In this issue of Blood, Izzi et al pair a platelet function–associated single-nucleotide polymorphism (SNP) with allele-specific methylation at a cytosine guanine dinucleotide (CpG) island and regulation of platelet endothelial aggregation receptor 1 (PEAR1) RNA expression in megakaryopoiesis, describing one of the first epigenetic-SNP links to platelet biology.1
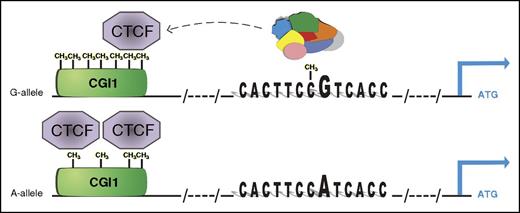
Epigenetically dependent regulatory mechanism of rs12041331 in PEAR1 expression. The major G allele of the CpG-SNP introduces a CpG site in the enhancer region that binds with higher affinity to several nuclear proteins (colored complex) than the A allele. At the same time, the G-methylated allele marks higher methylation at CGI1, the CpG island located at the UTR region of PEAR1 that contains several binding sites for CTCF, important for chromosome looping and architecture. When higher methylation is present, CTCF binds with lower affinity to the CGI1 region, liberating the enhancer activity of intron 1 where the CpG-SNP is also located. In contrast, lower methylation at CGI1 is associated with the minor A allele of the CpG-SNP, reinforcing CTCF binding and partially inhibiting gene expression. This model advances a positive effect of methylation on gene expression, and is in accordance with the association between increasing methylation and PEAR1 expression during early MK progenitor development. ATG, start codon; CH3, methyl group. See Figure 6 in the article by Izzi et al that begins on page 1003.
Epigenetically dependent regulatory mechanism of rs12041331 in PEAR1 expression. The major G allele of the CpG-SNP introduces a CpG site in the enhancer region that binds with higher affinity to several nuclear proteins (colored complex) than the A allele. At the same time, the G-methylated allele marks higher methylation at CGI1, the CpG island located at the UTR region of PEAR1 that contains several binding sites for CTCF, important for chromosome looping and architecture. When higher methylation is present, CTCF binds with lower affinity to the CGI1 region, liberating the enhancer activity of intron 1 where the CpG-SNP is also located. In contrast, lower methylation at CGI1 is associated with the minor A allele of the CpG-SNP, reinforcing CTCF binding and partially inhibiting gene expression. This model advances a positive effect of methylation on gene expression, and is in accordance with the association between increasing methylation and PEAR1 expression during early MK progenitor development. ATG, start codon; CH3, methyl group. See Figure 6 in the article by Izzi et al that begins on page 1003.
PEAR1 encodes a transmembrane tyrosine kinase receptor expressed in platelets and endothelial cells and plays a role in platelet cell activation.2 Discovered in 2005, PEAR1 was found to be phosphorylated following platelet aggregation in a proteomic screen.2 Subsequently, we and others found association of common genetic variants in or near PEAR1 with platelet aggregation in response to agonists including adenosine diphosphate and epinephrine,3,4 collagen,4 and thrombin.5 The genetic study with greatest statistical power and genomic coverage implicated 2 correlated variants, rs12041331 and rs12566888, with platelet aggregation, accounting for >1% of phenotypic variation.3 Both variants were intronic, leaving a functional mechanism unclear. Further resequencing of the PEAR1 gene in 104 individuals with extreme platelet aggregation responses did not implicate any coding variants, maintaining rs12041331 as the most strongly associated variant.6 That study further linked the G allele of rs12041331 to higher PEAR1 RNA expression in megakaryocytic (MEG-01) and endothelial (human umbilical vein endothelial cell) cell reporter gene assays and protein expression via enzyme-linked immunosorbent assay in platelet lysates.6 Until the work of Izzi and colleagues presented here, a potential mechanism of how rs12041331 influences PEAR1 expression and platelet function remained unknown.1
Located intronically between an upstream noncoding exon and the first coding exon of PEAR1, rs12041331 is a G to A change in a CpG. Such genomic alterations in a CpG site could potentially result in loss of a DNA methylation site. The authors observed that this particular SNP overlaps with histone marks and a DNase I hypersensitivity site in ENCODE project data from megakaryoblastic K562 and endothelial cells. Furthermore, chromatin immunoprecipitation sequencing experiments in megakaryocytic CHRF cells suggested that an active enhancer overlapped the rs12041331 region. These observations indicated to the authors that the variant may lie within and potentially modify an active enhancer influencing PEAR1 RNA expression.
To test this hypothesis, Izzi and colleagues performed a series of experiments examining the epigenetic and regulatory implications of rs12041331 on PEAR1. Bisulfite sequencing of leukocytes of healthy donors with GG, GA, and AA genotypes at rs12041331 showed full methylation of the nearby CpG site in individuals with the GG genotype. With evidence of allelic differences in DNA methylation, the authors next performed electromobility shift assays of nuclear extracts from megakaryoblastic, endothelial, and kidney cell lines to ascertain whether there were also allele-specific differences in protein interaction at rs12041331. In all cell lines, more binding was observed with the G allele of rs12041331 as opposed to the alternate A allele. Furthermore, through engineered assays, they demonstrated that DNA methylation at this site increased protein binding at this site. Luciferase assays also confirmed the prior findings linking the G allele to higher PEAR1 RNA expression.6
With evidence that rs12041331 influences PEAR1 expression, the authors next examined the SNP’s effects through in vitro megakaryocyte (MK) differentiation from hematopoietic stem cells as previous studies showed increased PEAR1 expression through this process. By tracking PEAR1 methylation and expression, Izzi and colleagues found that PEAR1 RNA expression is upregulated in early- to mid-megakaryocytic differentiation. The rs12041331 region and an upstream region (labeled CGI1; see figure) overlapping the first 5′ untranslated region (5′UTR) exon and a CTCF site were consistently methylated during differentiation. Such expression and methylation patterns suggest a genotypically driven model whereby increased methylation of these PEAR1 regulatory regions drives up expression early in MK differentiation (see figure). Although epigenetics dogma generally proposes that increased CpG methylation leads to decreased RNA expression, recent studies have found that genomic context is important and methylation can also lead to increased expression. Unfortunately, the authors were limited in their ability to fully characterize genetic effects during differentiation as only a single AA individual was studied. Low cell yields from differentiation protocols meant that the authors were unable to study ploidy. However, clonogenic cell culture assays showed smaller colony-forming unit MKs in GG individuals than A-allele carriers, indirectly suggesting effects on MK ploidy.
These results on PEAR1 during MK differentiation indicate that it may influence not only platelet aggregation but also the size and number of mature platelets formed. We recently provided evidence at a population scale associating the rs12566888 SNP in PEAR1 with platelet count in Exomechip genotyping of >157 000 individuals.7 The minor allele of rs12566888 is correlated with the G allele of rs12041331, with an r2 of ∼0.8 in European and Asian populations, suggesting consistency among these investigations, whereby genetic influences on methylation and PEAR1 expression in megakaryocytic differentiation may enhance MK and platelet formation. Notably, in nucleated cells, PEAR1 has been localized to the nucleus, centrosomes, and cell junctions (http://proteinatlas.org/), indicating it could have roles in endomitosis or demarcation membrane system formation. Taken together, the genetic evidence for PEAR1 effects on both platelet count7 and platelet aggregation,3-5 epigenetic evidence from Izzi et al for influences in megakaryopoiesis,1 and prior work on the role of PEAR1 in sustaining platelet activation signaling8 indicate PEAR1 likely has dual roles influencing platelet genesis and platelet aggregation potentiation.
Further work is needed to clarify mechanistic roles of PEAR1 throughout MK differentiation. The exact protein complex binding the rs12041331 region remains to be identified, as do the mechanisms of interaction with the upstream CTCF or other regulatory elements.
The current study not only advances knowledge of PEAR1 regulation but also provides a rare example and roadmap for understanding the interplay of genetic and epigenetic factors in complex traits such as platelet biology. Applications of this type of approach could provide mechanistic insight and therapeutic opportunities in thrombocytopenias and other platelet disorders. Treatment with decitabine, an inhibitor of methylation at position 5 of cytosine, enhances MK differentiation and platelet release, with potential clinical utility in myelodysplastic syndromes and immune thrombocytopenias.9 Targeting global methylation or specific epigenetic sites may improve efforts to increase platelet generation, which is a significant challenge. Recent studies have made advances in generating platelets in human pluripotent stem cell models.10 Future studies may be able to modify specific genetic or epigenetic states of cells in bioreactors to improve MK and platelet formation for therapeutic applications. To do so, further insights into key epigenetically modified genes in platelet biology will be needed. Izzi et al have taken an important step forward in this by pairing methylation in megakaryopoiesis with PEAR1 regulation and function.1
Conflict-of-interest disclosure: The author declares no competing financial interests.