products). Data represent results from 6 HDs (*P < .05; Wilcoxon matched-pairs signed rank test).
CD8+ T-cell subset composition of CD19-CAR–modified standard and TSCM-enriched products. (A) Flow cytometry analyses of cells from a representative healthy donor (HD) at different steps of the 9-day manufacturing processes for the generation of CD19-CAR–modified standard and TSCM-enriched products. Data are shown after gating on live cells (left panels) or live CD3+ T cells (right panels). Numbers indicate the percentage of cells in each gate. (B) Fold expansion of CD19-CAR–modified T cells generated under standard and TSCM-enriched culture conditions. Data represent results from 6 HD; mean value ± SEM is indicated (*P < .05; Wilcoxon matched-pairs signed rank test). (C) Percentage of CD19-CAR–transduced T cells in standard and TSCM-enriched products. Data represent results from 6 HDs; mean value ± SEM is indicated. (D) Percentage of CD8+ T-cell subsets in the CD19-CAR–modified standard and TSCM-enriched products. TSCM were defined as CD3+CD8+CD45RO−CCR7+CD45RA+CD62L+CD27+CD95+ cells; TCM as CD3+CD8+CCR7+CD45RO+; TEM as CD3+CD8+CCR7−CD45RO+; TTE as CD3+CD8+CCR7−CD45RO−; other cells as CD3+CD8+CCR7+CD45RO− not displaying the full TSCM phenotype. Data representing results from 6 HDs are shown as box-and-whisker plots extending to minimum and maximum values. Bands inside the box represent median values. (E) Theoretical number of CD19-CAR+CD8+ TSCM obtainable from stimulation of 1 × 108 PBMCs (standard products) or Fab-streptamer–enriched CD8+ TN (TSCM-enriched products). Data represent results from 6 HDs (*P < .05; Wilcoxon matched-pairs signed rank test).
As anticipated, cells grown in the presence of a CD3-specific antibody and IL-2 expanded significantly greater numbers than did cells grown under TSCM-favoring conditions (Figure 2B). Given the notorious ability of retroviruses to exclusively infect actively dividing cells42 and the striking differences in cell expansion between the 2 groups, we were surprised to observe only marginal differences in transduction efficiencies. The frequency of CD19-CAR+ T cells was relatively high for both culture conditions, averaging 73.3% ± 6.8% in TSCM cultures and 84.1% ± 4.3% in standard cultures (Figure 2C). Polychromatic flow cytometry analyses of the end products revealed major phenotypic differences between the 2 manufacturing procedures (Figure 2A,D). Consistent with prior reports, standard CD19-CAR CD8+ T cells were largely composed of highly differentiated T-cell subsets such as TEM (52.7% ± 14.9%) and TTE (12.8% ± 11.1%). Notably, this culture condition was inadequate to sustain the generation and maintenance of TSCM, which represented a median 1.36% of final cell products. Conversely, activation of naive CD8+ T cells by CD3/CD28 engagement in the presence of IL-7, IL-21, and TWS119 resulted in cell products primarily enriched with TSCM (52.2% ± 12.52%). Another large component of the final cell preparation (20.1% ± 4.7%) consisted of naive-like T cells (CCR7+CD45RO−), which did not express the full phenotypic traits of TSCM. Similar phenotypic differences were observed in the CD4+ CD19-CAR+ T-cell compartment with standard cultures predominantly yielding TEM whereas TSCM conditions gave rise to less-differentiated T-cell subsets (supplemental Figure 3). Despite restraining T-cell expansion and reducing the transduction efficiency, TSCM culture conditions generated >10-fold higher numbers of CD19-CAR+ TSCM (Figure 2E). A summary of the cell yields for each step of the 2 cell-manufacturing procedures is provided in supplemental Table 2. Theoretical CD19-CAR+ TSCM yields from our series of 6 healthy donors ranged from 8.2 × 107 to 1.1 × 109 cells with a median of 2.63 × 108 of CD19-CAR+ TSCM. By comparison, generation of equivalent numbers of CD19-CAR+ TSCM by standard methods would be far more expensive (supplemental Table 3). Altogether, these findings indicate that clinically relevant numbers of tumor-redirected TSCM can be efficiently obtained using this newly developed cell-manufacturing protocol.
Effector cells generated under TSCM culture conditions exhibit enhanced polyfunctionality
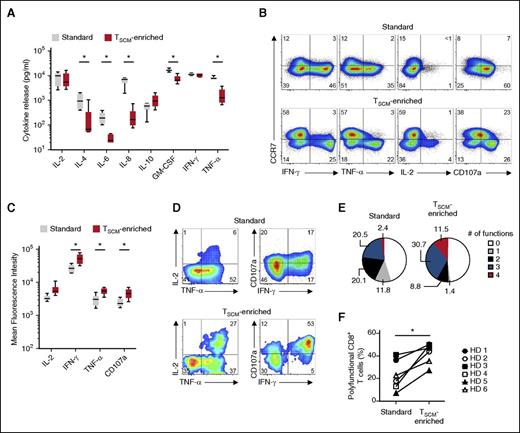
To compare the functionality of standard and TSCM-enriched products, we measured a variety of cytokines released by CD19-CAR–modified T cells after overnight cocultures with CD19+ SUDHL4 or CD19− CCRF-CEM cell lines. Standard CD19-CAR+ T cells released greater amounts of IL-4, IL-6, IL-8, granulocyte-macrophage colony-stimulating factor (GM-CSF), and tumor necrosis factor-α (TNF-α) than TSCM-enriched cells (Figure 3A). These results likely reflected differences in the frequency of CD4+ T cells, which are the main producers of IL-4, IL-6, and IL-8. Indeed, TSCM-enriched preparations were enriched with CD8+ T cells by protocol design. Surprisingly, despite a skewing in effector subsets in standard CD19-CAR+ T cells, we did not detect significant differences in the release of interferon-γ (IFN-γ) and IL-2 between the 2 groups (Figure 3A).
Effector CD8+ T cells generated under TSCM-enriched culture condition are polyfunctional. (A) Concentration of cytokine in the supernatant of CD19-CAR–modified T cells after 16-hour coculture with CD19+ SUDHL4 cells or CD19− CCRF-CEM cells. Values are shown after subtraction of background cytokine release values obtained after coculture with the CCRF-CEM. Data representing results from 6 healthy volunteer donors (HDs) are shown as box-and-whisker plots extending to minimum and maximum values. Bands inside the box represent median values (*P < .05; Wilcoxon matched-pairs signed rank test). (B) Intracellular cytokine staining of CD19-CAR–modified standard and TSCM-enriched products from a representative HD after coculture with CD19+ SUDHL4 cells. Data are shown after gating on live CD3+CD8+ cells. Numbers indicate the percentage of cells in each quadrant. (C) Mean fluorescence intensity of cytokines produced by CCR7− effector CD8+ T cells within standard and TSCM-enriched products after coculture with CD19+ SUDHL4 cells. Data representing results from 6 HDs are shown as box-and-whisker plots extending to minimum and maximum values. Bands inside the box represent median values (*P < .05; Wilcoxon matched-pairs signed rank test). (D) Intracellular cytokine staining of CD19-CAR–modified standard and TSCM-enriched products from a representative HD after coculture with CD19+ SUDHL4 cells. Data are shown after gating on live CD3+CD8+CCR7− cells. Numbers indicate the percentage of cells in each quadrant. (E) Pie charts depicting the quality of the cytokine response in CCR7− effector CD8+ T cells from 6 HDs after coculture with CD19+ SUDHL4 cells. Values are determined by the Boolean combination of gates identifying IFN-γ+, IL-2+, TNF-α+, and CD107a+ cells. Numbers indicate cell percentages. (F) Percentage of polyfunctional CCR7− effector CD8+ T cells from 6 HDs after coculture with CD19+ SUDHL4 cells (*P < .05; Wilcoxon matched-pairs signed rank test).
Effector CD8+ T cells generated under TSCM-enriched culture condition are polyfunctional. (A) Concentration of cytokine in the supernatant of CD19-CAR–modified T cells after 16-hour coculture with CD19+ SUDHL4 cells or CD19− CCRF-CEM cells. Values are shown after subtraction of background cytokine release values obtained after coculture with the CCRF-CEM. Data representing results from 6 healthy volunteer donors (HDs) are shown as box-and-whisker plots extending to minimum and maximum values. Bands inside the box represent median values (*P < .05; Wilcoxon matched-pairs signed rank test). (B) Intracellular cytokine staining of CD19-CAR–modified standard and TSCM-enriched products from a representative HD after coculture with CD19+ SUDHL4 cells. Data are shown after gating on live CD3+CD8+ cells. Numbers indicate the percentage of cells in each quadrant. (C) Mean fluorescence intensity of cytokines produced by CCR7− effector CD8+ T cells within standard and TSCM-enriched products after coculture with CD19+ SUDHL4 cells. Data representing results from 6 HDs are shown as box-and-whisker plots extending to minimum and maximum values. Bands inside the box represent median values (*P < .05; Wilcoxon matched-pairs signed rank test). (D) Intracellular cytokine staining of CD19-CAR–modified standard and TSCM-enriched products from a representative HD after coculture with CD19+ SUDHL4 cells. Data are shown after gating on live CD3+CD8+CCR7− cells. Numbers indicate the percentage of cells in each quadrant. (E) Pie charts depicting the quality of the cytokine response in CCR7− effector CD8+ T cells from 6 HDs after coculture with CD19+ SUDHL4 cells. Values are determined by the Boolean combination of gates identifying IFN-γ+, IL-2+, TNF-α+, and CD107a+ cells. Numbers indicate cell percentages. (F) Percentage of polyfunctional CCR7− effector CD8+ T cells from 6 HDs after coculture with CD19+ SUDHL4 cells (*P < .05; Wilcoxon matched-pairs signed rank test).
To better understand of the functional characteristics of distinct T-cell subsets within the final cell products, we measured the production of IFN-γ, IL-2, TNF-α, and the degranulation marker CD107a by intracellular cytokine staining. As expected, less-differentiated CCR7+CD8+ T cells exhibited reduced effector functions in both groups. Interestingly, CCR7− TEM and TTE generated under TSCM culture conditions exhibited an increased ability to degranulate and produce inflammatory cytokines as manifested by higher values of mean fluorescence intensities (Figure 3B-C). The capacity of T cells to produce large amounts of cytokines has been linked to an ability to simultaneously exert multiple effector functions.43 Consistent with this notion, a higher frequency of effector T cells generated under TSCM culture conditions displayed polyfunctional capacity, performing 3 or more functions (Figure 3D-F). Conversely, the majority of responding effector T cells generated with the standard protocol were mono or bifunctional (Figure 3D-F). No significant functional differences were observed in the CD4+ T-cell compartment (supplemental Figure 4). These findings revealed that, in addition to containing a large fraction of less-differentiated T cells with enhanced potential for reconstitution, the CD19-CAR–modified TSCM-enriched product comprises a small fraction of effector cells capable of immediately mediating multiple antitumor functions.
CD19-CAR–modified TSCM have a gene expression profile analogous to naturally occurring TSCM
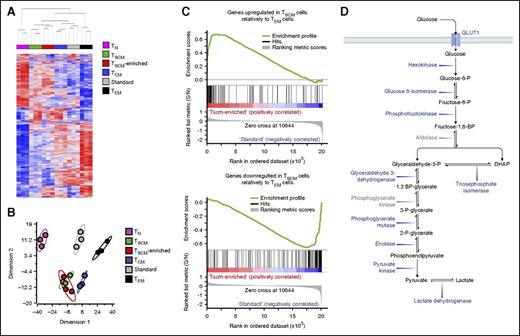
To determine whether in vitro–generated CD19-CAR–modified TSCM were equivalent to their naturally occurring counterpart, we compared their transcriptome profile with that of circulating TSCM. For this comparison, we focused on 900 transcripts found to be differentially expressed among TN, TSCM, TCM, and TEM.9 Unsupervised hierarchical clustering and principal component analysis revealed that TSCM-enriched products were closely related to naturally occurring TSCM (Figure 4A-B). Clustering analyses were further corroborated by gene set enrichment analyses,44 which showed that CD19-CAR–modified TSCM products were positively enriched with genes upregulated in TSCM compared with TEM whereas standard preparations were enriched with genes overexpressed in TEM compared with TSCM (Figure 4C).
CD19-CAR–modified TSCM have a transcriptome profile similar to their naturally occurring counterpart. (A) Hierarchical clustering of CD19-CAR–modified standard and TSCM-enriched products and naturally occurring CD8+ T-cell subsets from GSE23321 performed using a 900 gene list from Gattinoni et al.9 Red and blue colors indicate increased and decreased expression of 900 differentially regulated genes described by Gattinoni et al.9 Each column represents a sample and each row, a gene. (B) Principal component analysis (PCA) of the 900 differentially expressed genes described by Gattinoni et al9 in CD19-CAR–modified standard and TSCM-enriched products and naturally occurring CD8+ T-cell subsets from Gattinoni et al.9 (C). Gene set enrichment analysis (GSEA) on transcriptomes of CD19-CAR–modified standard and TSCM-enriched products using genes upregulated or downregulated in TSCM relative to TEM retrieved from Gattinoni et al9 as gene sets. (D) Schematic representation of the glycolytic pathway. In blue font, glycolytic enzymes and molecules whose genes were downregulated in CD19-CAR–modified TSCM-enriched cells compared with standard products (P < .05). In gray font, glycolytic enzymes not differentially expressed. DHAP, dihydroxyacetone phosphate; S/N, signal/noise.
CD19-CAR–modified TSCM have a transcriptome profile similar to their naturally occurring counterpart. (A) Hierarchical clustering of CD19-CAR–modified standard and TSCM-enriched products and naturally occurring CD8+ T-cell subsets from GSE23321 performed using a 900 gene list from Gattinoni et al.9 Red and blue colors indicate increased and decreased expression of 900 differentially regulated genes described by Gattinoni et al.9 Each column represents a sample and each row, a gene. (B) Principal component analysis (PCA) of the 900 differentially expressed genes described by Gattinoni et al9 in CD19-CAR–modified standard and TSCM-enriched products and naturally occurring CD8+ T-cell subsets from Gattinoni et al.9 (C). Gene set enrichment analysis (GSEA) on transcriptomes of CD19-CAR–modified standard and TSCM-enriched products using genes upregulated or downregulated in TSCM relative to TEM retrieved from Gattinoni et al9 as gene sets. (D) Schematic representation of the glycolytic pathway. In blue font, glycolytic enzymes and molecules whose genes were downregulated in CD19-CAR–modified TSCM-enriched cells compared with standard products (P < .05). In gray font, glycolytic enzymes not differentially expressed. DHAP, dihydroxyacetone phosphate; S/N, signal/noise.
Priming naive CD8+ T cells in the presence of IL-7 and IL-15 has been proposed to be an effective method to generate TSCM in vitro.20-22 Cieri and colleagues reported that although T cells obtained under these culture conditions do not fully display the TSCM phenotype originally described, they express a core of genes characteristic of naturally occurring TSCM.20 However, Cieri’s gene list consists only of 65 genes, about half of which are present in the 900 transcripts used in our analysis (supplemental Figure 5A). We sought to evaluate TSCM generated under these 2 protocols by comparing Cieri’s dataset with ours. Additionally, we used a large third-party dataset from Willinger et al45 that comprises genes differentially regulated in TN, TCM, TEM, and effector memory T cells that reacquired CD45RA expression (TEMRA). Because gene expression profiles in these reports were obtained using different platforms, we used 62 of 65,20 740 of 900,9 and 1383 of 209245 genes that matched across these studies (supplemental Figure 5A; supplemental Table 4). Results from these different platforms were comparable, as naturally occurring TN and TCM subsets from the 2 studies closely clustered together (supplemental Figure 5B-E). Consistent with Cieri’s results,20 TSCM-like cells generated with IL-7 and IL-15 (originally labeled as naive-derived T cells [T(TN)]) grouped closely with naturally occurring TSCM when we used the 62 gene set, although cells obtained using our protocol clustered even more tightly (supplemental Figure 5B). However, when we used a larger set of genes, T(TN) failed to cluster with naturally occurring TSCM and exhibited a gene expression profile more related to standard T cells (supplemental Figure 5C-E). Taken together, these findings underscore that naive T cells activated in the presence of IL-7, IL-21, and TWS119, but not IL-7 and IL-15, have a transcriptomic profile similar to naturally occurring TSCM.
CD19-CAR–modified TSCM have enhanced metabolic fitness
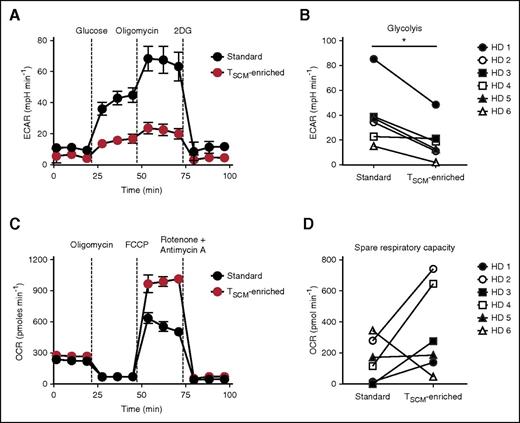
Pathway analysis performed on differentially regulated genes between standard and TSCM-enriched CD19-CAR–modified cell products (supplemental Table 5) revealed that glycolysis was the most significantly modulated pathway (supplemental Figure 6A-B). Accordingly, the vast majority of glycolytic enzymes as well as the gene encoding for glucose transporter 1 (SLC2A1) were significantly downregulated in TSCM-enriched compared with standard cells (Figure 4D). These findings were further validated by qRT-PCR (supplemental Figure 6C). Because glycolysis is a major metabolic pathway limiting the ability of CD8+ T cells to form long-lived memory cells,46 we sought to determine whether these changes in the expression of glycolytic genes were associated with functional metabolic differences. We measured the extracellular acidification rate (ECAR), which quantifies proton production as a surrogate for lactate production and glycolytic flux, in response to glucose supplementation and subsequent administration of the adenosine triphosphate (ATP) synthase inhibitor oligomycin, which drives cells to maximal glycolytic activity by shutting-down oxidative phosphorylation. Consistent with the gene expression results, TSCM-enriched cells exhibited only modest increases in ECAR levels compared with standard cells, which instead demonstrated robust glycolysis and glycolytic capacity after sequential administration of glucose and oligomycin (Figure 5A-B).
CD19-CAR–modified TSCM exhibit enhanced metabolic fitness. (A) ECAR of CD19-CAR–modified standard and TSCM-enriched products from a representative healthy donor (HD) under basal culture conditions and in response to the indicated molecules. Data are shown as mean values ± SEM. (B) Glycolytic response by CD19-CAR–modified standard and TSCM-enriched products of 6 HDs following glucose administration. ECAR values are shown after subtraction of basal ECAR measurements (*P < .05; Wilcoxon matched-pairs signed rank test). (C) OCR of CD19-CAR–modified standard and TSCM-enriched products from a representative HD under basal culture conditions and in response to the indicated molecules. Data are shown as mean values ± SEM. (D) SRC (maximal OCR − basal OCR) of CD19-CAR–modified standard and TSCM-enriched products of 6 HD. 2-DG, 2-Deoxyglucose.
CD19-CAR–modified TSCM exhibit enhanced metabolic fitness. (A) ECAR of CD19-CAR–modified standard and TSCM-enriched products from a representative healthy donor (HD) under basal culture conditions and in response to the indicated molecules. Data are shown as mean values ± SEM. (B) Glycolytic response by CD19-CAR–modified standard and TSCM-enriched products of 6 HDs following glucose administration. ECAR values are shown after subtraction of basal ECAR measurements (*P < .05; Wilcoxon matched-pairs signed rank test). (C) OCR of CD19-CAR–modified standard and TSCM-enriched products from a representative HD under basal culture conditions and in response to the indicated molecules. Data are shown as mean values ± SEM. (D) SRC (maximal OCR − basal OCR) of CD19-CAR–modified standard and TSCM-enriched products of 6 HD. 2-DG, 2-Deoxyglucose.
Memory T cells have also been shown to possess substantial mitochondrial spare respiratory capacity (SRC) compared with naive and effector cells.47 SRC is defined as the quantitative difference between maximal uncontrolled oxygen consumption rate (OCR) and basal OCR, and is thought to represent the extra mitochondrial capacity available in a cell to produce energy under conditions of increased work or stress.47 To determine whether TSCM-enriched products were endowed with high SRC, we measured OCR during a mitochondrial stress test. To trigger maximal uncontrolled OCR, we administered FCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone), which uncouples ATP synthesis from the electron transport chain. Standard CD19-CAR–modified T cells, which are enriched with TEM and TTE subsets, displayed poor SRC compared with TSCM-enriched products (Figure 5C-D). These findings were in general reproducible across the donors with the exception of donor 6 (Figure 5D), whose cells exhibited poor in vitro expansion (Figure 2B). Because naive T cells also have low SRC,47 the results obtained with donor 6 might have reflected an increased naivety of that cell product. Altogether, these findings indicate that TSCM-enriched CD19-CAR–modified T cells have enhanced metabolic fitness compared with standard cell preparations.
CD19-CAR–modified TSCM-mediated robust, long-lasting antitumor responses
Phenotypic, functional, and metabolic analyses suggest that CD19-CAR–modified TSCM-enriched products are better equipped than standard T cells to kill leukemic cells in vivo upon adoptive transfer. We sought to assess the antileukemic activity of CD19-CAR–modified standard and TSCM-enriched cells against systemic acute lymphoblastic leukemia xenografts in highly immunodeficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice. Because CD4+ T cells can favor the engraftment and function of CD8+ T cells in these mice by providing a variety of human cytokines,9,11 and given the uneven distribution of this subset within the 2 cell products, we compared the antitumor activity of CD19-CAR–modified CD8+ T-cell–enriched products. Two million NALM6-GL were injected IV, followed 3 days later by 2.5 × 105 CD19-CAR+CD8+ T cells. To support T-cell engraftment and survival, we intraperitoneally administered low doses of human IL-15 every other day. Consistent with the established clinical efficacy of CD19-CAR–modified T cells, both standard and TSCM-enriched products mediated significant antitumor responses as revealed by the virtual disappearance of tumor-derived bioluminescent signals in the mice 12 days after adoptive transfer (Figure 6A). Although we did not detect significant differences in the persistence of CD19-CAR T cells in the circulation and spleens at the time point analyzed (supplemental Figure 7), mice receiving CD19-CAR–modified TSCM-enriched cells exhibited a prolonged tumor control (Figure 6A-B) and increased survival (Figure 6C) indicating that transferred TSCM were capable of mediating long-lasting antitumor responses.
CD19-CAR–modified TSCM mediate long-lasting antitumor responses. (A) In vivo bioluminescent imaging, (B) radiance, and (C) survival of NSG mice bearing systemic NALM6-GL leukemia xenografts after adoptive transfer of CD19-CAR–modified standard or TSCM-enriched CD8+ T cells (2.5 × 105) in conjunction with intraperitoneal injections of recombinant human IL-15 every other day (****P < .0001; log-rank [Mantel-Cox]; n = 5). Data shown are representative of 2 independent experiments. d, day.
CD19-CAR–modified TSCM mediate long-lasting antitumor responses. (A) In vivo bioluminescent imaging, (B) radiance, and (C) survival of NSG mice bearing systemic NALM6-GL leukemia xenografts after adoptive transfer of CD19-CAR–modified standard or TSCM-enriched CD8+ T cells (2.5 × 105) in conjunction with intraperitoneal injections of recombinant human IL-15 every other day (****P < .0001; log-rank [Mantel-Cox]; n = 5). Data shown are representative of 2 independent experiments. d, day.
Discussion
Immunotherapy with gene-modified T cells expressing a tumor-specific TCR and CAR has emerged as a potent therapeutic weapon in the armamentarium of hematologists-oncologists.1,2 The field has mainly been driven by the success of CD19-CAR–modified T cells that have been shown to induce impressive responses in patients with B-cell malignancies.48,49 Complete responses in patients with refractory acute lymphoblastic leukemia can be observed in up to 90% of cases.37,50-52 However, the efficacy of CD19-CAR–modified T cells appears more limited against chronic lymphocytic leukemia,35,53,54 non-Hodgkin lymphoma,38 and follicular lymphoma,35 underscoring the need to further improve this type of treatment. Moreover, the follow-up in most trials is short, and the durability of reported complete responses remains to be determined. Objective responses have been associated with the level and duration of CD19-CAR T-cell engraftment and patients experiencing robust T-cell expansion and persistence are more likely to remain in remission.37-39,51,55 The proliferative potential and longevity of CD19-CAR–modified T cells can markedly differ from patient to patient because cell products are derived from PBMCs, which have patient-specific T-cell subset composition. Indeed, high frequencies of TN and TSCM in the PBMCs have been linked to the generation of CD19-CAR–modified products with enhanced fitness.16 Notably, in a recent study, expansion of CD19-CAR–modified T cells correlated with the frequency of CD8+CD45RA+CCR7+ cells, whose phenotype is consistent with that of TSCM, within the infused product.21 However, the frequency of these cells is mostly negligible in cell preparations currently used in clinical trials.21,37
Here, we present a feasible and robust clinical-grade platform for the generation of tumor-redirected allogeneic TSCM and provide evidence that these cells are therapeutically superior to CD8+ T-cell products generated with clinical protocols currently under investigation. Notably, our strategy was also effective in generating CD19-CAR TSCM from patients’ apheresis (supplemental Figure 8; supplemental Table 6), indicating that this cell-manufacturing procedure can be readily implemented in the autologous setting. CD19-CAR cell preparations obtained with the TSCM protocol present multiple advantages compared with current CD19-CAR T-cell products. First, they contain major fractions of TSCM and less-differentiated T-cell subsets, which have higher proliferative and survival capacities.9,10,56-58 Second, TEM and TTE obtained under conditions favoring TSCM generation have better functionality than those produced with current protocols. The capacities of effector cells to produce large amounts of effector cytokines and display polyfunctionality is known to be a crucial determinant of vaccine protection43,59 and productive antitumor responses.60,61 These qualities might also be key ingredients to the effectiveness of the TSCM-enriched products. Finally, because our cell products are derived from defined CD8+ TN, they display relatively homogenous characteristics across multiple individuals rendering results from clinical trials more interpretable and perhaps more consistent.
Gene modification of long-lived stem cell–like populations using retroviral vectors might raise concern for potential induction of cell transformation.62 For instance, the longevity of TSCM has been shown to be exploited by human T-cell lymphotropic virus type 1 to initiate adult T-cell leukemia/lymphoma.63,64 On the other hand, high-resolution tracking of individual gene-engineered TSCM clones over a decade did not show the emergence of clonal dominance, indicating that gene modification of TSCM is relatively safe.65 Another safety concern is related to the potential insurgence of cytokine release syndrome (CRS), a known side effect of CD19-CAR T-cell therapies.37,52,66,67 Indeed, T-cell proliferation and expansion, which can be particularly prominent in hosts receiving TSCM,9,10 are linked to the severity of CRS.37,52,66,67 However, it should be noted that IL-6, a cytokine implicated as a central mediator of toxicity in CRS,66,67 was poorly released by CD19-CAR–modified TSCM-enriched cells after recognition of leukemic cells.
In summary, in this study we provide a unique protocol for the generation of clinically relevant numbers of tumor-redirected TSCM that exhibit phenotypic traits, functional properties, and a gene signature analogous to their naturally occurring counterpart. To our knowledge, this is the first clinical-grade platform developed that can efficiently generate bona fide TSCM. Based on the enhanced therapeutic efficacy of this cell product compared with standard CD19-CAR T cells and relatively cautioned by minor safety concerns, we have recently amended our allogeneic CD19-CAR T-cell trial, NCT01087294, to test these newly developed TSCM-enriched products.
The microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE68003).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank C. Stemberger and L. Germeroth (Stage Cell Therapeutics, now Juno Therapeutics) for the scientific discussions and the support of streptamer reagents.
This work was supported by the 2014 National Institutes of Health (NIH) Bench-to-Bedside Award and the Intramural Research Program of the US NIH, National Cancer Institute, Center for Cancer Research (grant ZIABC011480).
Authorship
Contribution: M. Sabatino and L.G. conceptualized the overall cell-manufacturing strategy; M. Sabatino, V.F., and Y.J. performed manufacturing of T cells; S.D., J.D.H., Y.J., and L.G. performed flow cytometry and intracellular cytokine staining; M. Sommariva performed microarray analysis, cytokine release; Y.J. and M. Sommariva performed qRT-PCR; Y.J. prepared western blots; S.G. performed metabolic studies; J.H., Y.J., and H.Q. performed animal experiments; M. Sommariva, Y.J. and L.G. performed statistical analyses; C.A.K., J.N.K., and T.J.F. provided critical reagents; J.N.K., T.J.F, R.E.G., D.F.S., Y.J., and L.G. supervised the experiments and discussed and interpreted the results; L.G. wrote the manuscript; and all authors edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luca Gattinoni, Experimental Transplantation and Immunology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Room 3E 3510, 10 Center Dr, Bethesda, MD 20892; e-mail: gattinol@mail.nih.gov.
References
Author notes
M. Sabatino, J.H., and M. Sommariva contributed equally to this work.

![Figure 6. CD19-CAR–modified TSCM mediate long-lasting antitumor responses. (A) In vivo bioluminescent imaging, (B) radiance, and (C) survival of NSG mice bearing systemic NALM6-GL leukemia xenografts after adoptive transfer of CD19-CAR–modified standard or TSCM-enriched CD8+ T cells (2.5 × 105) in conjunction with intraperitoneal injections of recombinant human IL-15 every other day (****P < .0001; log-rank [Mantel-Cox]; n = 5). Data shown are representative of 2 independent experiments. d, day.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/4/10.1182_blood-2015-11-683847/4/m_519f6.jpeg?Expires=1768219886&Signature=BTj43FtuE4dDGObyAxNGVW5sg-eBHdNN8vkSlNYZMtThuXQPrmyHL81maBH85b5hrg1VdKGfO2NAw6CaRPer8QBk5F2-Yppv-2I4oWSK0nQgId-VwWG6X4VCqxXNJAJ94Q5RDFMQg7yL6eTz5CKygtFKe-weHnVGo0YrQohHjNni4513-a6or9jOdpKSHwiEKO1RJ55sZJ4Y0PfbUtiQDz8TYMR~S2UFm9OUcwedxFq2IqgNbFleOWNPiGCEOsBgXpFNyHzKC~fbraXmhd29scxSBdloF1ygd37GFgsSbdMPCS9yUST96D7K5z7wNubWDNJ8VaKmOtkrMeg072gNIA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)