Key Points
Bone marrow adipocytes support AML survival.
AML induces adipocyte lipolysis of triglyceride to free fatty acids and subsequent transport by FABP4.
Abstract
Despite currently available therapies, most patients diagnosed with acute myeloid leukemia (AML) die of their disease. Tumor-host interactions are critical for the survival and proliferation of cancer cells; accordingly, we hypothesize that specific targeting of the tumor microenvironment may constitute an alternative or additional strategy to conventional tumor-directed chemotherapy. Because adipocytes have been shown to promote breast and prostate cancer proliferation, and because the bone marrow adipose tissue accounts for up to 70% of bone marrow volume in adult humans, we examined the adipocyte-leukemia cell interactions to determine if they are essential for the growth and survival of AML. Using in vivo and in vitro models of AML, we show that bone marrow adipocytes from the tumor microenvironment support the survival and proliferation of malignant cells from patients with AML. We show that AML blasts alter metabolic processes in adipocytes to induce phosphorylation of hormone-sensitive lipase and consequently activate lipolysis, which then enables the transfer of fatty acids from adipocytes to AML blasts. In addition, we report that fatty acid binding protein-4 (FABP4) messenger RNA is upregulated in adipocytes and AML when in coculture. FABP4 inhibition using FABP4 short hairpin RNA knockdown or a small molecule inhibitor prevents AML proliferation on adipocytes. Moreover, knockdown of FABP4 increases survival in Hoxa9/Meis1-driven AML model. Finally, knockdown of carnitine palmitoyltransferase IA in an AML patient-derived xenograft model improves survival. Here, we report the first description of AML programming bone marrow adipocytes to generate a protumoral microenvironment.
Introduction
Survival of patients with acute myeloid leukemia (AML) is poor; two-thirds of young adults and 90% of older adults die of their disease.1 Improved outcomes will now only come from novel treatment strategies derived from an improved understanding of the biology of the disease.
All mammals generate blood within their bones, and AML arises from myeloid progenitors within the bone marrow (BM) microenvironment. AML blasts exhibit high levels of spontaneous apoptosis when cultured in vitro but have a prolonged survival time in vivo, further indicating that the tissue microenvironment plays a critical role in promoting and maintaining AML cell survival.2-5 In human leukemias, the microenvironment provides a number of soluble factors whose primary functions are to promote survival and homing,6,7 which implies that the apoptotic defect in AML is not cell-autonomous but highly dependent on extrinsic signals derived from the cellular microenvironment. These complex cell-cell interactions between AML blasts and the cells that support them within the BM may therefore provide an attractive target for novel drug therapies.
The BM microenvironment is defined by cell types not directly involved in hematopoiesis and include macrophages; endothelial cells; osteoclasts; osteoblasts; adipocytes; and fibroblasts. BM adipocytes were identified over a century ago, and bone marrow adipose tissue (MAT) accounts for up to 70% of BM volume of the axial skeleton in adult humans.8-10 MAT is an energy storage organ and consists primarily of triglycerides, which can be broken down to release free fatty acids (FAs), which in turn can be used to generate adenosine triphosphate. In cancer, a recent study by Cawthorn et al demonstrated that BM adiposity and circulating adiponectin increase in cancer therapy.8 Moreover, TNF and adiponectin from the BM of patients with AML inhibit normal hematopoiesis.11 However, although AML blasts are highly proliferative and grow within an adipocyte-rich environment, to date there is limited knowledge regarding the specific functions of BM adipocytes in relation to the proliferation and survival of AML.
Adipocytes, in the context of solid malignancies, support tumor survival, proliferation, and metastasis.12-14 In a study on breast cancer and the associated adipocytes therein, Dirat et al showed that these cancer-associated adipocytes are greatly influenced by the invasive cancer cells with which they are associated.12 This study supports the concept of cancer cells orchestrating an environment that favors their progression through complex mechanisms by obtaining adipocyte participation. Furthermore, studies carried out in prostate cancer have demonstrated lipid transport between adipocytes and prostate cancer cells.13,15 In models of bone metastasis following prostate cancer, Herroon et al15 show functional evidence of BM adipocyte support of tumor growth and identify fatty acid binding protein-4 (FABP4) as a key protein involved in the mechanism. Although this study investigates FABP4 expression in the cancer cells, a separate study has investigated FABP4 expression in ovarian cancer–associated adipocytes.14 FABP4 upregulation has been shown in the fat-rich omental metastases compared with the primary tumor site, and mice lacking FABP4 show significant reduction in metastatic tumor growth, indicating the role of FABP4 in cancer proliferation and metastasis. Taken together, these studies identify a functional role for adipocytes and FA transporters in the support of solid tumor metabolism and subsequent metastatic spread.
In the present study, we look to identify whether adipocytes directly support the proliferation of human AML. Furthermore, we evaluate the mechanisms controlling the interaction between AML blasts and MAT as well as the downstream metabolic consequences in both the adipocytes and leukemia cells. Finally, we identify key regulators in the AML/MAT interaction and assess AML survival following targeted inhibition of this interaction.
Materials and methods
Materials
Anti–phosphorylated hormone-sensitive lipase (pHSL), anti–hormone-sensitive lipase (HSL), anti-histidine (anti-His), and anti-FABP4 were purchased from Cell Signaling Technology (Cambridge, MA; Catalog numbers 4126, 4107, 2365, and 3544, respectively). Control-immnunoglobulin G (IgG)-fluorescein isothiocyanate (FITC), control-IgG-phycoerythrin (PE), control-IgG-antigen-presenting cell (APC), anti-CD34-PE, anti-CD90-FITC, anti-CD73-PE, anti-CD105-APC, anti-CD33-APC, and anti-CD45-FITC antibodies were from Miltenyi Biotec (Bergisch Gladbach, Germany; Catalog numbers 130098847, 130098845, 130092214, 130098139, 130095403, 130095182, 130094926, 130098043, and 130098864). Lineage Cell Depletion Kit (mouse) was also purchased from Miltenyi Biotech (Catalog number 130090858). Acipomox, etomoxir (ETX), and FABP4 inhibitor were purchased from R&D Systems (Abingdon, UK; Catalog numbers 2784 and 4539). Recombinant FABP4-His was purchased from Abcam (Cambridge, UK; Catalog number 204760). All other reagents were obtained from Sigma-Aldrich (St Louis, MO), unless otherwise indicated.
Primary cell culture and differentiation
Primary AML blasts and nonmalignant CD34+ cells were obtained from patient BM or blood following informed consent and under approval from the UK National Research Ethics Service (LRECref07/H0310/146). Table 1 shows AML patient sample information used in this study. Cell isolation was carried out by density gradient centrifugation using Histopaque (Sigma-Aldrich; Catalog number 1077), and cell type was confirmed by flow cytometry. All cells were grown in normal media (Dulbecco’s modified Eagle medium [DMEM; ThermoFisher; Catalog number 21885-025] supplemented with 20% fetal bovine serum [FBS; ThermoFisher; Catalog number 10082139]) with or without the addition of cytokines purchased from Peprotech, UK, 100 ng/mL stem cell factor (SCF; 300-07), 50 ng/mL FMS-like tyrosine kinase 3 ligand (300-19), 20 ng/mL interleukin-3 (IL-3; 200-03), and 20 ng/mL granulocyte colony-stimulating factor (300-23). Bone marrow stromal cells (BMSCs) were isolated from AML BM samples by adherence to tissue culture plastic and were then expanded in DMEM containing 20% FBS and supplemented with 1% penicillin-streptomycin (ThermoFisher; Catalog number 15140122). BMSC markers were confirmed using flow cytometry for expression of CD90+, CD73+, CD105+, and CD45−.16 To induce adipocyte differentiation of BMSC (passage 2-4), a cocktail of dexamethasone (1 μM), indomethacin (0.2 mM), insulin (100 nM), and 3-isobutyl-1-methylxanthine (0.5 mM) (Catalog numbers D4902, 17378, I9278, and I7018) in DMEM containing 10% FBS was prepared. The differentiation of BMSC to adipocytes was between 75% and 95% as measured by neutral lipid-specific BODIPY 493/503 dye (supplemental Figure 1A; supplemental Table 1, available on the Blood Web site). After differentiation, adipocytes and control BMSC were analyzed for markers of adipocytes (adiponectin, FABP4, and CEBPa). Figure 3A and supplemental Figure 1B show that differentiated adipocytes express higher levels of adiponectin, FABP4, and CEBPa than BMSC.
AML patient sample information used in this study
| Number . | Age . | Sex . | WHO diagnosis . | Cytogenetics . | % Blasts . |
|---|---|---|---|---|---|
| AML#1 | 69 | M | AML NOS | N/A | 60 |
| AML#2 | 66 | F | AML without maturation | Trisomy 9 | 90 |
| AML#3 | 41 | F | AML with t(6;9)(p23;q34); DEK-NUP214 | t(6;9) | 95 |
| AML#4 | 62 | M | AML with maturation | Complex | 55 |
| AML#5 | 70 | M | AML with maturation | Normal | 80 |
| AML#6 | 70 | M | AML without maturation | Complex | 95 |
| AML#7 | 91 | F | AML NOS | N/A | 60 |
| AML#8 | 55 | F | AML | Not available | 70 |
| AML#9 | 59 | F | AML with t(8;21)(q22;q22) RUNX1-RUNX1T1 | t(8;21) | 80 |
| AML#10 | 78 | F | AML without maturation | Normal | 95 |
| AML#11 | 58 | F | AML with maturation | Normal | 80 |
| AML#12 | 65 | M | AML with maturation | Trisomy 13 | 30 |
| AML#13 | 73 | F | AML without maturation | Normal | 95 |
| AML#14 | 49 | M | AML with myelodysplasia-related changes | N/A | 55 |
| AML#15 | 64 | F | AML with myelodysplasia-related changes | Normal | 45 |
| AML#16 | 65 | M | AML with minimal differentiation | Normal | 95 |
| AML#17 | 23 | F | AML without maturation | t(5;12) | 95 |
| AML#18 | 37 | M | AML without maturation | Normal | 90 |
| AML#19 | 59 | M | AML with t(8;21)(q22;q22) RUNX1-RUNX1T1 | t(8;21) | 60 |
| AML#20 | 65 | M | AML with maturation | Not available | 70 |
| AML#21 | 68 | M | Therapy-related AML | Not available | 40 |
| AML#22 | 63 | M | AML with myelodysplasia related changes | Not available | 30 |
| AML#23 | 76 | M | AML without maturation | Not available | 95 |
| AML#24 | 75 | M | AML with maturation | Complex | 55 |
| AML#25 | 88 | M | AML with maturation | Trisomy 8 | 30 |
| AML#26 | 35 | M | AML without maturation | 46 XY | 80 |
| AML#27 | 72 | M | AML with myelodysplasia-related changes | 46,XY isochrome (17)(q10) | 90 |
| Number . | Age . | Sex . | WHO diagnosis . | Cytogenetics . | % Blasts . |
|---|---|---|---|---|---|
| AML#1 | 69 | M | AML NOS | N/A | 60 |
| AML#2 | 66 | F | AML without maturation | Trisomy 9 | 90 |
| AML#3 | 41 | F | AML with t(6;9)(p23;q34); DEK-NUP214 | t(6;9) | 95 |
| AML#4 | 62 | M | AML with maturation | Complex | 55 |
| AML#5 | 70 | M | AML with maturation | Normal | 80 |
| AML#6 | 70 | M | AML without maturation | Complex | 95 |
| AML#7 | 91 | F | AML NOS | N/A | 60 |
| AML#8 | 55 | F | AML | Not available | 70 |
| AML#9 | 59 | F | AML with t(8;21)(q22;q22) RUNX1-RUNX1T1 | t(8;21) | 80 |
| AML#10 | 78 | F | AML without maturation | Normal | 95 |
| AML#11 | 58 | F | AML with maturation | Normal | 80 |
| AML#12 | 65 | M | AML with maturation | Trisomy 13 | 30 |
| AML#13 | 73 | F | AML without maturation | Normal | 95 |
| AML#14 | 49 | M | AML with myelodysplasia-related changes | N/A | 55 |
| AML#15 | 64 | F | AML with myelodysplasia-related changes | Normal | 45 |
| AML#16 | 65 | M | AML with minimal differentiation | Normal | 95 |
| AML#17 | 23 | F | AML without maturation | t(5;12) | 95 |
| AML#18 | 37 | M | AML without maturation | Normal | 90 |
| AML#19 | 59 | M | AML with t(8;21)(q22;q22) RUNX1-RUNX1T1 | t(8;21) | 60 |
| AML#20 | 65 | M | AML with maturation | Not available | 70 |
| AML#21 | 68 | M | Therapy-related AML | Not available | 40 |
| AML#22 | 63 | M | AML with myelodysplasia related changes | Not available | 30 |
| AML#23 | 76 | M | AML without maturation | Not available | 95 |
| AML#24 | 75 | M | AML with maturation | Complex | 55 |
| AML#25 | 88 | M | AML with maturation | Trisomy 8 | 30 |
| AML#26 | 35 | M | AML without maturation | 46 XY | 80 |
| AML#27 | 72 | M | AML with myelodysplasia-related changes | 46,XY isochrome (17)(q10) | 90 |
This table defines the nature of the AML disease, including WHO diagnosis and cytogenetics.
F, female; M, male; N/A, not applicable; NOS, not otherwise specified; WHO, World Health Organization.
Flow cytometry
For this study, we used the Accuri C6 (Becton Dickinson, Oxford, UK) and the CyFlow Cube 6 (Sysmex, Milton Keynes, UK) for flow cytometry analysis. Cells were incubated for 5 minutes with the FCR receptor blocker (Miltenyi Biotec; Catalog number 130-059-901) and then stained with isotype controls or test antibodies (Miltenyi Biotec). Gates were set to the appropriate isotype control.
Coculture assay
BMSC were seeded at 3 × 104 cells per well of a 12-well plate and 1 × 104 for a 24-well plate in normal growth media. Cells were left to adhere and proliferate until maximum confluency. Growth media were replaced with differentiation media, which was replaced every 4 days until day 21. Differentiated adipocytes were washed twice with normal growth media and then placed in normal growth media until coculture experiment. For coculture experiments, AML cells were placed on the adipocytes at 5 × 105 (per well of 12-well plate) and 1 × 105 (per well of a 24-well plate) in normal growth media. Adipocyte/AML cocultures were then incubated for indicated time points. Supplemental Table 2 shows the combination of different samples used for each experiment. All cocultures were from different patients; at no point were autologous cocultures performed. To isolate adipocytes from coculture experiments, all suspension cells were removed by gentle pipetting and washing with phosphate-buffered saline (PBS). Following removal of PBS, cells were subject to light trypsinization where diluted typsin with PBS (1:1) for 40 seconds followed by gentle tapping. All suspension cells were removed, and adhered adipocytes were washed with PBS and then lysed using RNA or protein lysis buffer.
Lentiviral knockdown
MISSION pLKO.1-puro Control Vector (Catalog number SHC001) was used as the lentivirus control (control short hairpin RNA [shRNA]). Plasmids containing MISSION shRNA TRCN0000418254 (human FABP4 shRNA), TRCN0000105185 (mouse FABP4 shRNA), and TRCN0000036279 (human carnitine palmitoyltransferase IA [CPT1A] shRNA) were purchased from Sigma-Aldrich, and viruses were produced as previously described.17 Lentiviral stocks were concentrated using Amicon Ultra centrifugal filters (Catalog number UFC910024), and titers were determined using Lenti-Xquantitative real-time polymerase chain reaction (RT-PCR) titration kit (CloneTech; Catalog number 631235). Stromal cells were plated at a density of 3 × 104/well in a 12-well plate, expanded, and differentiated into adipocytes. Adipocytes were then infected with FABP4 shRNA lentiviral stock at a multiplicity of infection of 20. Human and mouse AML cells were infected with respective lentiviral stocks at a multiplicity of infection of 20 in the presence of polybrene (10 µg/mL final). Infected cells were analyzed using RT-PCR (Roche) and western blotting.
Patient-derived AML xenograft
For this study, the NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ (NSG) mice from The Jackson Laboratory (Bar Harbor, ME) were used. The NSG mice were maintained under specific pathogen-free conditions in the research animal facility of The Disease Modeling Unit, The University of East Anglia, Norwich, UK. All animal experiments were performed in accordance with UK Home Office regulations.
Engraftment of primary AML cells in NSG mice
Isolated AML blasts amounting to 6-8 × 106 were grown in mono culture; 2 × 106 isolated AML blasts were grown on adipocytes for 6 days, and 2 × 106 viable AML cells were washed and subsequently resuspended in PBS. Cells were then injected into the tail vein of nonirradiated 6- to 8-week-old female NSG mice. When clinical signs of illness became apparent (rough fur, hunchback, or reduced motility) or if 12 weeks postinjection was reached, mice were euthanized by exposure to CO2. BM and spleen were harvested and analyzed for human CD33 and CD45. If >1% of human CD45/CD33 cells were detected in the BM or spleen, the AML sample was said to be engrafted. For AML grown in cytokine-supplemented media, 2 × 106 viable AML were grown in normal media with the addition of cytokines (100 ng/mL SCF [Peprotech 300-07], 50 ng/mL FMS-like tyrosine kinase 3 ligand [Peprotech 300-19], 20 ng/mL IL-3 [Peprotech 200-03], and 20 ng/mL granulocyte colony-stimulating factor [Peprotech 300-23]).
For the carnitine palmitoyltransferase I-knockdown (CPT1-KD) experiment, isolated primary AML cells were grown on adipocytes for 6 days. The AML cells were removed from the adipocytes and then infected with a control-shRNA or CPT1-shRNA lentivirus, and after 96 hours, cells were expanded on the adipocytes; 2 × 106 primary AML cells were then injected into the tail vein of nonirradiated 6- to 8-week-old female NSG mice.
Retroviral AML transplantation mouse model
C57BL/6J mice were obtained from Janvier Labs (Le Genest-Saint-Isle, France). All animal experiments were performed according to national and international standards and had been approved. BM cells were harvested from mice, and lineage-negative cells were obtained by negative selection using the Lineage Cell Depletion Kit (mouse) as recommended by the manufacturer. Lineage-negative cells derived from C57BL/6J were retrovirally infected by coculture with GP+E86 cells in the presence of polybrene (10 µg/mL; Sigma-Aldrich, Munich, Germany). Coculture with GP+E86 packaging MSCV-Hoxa9-PGK-neo was performed for 3 days followed by coincubation with GP+E86 MSCV-Meis1-IRES-YFP for 1 day. Hoxa9 cells were selected with 0.6 mg/mL G418 (Sigma-Aldrich) for at least 5 days. After selection, cells were sorted with a fluorescence-activated BD FACSARIA III cell sorter. Lentiviral transductions of cultured cells with vectors encoding FABP4-specific shRNA and control shRNA were performed as described previously.18 Transplanted were 8 × 104 cells together with 2 × 105 support cells by injection into the tail vein of lethally irradiated (9.5 Gy) recipient mice (C57BL/6J). Support cells were isolated from C57BL/6J mice and purified on a Ficoll gradient. To perform in vitro assays with Hoxa9/Meis1-expressing cells, cells were cultured in DMEM supplemented with 20% FBS and supplemented with the following cytokines: murine SCF (Catalog number 250-03; 100 ng/mL final), IL-6 (Catalog number 200-06; 10 ng/mL final), and IL-3 (Catalog number 213-13; 10 ng/mL final).
Immunocytochemistry
AML-adipocyte niches formed in the BM were isolated from patient samples and grown on chambered tissue culture–treated slides (12-well chamber, removable;– Ibidi; Catalog number 81201). Niches were then fixed with 4% paraformaldehyde, permeabilized with 0.01% Triton X-100, and blocked with goat serum. Cells were stained with neutral lipid-specific BODIPY 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene) dye, CD90, and 4′,6-diamidino-2-phenylindole. AML blasts were removed and stained with neutral lipid-specific BODIPY 493/503 and CD34 antibody. Images were visualized with secondary Alexa Fluor 568 or 488 conjugate immunoglobulin G (Invitrogen). Nuclei were stained with 4′6-diamidino-2-phenylindole before samples were mounted with Fluoromount aqueous mounting medium (Sigma; Catalog number F4680). Cells were imaged by an AxioCam ICm1 monochrome charge-coupled device camera attached to the Apotome.2 Imaging System and confocal microscopy (Zeiss LSM 800 with Airyscan) using Axiovision 4.8.2 software (Carl Zeiss). Image staining intensities were analyzed with ImageJ software.
Proliferation, cell cycle, and death assays
Treated primary AML blasts were incubated with 5-bromo-2′-deoxyuridine (BrdU staining kit; eBioscience; Catalog number 8811-6600) and CD34 antibody. The proliferation percentage of cycling AML blasts was analyzed using flow cytometry. Cell-cycle analysis of AMLs from monoculture and coculture was carried out by ethanol fixation and propidium iodide (PI) staining and quantified using flow cytometry. AML apoptosis was measured using PI/AnnexinV (eBiosciences; Catalog number 88-8005-72) and was also quantified using flow cytometry.
Free FA and glycerol detection
Free FA and glycerol detection was performed using a lipolysis assay kit for detection of both free glycerol and FAs (Zenbio; Research Triangle, NC; Catalog number LIP-2-NC). AMLs were cultured with adipocytes in LIP2/3 assay buffer and incubated for 24 hours at 37°C. Media were assayed to detect free FA and glycerol according to manufacturer’s specifications.
Free FA uptake assay
For lipid visualization and transfer, adipocytes were incubated with dodecanoic acid fluorescent FA analog (DAA; QBT Fatty acid uptake assay kit; Molecular Devices; Catalog number R6138) for 3 hours and washed 3 times in PBS. Primary AML blasts were then cultured alone or with labeled adipocytes for 24 hours. AML blasts were then removed, and fluorescence was subsequently measured by flow cytometry to indicate transfer of fluorescent-labeled FA from adipocytes to AML.
RT-PCR
Total RNA was extracted from cocultured BMSC, adipocytes ,and AMLs and from monoculture of the same using Nucleic Acid PrepStation from Applied Biosystems (Catalog number 6100). The RNA PCR core kit was used for reverse transcription. SYBR green I dye (Roche; Catalog number 04887352001) was used for quantitative RT-PCR using human FABP4 (F: CCACCATAAAGAGAAAACGAG; R: AGTTGCTTGCTAAATCATGG), human glyceraldehyde-3-phosphate dehydrogenase (F: CTTTTGCGTCGCCAG; R: TTGATGGCAACAATATCCAC), human CPT1 (F: TGGATCTGCTGTATATCCTTC; R: AATTGGTTTGATTTCCTCCC), mouse FABP4 (F: GTAAATGGGGATTTGGTCAC; R: TATGATGCTCTTCACCTTCC), mouse glyceraldehyde-3-phosphate dehydrogenase (F: AAGGTCATCCCAGAGCTGAA; R: CTGCTTCACCACCTTCTTGA) primers.
Western immunoblotting
Western analyses following sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis were carried out as described. Whole-cell lysates were extracted from AML/adipocyte coculture and monocultures by using radioimmunoprecipitation assay buffer containing 1% Nonidet-40 (Sigma-Aldrich, Gillingham, UK), 50 mmol/L Tris, 10% glycerol, 0.02% NaN3, 150 mmol/L NaCl, and a cocktail of phosphatase and protease inhibitors (Sigma-Aldrich, Gillingham, UK). Sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis was performed as previously described.17,19
Bioinformatic analysis
Publicly available RNA sequencing data were downloaded for a panel of 43 AML patients, which comprised 22 AML samples obtained from peripheral blood and 21 AML samples obtained from BM aspirate and for 17 nonmalignant CD34+ from 17 nonpooled individuals (Gene Expression Omnibus Accession ID GSE49642 and GSE48846).20 Data were available as reads per kilobase per million mapped reads (RPKM). RPKM data for FABP4 were extracted and processed further by first replacing zero-valued entries with one followed by logarithmic transformation to the base 2.21 FABP4 RPKM values for blood, BM samples, and nonmalignant CD34+ cells were compared with a Wilcoxon rank-sum test.
Clonogeneic methylcellulose assay
Primary AML blasts from adipocyte coculture and monoculture were plated in methylcellulose medium (R&D Systems) at a density of 1 × 104 in all experiments. Colonies were visualized and counted after 14 days.
FA oxidation
β-Oxidation of FAs was assessed using the Seahorse XFp Analyzer and the Seahorse XF Palmitate-BSA (bovine serum albumin) FAO Substrate kit (Agilent Seahorse Bioscience) according to manufacturer’s specifications. Briefly, AML blasts or nonmalignant CD34+ cells were cultured with or without adipocytes and then removed from coculture and placed in substrate-limited media (supplemented with 0.5 mM glucose, 0.5 mM carnitine, 1 mM glutamine, and 1% FBS) for 4 hours before assaying. AML blasts were then plated in poly-d-lysine (Sigma) –coated assay wells at a density of 2 × 105 per well in base media containing 2.5 mM glucose, 0.5 mM carnitine, and 5 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) and adjusted to pH 7.4 with 1 N NaOH. ETX (40 µM) or BSA and Palmitate:BSA were added into the injection ports. The experimental template was designed using Wave software for desktop from Seahorse Bioscience.
Statistical analysis
We used the Mann-Whitney U test to compare results between groups unless otherwise stated in the legends. Results with P < .05 were considered statistically significant (denoted by *). Results represent the mean ± standard deviation of 4 or more independent experiments. For western blotting, data are representative images of 3 independent experiments. We generated statistics with GraphPad Prism 5 software (GraphPad, San Diego, CA).
Results
BM adipocytes support the survival and proliferation of primary AML
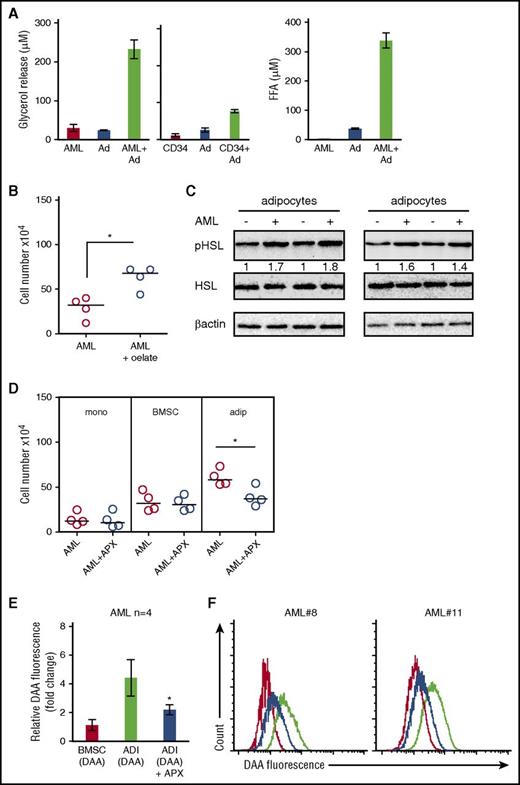
Cellular proliferation is a highly energy-intensive process. Because adipocytes store energy in the form of triglycerides, we hypothesized that adipocytes provide energy in the form of FAs to AML blasts to support rapid growth. Initially, to determine the presence of lipids in AML, we examined freshly isolated AML cells for lipid stores using the neutral lipid dye (BODIPY 493/503) and found that freshly isolated AML cells (CD34+) contain abundant lipids (Figure 1A). Figure 1B-C shows that lipid stores using the neutral lipid dye are depleted in AML and nonmalignant CD34+ cells when cultured in vitro. Supplemental Figure 2, available on the Blood Web site, shows that AML cells lose lipid content even when cocultured with BMSC. As primary AML cells undergo spontaneous apoptosis when cultured in vitro, to determine if adipocytes can support the survival of primary AML, we cultured primary AML blasts, either alone or in coculture with adipocytes derived from BM mesenchymal progenitor cells. Figure 1D and supplemental Figure 3A show that primary AML blasts cocultured with adipocytes (n = 11) are protected from undergoing apoptosis for up to 6 days when compared with AML blasts cultured alone.
BM adipocytes support the survival and proliferation of primary AML. (A) Immunofluorescence of primary AML blasts stained for CD34+ (Alexa Fluor 568 [red]) and neutral lipids (green) using BODIPY 493/503. All images are representative of 6 AML patient samples. Original magnification ×63. Scale bar = 10 μm. (B) Freshly isolated AML, AML cultured for 1 day, and AML cultured for 2 days stained with the neutral lipid BODIPY 493/503 dye and analyzed by flow cytometry. Data are represented as mean ± standard deviation. (C) Freshly isolated nonmalignant CD34+ cells and freshly isolated AML samples stained with the neutral lipid BODIPY 493/503 dye and analyzed by flow cytometry. (D) AML patients samples in monoculture and cocultured with BM-derived adipocytes for 6 days and then stained with PI/Annexin V. The line through the data indicates the median. (E) AML blasts incubated alone or with adipocytes or BMSC for 6 days and AML blasts counted using flow cytometry and Trypan blue exclusion (n = 12). The line through the data indicates the median. (F) Nonmalignant CD34+ cells cultured alone or in coculture with BMSC or adipocytes and CD34+ cell counted using flow cytometry and Trypan blue exclusion (n = 5). The line through the data indicates the median. (G) AML blasts from 3 different patients were cultured alone or with adipocytes or BMSC for 6 days and then placed in a CFC assay for 15 days. Colonies were then counted. Data are represented as mean ± standard deviation. (H) primary AML cells (2 × 106; 4 individual patient AML) cultured on BM adipocytes or cultured alone and then 2 × 106 viable cells were injected into NSG mice. Engraftment was measured using human CD33 and human CD45. Shown in the flow figure are the characteristics of AML#12 engraftment into BM and spleen. In the dot plot, each AML engraftment into NSG mice is shown for BM and spleen, and the engraftment of the AML cultured alone is shown by a shaded circle. The line through the data indicates the median. *P < .05. Ad, adipocytes; DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast; MFI, mean fluorescent intensity.
BM adipocytes support the survival and proliferation of primary AML. (A) Immunofluorescence of primary AML blasts stained for CD34+ (Alexa Fluor 568 [red]) and neutral lipids (green) using BODIPY 493/503. All images are representative of 6 AML patient samples. Original magnification ×63. Scale bar = 10 μm. (B) Freshly isolated AML, AML cultured for 1 day, and AML cultured for 2 days stained with the neutral lipid BODIPY 493/503 dye and analyzed by flow cytometry. Data are represented as mean ± standard deviation. (C) Freshly isolated nonmalignant CD34+ cells and freshly isolated AML samples stained with the neutral lipid BODIPY 493/503 dye and analyzed by flow cytometry. (D) AML patients samples in monoculture and cocultured with BM-derived adipocytes for 6 days and then stained with PI/Annexin V. The line through the data indicates the median. (E) AML blasts incubated alone or with adipocytes or BMSC for 6 days and AML blasts counted using flow cytometry and Trypan blue exclusion (n = 12). The line through the data indicates the median. (F) Nonmalignant CD34+ cells cultured alone or in coculture with BMSC or adipocytes and CD34+ cell counted using flow cytometry and Trypan blue exclusion (n = 5). The line through the data indicates the median. (G) AML blasts from 3 different patients were cultured alone or with adipocytes or BMSC for 6 days and then placed in a CFC assay for 15 days. Colonies were then counted. Data are represented as mean ± standard deviation. (H) primary AML cells (2 × 106; 4 individual patient AML) cultured on BM adipocytes or cultured alone and then 2 × 106 viable cells were injected into NSG mice. Engraftment was measured using human CD33 and human CD45. Shown in the flow figure are the characteristics of AML#12 engraftment into BM and spleen. In the dot plot, each AML engraftment into NSG mice is shown for BM and spleen, and the engraftment of the AML cultured alone is shown by a shaded circle. The line through the data indicates the median. *P < .05. Ad, adipocytes; DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast; MFI, mean fluorescent intensity.
Next, we wanted to determine if BM adipocytes can support the proliferation of primary AML blasts. Primary AML blasts increase in number when cocultured with BM adipocytes for 6 days, compared with AML monoculture and AML cultured on BMSC (n = 12) (Figure 1E). Figure 1F shows that adipocytes also support nonmalignant CD34+ cell survival. Supplemental Figure 3B-C shows that adipocytes support AML proliferation as measured by 5-bromo-2′-deoxyuridine incorporation. Supplemental Figure 4 shows AML blasts on BMSC and adipocytes from the same patient. In AML blast colony-forming-cell (CFC) assays, adipocyte coculture promoted CFC growth when compared with AML monoculture (Figure 1G). Finally, we showed that primary AML blasts from patient samples (n = 4) that had been cultured on BM-derived adipocytes for 6 days reliably engrafted NSG mice (Figure 1H). The same AML cells cultured in isolation did not engraft NSG mice after 12 weeks. AML were stained for CD33 and CD45 to confirm cell identity.16,22 We also examined the effect of cytokines on AML survival compared with coculture with adipocytes. Supplemental Figure 5A shows that there is a significant increase in survival of AML when cultured on adipocytes compared with media supplemented with cytokines. Supplemental Figure 5B shows that adipocytes support the engraftment of AML blasts compared with AML grown with cytokines. Taken together, these results show that adipocytes maintain AML progenitor cells.
AML blasts induce adipocyte lipolysis
Our results indicate that adipocytes confer a proliferative advantage to primary AML. We hypothesized that this could be due to transfer of FA from the adipocytes to AML. Therefore, to understand the effect AML cells have on adipocytes, we assessed FA and glycerol release. Coculture of AML with adipocytes increases FA and glycerol release compared with adipocyte and AML alone (Figure 2A). Moreover, nonmalignant CD34+ cells could also induce lipolysis of adipocytes, albeit at a much reduced quantity compared with AML (Figure 2A). Supplemental Figure 6A shows that conditioned media from AML can also induce adipocyte lipolysis. Moreover, Figure 2B shows that when we culture AML blasts with the FA, oleate, this supports the survival of AML. Lipolytic activation of adipocytes results from the phosphorylation of HSL. Figure 2C shows that AML induced pHSL in adipocytes. Conversely, acipomox (which is known to inhibit lipolysis by inactivation of AMP-activated protein kinase and downstream pHSL) inhibited AML proliferation when cultured on adipocytes but not alone or on BMSC (Figure 2D). Supplemental Figure 7 shows that adipocytes have significantly more HSL messenger RNA (mRNA) expression compared with BMSC and AML. To determine if lipids detected in the tumor cells after coculture were derived from adipocytes, we cultured primary AML blasts with either adipocytes or BMSC that had previously been loaded with a fluorescent lipid dye (DAA). Supplemental Figure 8 shows loading of DAA into both BMSC and adipocytes. During coculture, we observed that lipids were transferred from adipocytes to AML (Figure 2E-F; supplemental Figure 9); moreover, this transfer was inhibited by acipomox, thus supporting a model in which adipocytes provide lipids to support tumor growth.
AML blasts induce adipocyte lipolysis. (A) Primary AML blasts or nonmalignant CD34+ cells were cultured alone or in coculture with adipocytes for 24 hours. Media were removed and used to detect free FA and glycerol. Data are represented as mean ± standard deviation. (B) AML blasts incubated with media supplemented with BSA or media supplemented with 100 µM of oleate-BSA conjugate for 2 days and AML blasts counted using flow cytometry and Trypan blue exclusion (n = 4). The line through the data indicates the median. (C) Immunoblot for pHSL from adipocytes cultured with and without AML blasts (n = 4). Blots were reprobed for total HSL and β-actin to show equal sample loading. (D) Primary AML blasts in monoculture or cultured with adipocytes (adip) or BMSC with and without treatment with acipomox (APX) (10 μM) for 72 hours. AML blasts were counted using flow cytometry and Trypan blue exclusion (n = 4). The Mann-Whitney U test was used to determine statistical significance between treatment groups. Data are represented as mean ± standard deviation. (E-F) AML blasts cultured on adipocytes (ADI; with and without acipomox, 10 μM) or BMSC that had been preincubated with fluorescent DDA for 24 hours (n = 4). Blasts were analyzed for uptake of the fluorescent DDA by flow cytometry. Data are represented as mean ± standard deviation. (E) The red line is AML cultured on BMSC; the black line is AML cultured on adipocytes, and the blue line is AML cultured on adipocytes treated with acipomox. *P < .05. FFA, free fatty acid.
AML blasts induce adipocyte lipolysis. (A) Primary AML blasts or nonmalignant CD34+ cells were cultured alone or in coculture with adipocytes for 24 hours. Media were removed and used to detect free FA and glycerol. Data are represented as mean ± standard deviation. (B) AML blasts incubated with media supplemented with BSA or media supplemented with 100 µM of oleate-BSA conjugate for 2 days and AML blasts counted using flow cytometry and Trypan blue exclusion (n = 4). The line through the data indicates the median. (C) Immunoblot for pHSL from adipocytes cultured with and without AML blasts (n = 4). Blots were reprobed for total HSL and β-actin to show equal sample loading. (D) Primary AML blasts in monoculture or cultured with adipocytes (adip) or BMSC with and without treatment with acipomox (APX) (10 μM) for 72 hours. AML blasts were counted using flow cytometry and Trypan blue exclusion (n = 4). The Mann-Whitney U test was used to determine statistical significance between treatment groups. Data are represented as mean ± standard deviation. (E-F) AML blasts cultured on adipocytes (ADI; with and without acipomox, 10 μM) or BMSC that had been preincubated with fluorescent DDA for 24 hours (n = 4). Blasts were analyzed for uptake of the fluorescent DDA by flow cytometry. Data are represented as mean ± standard deviation. (E) The red line is AML cultured on BMSC; the black line is AML cultured on adipocytes, and the blue line is AML cultured on adipocytes treated with acipomox. *P < .05. FFA, free fatty acid.
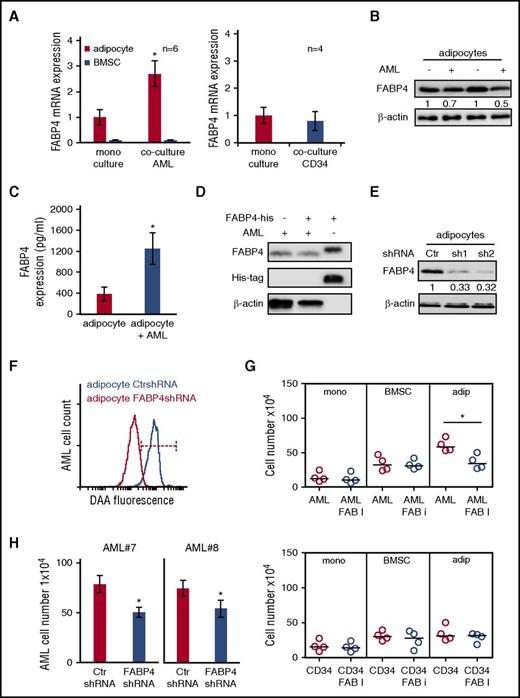
FABP4 is important for the transfer of lipids from adipocytes to AML
FABP4 is a carrier protein for FA expressed in adipocytes23 and supports the movement of long-chain FAs generated by lipolysis to the extracellular membrane and beyond.24,25 In addition, increased FABP4 has been shown to be involved in the transfer of FA from adipocytes to breast and ovarian cancer cells.12,14 We therefore examined the expression of FABP4 in human adipocytes and BMSC cultured with AML blasts or nonmalignant CD34+ cells. RNA expression data show that FABP4 levels are increased in adipocytes but not BMSC when cultured with AML; moreover, nonmalignant CD34+ cells had no effect on adipocyte FABP4 mRNA expression (Figure 3A). We also found that AML-conditioned media could induce FABP4 upregulation in adipocytes (supplemental Figure 10A). Next, we examined if other genes associated with lipolysis were also induced in adipocytes when cultured with AML. Supplemental Figure 11A shows that ATLG and MGLL mRNA expression is not changed in adipocytes when cultured with AML. In an apparent paradox, when the expression of FABP4 protein was examined, a reduction was observed in adipocytes cultured with AML compared with adipocytes alone (Figure 3B). FABP4 is known to be released from adipocytes as they transport the FA26 ; therefore, FABP4 expression in the culture media was measured by a FABP4-specific enzyme-linked immunosorbent assay. Figure 3C shows that culturing AML with adipocytes induces a release of FABP4 into the culture media. To determine if the FABP4 released by the adipocyte is taken up by the AML, we used a recombinant FABP4 tagged to a His sequence. AML were cultured with and without the recombinant FABP4 for 4 hours. Figure 3D shows that no recombinant FABP4 was detected in AML. Next, we examined if we could prevent the increase of lipids in AML blast by knockdown of FABP4 in the adipocyte. Figure 3E shows the knockdown FABP4 protein in adipocytes, and Figure 3F demonstrates that knockdown of adipocyte FABP4 inhibited the transfer of lipids to AML. Furthermore, FABP4 chemical inhibitor (BMS309403) inhibits AML blast survival but had no effect on nonmalignant CD34+ cells (Figure 3G). Finally, FABP4 lentiviral knockdown in adipocytes (Figure 3H) inhibits AML blast survival. Together, these results demonstrate that FABP4 is critical for the transfer of FA from adipocytes to AML blasts.
FABP4 controls the transfer of lipids from adipocytes to AML. (A) AML blasts were cultured alone or with adipocytes or BMSC for 48 hours and then the adipocytes and BMSC were assessed for FABP4 mRNA expression using RT-PCR (n = 6). Nonmalignant CD34+ cells were cocultured alone or with adipocytes, and FABP4 mRNA expression was measured. Data are represented as mean ± standard deviation. (B) Immunoblot for FABP4 from adipocytes cultured with and without AML blasts. Blots were reprobed for β-actin to show equal sample loading. (C) Enzyme-linked immunosorbent assay to detect FABP4 in media from adipocytes cultured alone or with AML blasts. Data are represented as mean ± standard deviation. (D) AML blasts were cultured alone or with the addition of 2 μg/mL of recombinant FABP4 (his-tagged) for 4 hours. Immunoblots were performed for FABP4 and His. Blots were reprobed for β-actin to show equal sample loading. (E) Adipocytes were infected with FABP4-targeted shRNA or control shRNA lentivirus, and after 72 hours, analyzed for FABP4 protein expression using western blotting. Blots were reprobed for β-actin to show equal sample loading. (F) Adipocytes were infected with FABP4-targeted shRNA or control shRNA lentivirus and after 72 hours preloaded with fluorescent FA DAA and incubated with AML for 24 hours. AML blasts were analyzed for fluorescence using flow cytometry (n = 4). (G) Primary AML blasts or nonmalignant CD34+ cells were cultured alone or cocultured with adipocytes or BMSC with and without treatment with FABP4 inhibitor for 72 hours. AML blasts were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (H) Adipocytes were infected with FABP4 targeted shRNA or control shRNA lentivirus, and after 72 hours, were incubated with AML for 72 hours. AML blasts and nonmalignant CD34+ cells were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. *P < .05. Ctr, control; FAB, fatty acid binding.
FABP4 controls the transfer of lipids from adipocytes to AML. (A) AML blasts were cultured alone or with adipocytes or BMSC for 48 hours and then the adipocytes and BMSC were assessed for FABP4 mRNA expression using RT-PCR (n = 6). Nonmalignant CD34+ cells were cocultured alone or with adipocytes, and FABP4 mRNA expression was measured. Data are represented as mean ± standard deviation. (B) Immunoblot for FABP4 from adipocytes cultured with and without AML blasts. Blots were reprobed for β-actin to show equal sample loading. (C) Enzyme-linked immunosorbent assay to detect FABP4 in media from adipocytes cultured alone or with AML blasts. Data are represented as mean ± standard deviation. (D) AML blasts were cultured alone or with the addition of 2 μg/mL of recombinant FABP4 (his-tagged) for 4 hours. Immunoblots were performed for FABP4 and His. Blots were reprobed for β-actin to show equal sample loading. (E) Adipocytes were infected with FABP4-targeted shRNA or control shRNA lentivirus, and after 72 hours, analyzed for FABP4 protein expression using western blotting. Blots were reprobed for β-actin to show equal sample loading. (F) Adipocytes were infected with FABP4-targeted shRNA or control shRNA lentivirus and after 72 hours preloaded with fluorescent FA DAA and incubated with AML for 24 hours. AML blasts were analyzed for fluorescence using flow cytometry (n = 4). (G) Primary AML blasts or nonmalignant CD34+ cells were cultured alone or cocultured with adipocytes or BMSC with and without treatment with FABP4 inhibitor for 72 hours. AML blasts were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (H) Adipocytes were infected with FABP4 targeted shRNA or control shRNA lentivirus, and after 72 hours, were incubated with AML for 72 hours. AML blasts and nonmalignant CD34+ cells were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. *P < .05. Ctr, control; FAB, fatty acid binding.
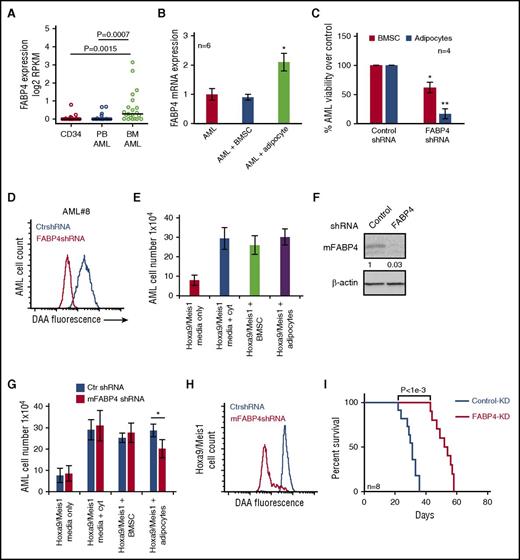
As Herroon and colleagues have shown that FABP4 in prostate cancer cells is upregulated in response to culture with adipocytes,15 we next examined whether FABP4 expression levels were elevated in AML cells extracted from BM as compared with AML cells extracted from peripheral blood or nonmalignant CD34+ cells. We found FABP4 expression to be significantly higher in AML BM samples than in AML from peripheral blood or nonmalignant CD34+ cells (P < .001; Wilcoxon rank-sum test) (Figure 4A). To confirm that this upregulation of FABP4 was a result of AML interacting with adipocytes rather than BMSC, we examined FABP4 expression in AML cultured alone, with adipocytes or BMSC. Supplemental Figure 12 shows the purity of AML after separation from adipocytes coculture. Figure 4B shows that FABP4 RNA is upregulated in AML in response to coculture with adipocytes but not BMSC. We determined whether knockdown of FABP4 in AML cells could reduce survival when cultured on adipocytes. Figure 4C shows that FABP4-KD AML cells have reduced viability when cocultured with adipocytes compared with coculture with BMSC. Furthermore, knockdown of AML FABP4 inhibited the transfer of lipids from adipocytes to AML (Figure 4D).
AML-derived FABP4 is crucial for blast survival in vivo. (A) FABP4 gene expression (expressed in log2 RPKM values) was obtained from GSE49642 and GSE48846 for nonmalignant CD34+ cells, 22 blood AML, and 21 BM AML patient samples. P value was obtained by Wilcoxon rank-sum test. Middle band denotes the median value with lower and upper bands denoting the first and third quartiles, respectively. (B) AML blasts were cultured alone or with adipocytes or BMSC for 48 hours before the AML were assessed for FABP4 mRNA expression using RT-PCR (n = 6). Data are represented as mean ± standard deviation. (C) Primary AML blasts were infected with FABP4 shRNA1 and shRNA2 and control shRNA, and after 96 hours, were subsequently cultured on adipocytes or BMSC for a further 72 hours. AML blasts counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (D) Primary AML were infected with FABP4-targeted shRNA1 or control shRNA lentivirus, and after 96 hours, were cultured with adipocytes preloaded with fluorescent FA DAA for 24 hours. AML blasts were analyzed for fluorescence using flow cytometry. (E) Hoxa9/Meis1-transformed cells (1 × 105/mL) were cultured as normal media or normal media with IL-3, IL-6, and SCF supplemented or cocultured on BMSC with normal media or on adipocytes with normal media. Hoxa9/Meis1-expressing cells were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (F) Hoxa9/Meis1-expressing cells were infected with mouse FABP4 shRNA or control shRNA, and after 72 hours, analyzed for FABP4 protein expression using western blotting. Blots were reprobed for β-actin to show equal sample loading. (G) Hoxa9/Meis1-expressing cells were infected with FABP4-targeted shRNA or control shRNA, and after 72 hours, incubated either alone, with cytokines, with BMSC, or with adipocytes. Hoxa9/Meis1-expressing cells counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (H) Hoxa9/Meis1 expressing cells were infected with FABP4-targeted shRNA or control shRNA lentivirus, and after 72 hours, incubated for 24 hours with adipocytes preloaded with fluorescent FA DAA. Hoxa9/Meis1-expressing cells were analyzed for fluorescence using flow cytometry (n = 4). (I) Kaplan-Meier survival curves for C57BL/6 mice injected with Hoxa9/Meis1 FABP4-KD cells or Hoxa9/Meis1 AML control-KD cells. *P < .05; **P < .01. PB, peripheral blood.
AML-derived FABP4 is crucial for blast survival in vivo. (A) FABP4 gene expression (expressed in log2 RPKM values) was obtained from GSE49642 and GSE48846 for nonmalignant CD34+ cells, 22 blood AML, and 21 BM AML patient samples. P value was obtained by Wilcoxon rank-sum test. Middle band denotes the median value with lower and upper bands denoting the first and third quartiles, respectively. (B) AML blasts were cultured alone or with adipocytes or BMSC for 48 hours before the AML were assessed for FABP4 mRNA expression using RT-PCR (n = 6). Data are represented as mean ± standard deviation. (C) Primary AML blasts were infected with FABP4 shRNA1 and shRNA2 and control shRNA, and after 96 hours, were subsequently cultured on adipocytes or BMSC for a further 72 hours. AML blasts counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (D) Primary AML were infected with FABP4-targeted shRNA1 or control shRNA lentivirus, and after 96 hours, were cultured with adipocytes preloaded with fluorescent FA DAA for 24 hours. AML blasts were analyzed for fluorescence using flow cytometry. (E) Hoxa9/Meis1-transformed cells (1 × 105/mL) were cultured as normal media or normal media with IL-3, IL-6, and SCF supplemented or cocultured on BMSC with normal media or on adipocytes with normal media. Hoxa9/Meis1-expressing cells were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (F) Hoxa9/Meis1-expressing cells were infected with mouse FABP4 shRNA or control shRNA, and after 72 hours, analyzed for FABP4 protein expression using western blotting. Blots were reprobed for β-actin to show equal sample loading. (G) Hoxa9/Meis1-expressing cells were infected with FABP4-targeted shRNA or control shRNA, and after 72 hours, incubated either alone, with cytokines, with BMSC, or with adipocytes. Hoxa9/Meis1-expressing cells counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (H) Hoxa9/Meis1 expressing cells were infected with FABP4-targeted shRNA or control shRNA lentivirus, and after 72 hours, incubated for 24 hours with adipocytes preloaded with fluorescent FA DAA. Hoxa9/Meis1-expressing cells were analyzed for fluorescence using flow cytometry (n = 4). (I) Kaplan-Meier survival curves for C57BL/6 mice injected with Hoxa9/Meis1 FABP4-KD cells or Hoxa9/Meis1 AML control-KD cells. *P < .05; **P < .01. PB, peripheral blood.
Next, we sought to determine the functional role of FABP4 in an AML mouse model. To do this, we used a retroviral AML transplantation model where Hoxa9 and Meis1 are used to transform myeloid progenitor cells, as described by Kroon et al.27 Initially, we determined if Hoxa9/Meis1-expressing cells could proliferate on adipocytes compared with BMSC or monoculture or monoculture without cytokine supplements. Figure 4E shows that Hoxa9/Meis1-expressing cells proliferate on adipocytes and BMSC. Next, we knocked down FABP4 in Hoxa9/Meis1-expressing cells (Figure 4F) and found that FABP4 KD reduced AML survival when cultured with adipocytes compared with media supplemented with cytokines or BMSC (Figure 4G). Moreover, knockdown of FABP4 in Hoxa9/Meis1-expressing cells also reduces FA uptake, quantified by DAA fluorescence, when tumor cells were cultured on adipocytes (Figure 4H). Finally, we show in vivo that FABP4-KD in Hoxa9/Meis1-expressing blasts significantly increases animal survival (Figure 4I). Taken together, these results demonstrate that FABP4 in AML is essential for FA transport and tumor proliferation in the BM microenvironment.
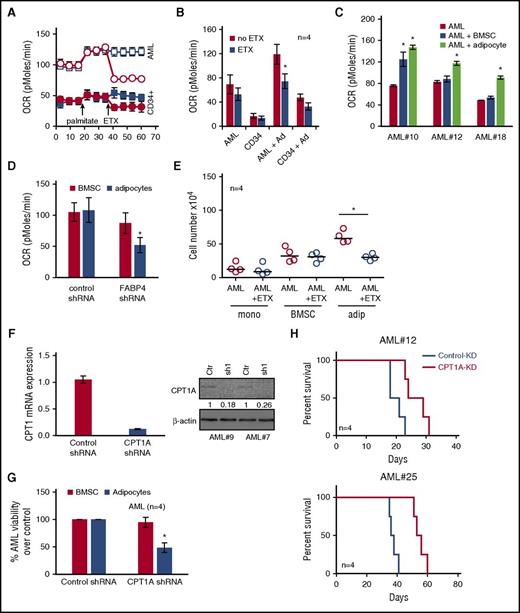
Coculture of AML with adipocytes activates β-oxidation in AML blasts
In order to study the reliance of AML blasts on adipocytes for their energy, we first measured the cellular oxygen consumption rate (OCR) in AML blasts to determine levels of fatty acid oxidation (FAO). To confirm that AML blasts can use oxidation of exogenous FA, we assessed FAO by monitoring OCR upon addition of palmitate (FAO substrate) followed by addition of ETX (FAO inhibitor). Figure 5A shows that primary AML have increased OCR in the presence of palmitate, which is inhibited by ETX; this is compared with nonmalignant CD34+ cells. Oxidation of exogenous FA can be measured by examining OCR of AML blasts and nonmalignant CD34+ cells in response to ETX. Figure 5B and supplemental Figure 13 show that AML blasts cultured with adipocytes have increased FAO compared with AML blasts cultured alone or with BMSC and nonmalignant CD34+ cells. In addition, we observed that AML blasts cultured on adipocytes had greater basal OCR compared with AML cultured alone or on BMSC (Figure 5C). OCR was reduced in AML cocultured on adipocytes compared with BMSC with FABP4 knocked down compared with control KD cells (Figure 5D). Moreover, because of the nature of the OCR experiment in which AML were serum starved for 4 hours prior to being loaded onto the Seahorse Bioanalyzer, we show that AML FABP4 mRNA levels were stable compared with non–serum-starved AML cells (supplemental Figure 10B). In addition, the FAO inhibitor ETX reduced AML blast survival when grown on adipocytes but not on BMSC (Figure 5E).
Coculture of AML with adipocytes activates β-oxidation in AML cells. (A) Primary AML blasts or nonmalignant CD34+ cells were cultured on adipocytes for 3 days and then starved for 4 hours before measuring OCR (pMoles/min) using the Seahorse XFp Analyzer, at baseline and then after injection of palmitate (18 minutes) and ETX (36 minutes). Circles represent ETX (40 μM) treatment, and squares represent no ETX treatment. (B) Primary AML blasts or nonmalignant CD34+ cells were cultured alone for 3 days, and AML blasts cultured on adipocytes for 3 days and then starved for 4 hours. AML blasts were then treated with ETX (40 μM), and OCR was measured as above (n = 4). Data are represented as mean ± standard deviation. (C) Primary AML blasts cultured alone for 3 days and AML blasts cultured on adipocytes or BMSC for 3 days and then starved for 4 hours. Data are represented as mean ± standard deviation. (D) AML were infected with FABP4-targeted shRNA or control shRNA lentivirus, and after 72 hours, incubated with adipocytes or BMSC for 24 hours, and OCR was measured in the AML (n = 4). Data are represented as mean ± standard deviation. (E) Primary AML blasts were in monoculture or cocultured on adipocytes or BMSC with and without treatment with ETX for 72 hours. AML blasts were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (F) AML blasts were infected with CPT1A shRNA or control shRNA lentivirus, and after 72 hours, analyzed for CPT1A mRNA and protein expression using RT-PCR and western blotting. Blots were reprobed for β-actin to show equal sample loading. Data are represented as mean ± standard deviation. (G) AML were infected with CPT1A shRNA or control shRNA lentivirus and then cocultured on BMSC or adipocytes. AML blasts were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (H) Two primary AML samples were infected with CPT1A shRNA or control shRNA lentivirus, and after 96 hours, were then grown on adipocytes for 48 hours; subsequently, 2 × 106 primary AML cells (n = 4) were IV injected into NSG mice. Survival of the NSG mice is represented by a Kaplan-Meier plot. P = .025 for mice injected with AML CPT1-KD compared with AML control-KD mice. *P < .05.
Coculture of AML with adipocytes activates β-oxidation in AML cells. (A) Primary AML blasts or nonmalignant CD34+ cells were cultured on adipocytes for 3 days and then starved for 4 hours before measuring OCR (pMoles/min) using the Seahorse XFp Analyzer, at baseline and then after injection of palmitate (18 minutes) and ETX (36 minutes). Circles represent ETX (40 μM) treatment, and squares represent no ETX treatment. (B) Primary AML blasts or nonmalignant CD34+ cells were cultured alone for 3 days, and AML blasts cultured on adipocytes for 3 days and then starved for 4 hours. AML blasts were then treated with ETX (40 μM), and OCR was measured as above (n = 4). Data are represented as mean ± standard deviation. (C) Primary AML blasts cultured alone for 3 days and AML blasts cultured on adipocytes or BMSC for 3 days and then starved for 4 hours. Data are represented as mean ± standard deviation. (D) AML were infected with FABP4-targeted shRNA or control shRNA lentivirus, and after 72 hours, incubated with adipocytes or BMSC for 24 hours, and OCR was measured in the AML (n = 4). Data are represented as mean ± standard deviation. (E) Primary AML blasts were in monoculture or cocultured on adipocytes or BMSC with and without treatment with ETX for 72 hours. AML blasts were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (F) AML blasts were infected with CPT1A shRNA or control shRNA lentivirus, and after 72 hours, analyzed for CPT1A mRNA and protein expression using RT-PCR and western blotting. Blots were reprobed for β-actin to show equal sample loading. Data are represented as mean ± standard deviation. (G) AML were infected with CPT1A shRNA or control shRNA lentivirus and then cocultured on BMSC or adipocytes. AML blasts were counted using flow cytometry and Trypan blue exclusion (n = 4). Data are represented as mean ± standard deviation. (H) Two primary AML samples were infected with CPT1A shRNA or control shRNA lentivirus, and after 96 hours, were then grown on adipocytes for 48 hours; subsequently, 2 × 106 primary AML cells (n = 4) were IV injected into NSG mice. Survival of the NSG mice is represented by a Kaplan-Meier plot. P = .025 for mice injected with AML CPT1-KD compared with AML control-KD mice. *P < .05.
ETX is a selective inhibitor of CPT1A, which transports fatty acyl chains from the cytosol into the mitochondria, a process essential for the production of adenosine triphosphate from FA oxidation.28 We therefore examined the expression of various genes including CPT1A in AML when cultured with adipocytes. Supplemental Figure 11B shows that CPT1A, CPT2, and ACADL are all upregulated in AML when cultured with adipocytes. Accordingly, we knocked down CPT1A in AML blasts. Figure 5F shows a reduction in CPT1A mRNA and protein in human AML cells after infection with CPT1A shRNA lentivirus. Following successful knockdown of CPT1A in AML cells, we found that CPT1A-KD AML cells have reduced viability when cultured with adipocytes compared with cells cultured on BMSC (Figure 5G). Finally, we tested the effect of CPT1A-KD in an AML patient-derived xenograft model. Figure 5H shows that NSG mice engrafted with CPT1A-KD AML blasts from 2 patient samples have increased survival compared with control-KD NSG mice (n = 4). Supplemental Figure 14 shows the BM and spleen engraftment data for Figure 5H.
Discussion
Here, we report that BM adipocytes support the survival and proliferation of AML blasts. We find that AML induces lipolysis of triglyceride stored within BM-derived adipocytes. Subsequently, the FA released by triglyceride lipolysis within adipocytes is transported out of the adipocyte in a process dependent on the chaperone protein FABP4. Proximity to adipocytes also results in upregulation of the same chaperone protein, FABP4, within the blasts, which is then used to transport adipocyte-derived FA to the mitochondria within the tumor cells. The AML mitochondria use the FA as a substrate for β-oxidation, generating the energy required for leukemic growth and proliferation.
AML arises from malignant transformation and proliferation of hematopoietic progenitor cells in the BM microenvironment. AML is colocated with BM adipose tissue (MAT), which is a biologically active energy storage and endocrine organ and accounts for ∼70% of BM volume in adult humans.8,29 Like the prevalence of AML, BM adipocytes are known to increase with age and furthermore are not merely passive occupants of the BM but are now appreciated to be actively involved in processes linked to bone metabolism, osteoporosis, inflammation, and regulation of the hematopoietic niche.30 Moreover, adipocytes have been shown to support the proliferation of solid tumor cells in studies of breast, ovarian, and prostate cancer metastases.12,14,15 Here, we describe the protumoral function of BM-derived adipocytes in AML.
We report that leukemia cells cause functional changes in nonmalignant adipocytes, which ultimately lead to the transfer of free FAs from adipocytes to the AML blast. FA transfer is a FABP4-dependent process. FABP4 has intracellular functions in adipocytes but is also known to be actively secreted by adipocytes where it can exert specific biological functions in tissues other than its origin.31 Coculture of AML and adipocytes results in transcriptional upregulation of FABP4, in both the adipocyte and the leukemic blast. Curiously, however, we found that despite this transcriptional increase of FABP4 in the adipocytes, the intra-adipocyte protein level of FABP4 is decreased upon coculture with AML. The inverse relationship between transcription and protein content led us to hypothesize that (1) adipocyte FABP4 rapidly chaperones the FA from the adipocyte into the microenvironment, (2) adipocyte FABP4 rapidly chaperones the FA from the adipocyte into the AML cells, or (3) excess intracellular FABP4 is simply degraded within the adipocyte. To consider this further, we investigated the levels of FABP4 in the media of AML adipocyte cocultures and confirmed FABP4 was released into the medium. In models of prostate cancer, recombinant FABP4 is taken up by the malignant cells.32 In our studies, we also conducted experiments using recombinant FABP4 to determine if AML blasts actively take up this protein. We showed that no recombinant FABP4 was detected in AML after 4 hours of culture. These data suggest that unlike prostate cancer FA/FABP4 is not directly transferred from adipocyte to the AML cell but may be involved in an extracellular-to-membrane transport whereby it is degraded after functionality outside of the cell. Further studies are needed to confirm this hypothesis.
Maintenance of AML appears to depend on both the adipocyte and the leukemic blast being able to generate FABP4. Pharmacological inhibition or lentiviral knockdown of FABP4 in the adipocytes showed a significant inhibitory effect on AML survival in coculture experiments. FA-binding proteins bind free FA, which otherwise in their free form are relatively insoluble and potentially toxic. Furthermore, FABP4 binds and donates its FA ligand via collisional interactions with membranes and is actively secreted from adipocytes.31,33 We suggest that the reduced AML survival on FABP4 KD adipocytes is due to the inability of FA to be secreted from the adipocytes in the absence of FABP4 protein. In addition, we observed that FABP4 knockdown in the AML cells improved survival in our in vivo models (although was unable to ultimately prevent leukemic engraftment). We also compared FABP4 transcript levels between AML blasts taken from the BM and the peripheral blood and found that FABP4 was enriched in AML cells collected from the adipose-rich environment of the BM, indicating that FABP4 expression in AML blasts requires an external BM-derived signal. Taken together, we propose that in AML, FA is transported in the BM adipocyte by adipocyte-derived FABP4, then secreted and taken up by the AML and transported within the AML by AML-derived FABP4. We conclude that delivery and intracellular transport of FA to and in AML blasts are dependent on the presence of functional BM adipocytes.
When we compare our in vitro FABP4 knockdown experiments in primary human AML cells and HoxA9/Meis1 cells, the effect in the HoxA9/Meis1 experiments is comparatively modest. However, in contrast, when FABP4 was knocked down in HoxA9/Meis1 cells, which were then injected into mice, we observed a more demonstrable antitumor effect. In addition, we also found that our AML blasts maintained on adipocytes appeared to engraft (range 4-6 weeks) earlier than expected when compared with other reports in which primary AML blasts had not been previously cultured on adipocytes.34-36 We have not investigated this observation formally but postulate that adipocytes may support the maintenance of primary leukemic progenitor cells. Overall, we suggest that our in vivo model more closely recapitulates the complexity of a tumor growing within a patient than the in vitro assays and also propose that adipocytes may improve efficiency of ex vivo culture systems for primary human AML.
Blocking lipolysis using Acipomox or inhibiting transfer of free FAs using a FABP4 inhibitor only decreased AML viability between 40% and 60%. The reduction in cell viability could in part be due to the efficiency of the drug to inhibit its target. Alternatively, perhaps this implies that adipocytes contribute other pro-AML survival factors in addition to free FAs. Adipocytes have been shown to secrete proinflammatory cytokines in the presence of malignant cells, which may have an impact on tumor cell migration and survival.37 The cytokine secretion by adipocytes has specifically been shown in the context of leukemias by Ye and colleagues, who report that BM and gonadal adipose tissues show high levels of tumor-associated proinflammatory cytokine secretion, several of which had previously been implicated in tumor cell migration.37-39 Therefore, it is likely that BM adipocytes provide more than just FA to AML to facilitate the proliferative capacity of this disease.
We found that the OCR of AML cells increased in adipocyte coculture experiments and that inhibition of CPT1A in the leukemic cells then significantly reduced AML OCR and survival. CPT1A is essential for the transfer of FA to the inner mitochondrial membrane for acetyl-Coenzyme A generation via β-oxidation,40 and these data indicate that the BM adipocytes are a source for the FA necessary for AML metabolism. Our observations are also consistent with the work of others who have previously reported the importance of β-oxidation in AML survival and proliferation.41,42 Conversely, in control experiments, nonmalignant CD34+ cells did not appear to significantly decrease their OCR upon treatment with the CPT1A inhibitor, ETX, indicating that this FA oxidation signature is particularly associated with leukemic progenitors.
In summary, although adipocytes in the nonmalignant setting appear to be negative regulators of hematopoietic microenvironment,43 in the context of disease, adipocytes, like other cells, can undergo distinct pathologic changes.44 Here, we report that in the setting of AML, BM-derived adipocytes support tumor proliferation and survival. Specifically, AML blasts modulate intracellular adipocyte metabolism into a lipolytic state, resulting in the release of FA into the microenvironment. Ultimately, this free FA is metabolically beneficial to the leukemia. Accordingly, we hypothesize that identification of this protumoral interaction will open up potential novel therapeutic strategies in the treatment of human AML.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession numbers GSE49642 and GSE48846).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Richard Ball, Mark Wilkinson, Iain Sheriffs, and Norwich Biorepository for help with sample collection and storage.
The authors thank the The Big C, The Rosetrees Trust, The Norwich Research Park, the National Institutes for Health Research (UK), and the Ministry of Higher Education and Scientific Research of the Libyan Government for funding.
Authorship
Contribution: M.S.S., T.O., K.M.B., and S.A.R. designed the research; M.S.S., S.M., L.Z., and A.A.-A. conducted the research; M.S.S., S.D.R., S.M., C.R.M., and R.E.P. carried out the in vivo work; D.R.E., M.F., J.T., M.L., and K.M.B. provided essential reagents and knowledge; J.A.W. performed the bioinformatics; and M.S.S., K.M.B., T.O., and S.A.R. wrote the paper.
Conflict-of-interest disclosure: S.A.R. reports research funding from Infinity Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Stuart A. Rushworth, Department of Molecular Haematology, Norwich Medical School, University of East Anglia, Norwich NR4 7UQ, United Kingdom; e-mail: s.rushworth@uea.ac.uk.

![Figure 1. BM adipocytes support the survival and proliferation of primary AML. (A) Immunofluorescence of primary AML blasts stained for CD34+ (Alexa Fluor 568 [red]) and neutral lipids (green) using BODIPY 493/503. All images are representative of 6 AML patient samples. Original magnification ×63. Scale bar = 10 μm. (B) Freshly isolated AML, AML cultured for 1 day, and AML cultured for 2 days stained with the neutral lipid BODIPY 493/503 dye and analyzed by flow cytometry. Data are represented as mean ± standard deviation. (C) Freshly isolated nonmalignant CD34+ cells and freshly isolated AML samples stained with the neutral lipid BODIPY 493/503 dye and analyzed by flow cytometry. (D) AML patients samples in monoculture and cocultured with BM-derived adipocytes for 6 days and then stained with PI/Annexin V. The line through the data indicates the median. (E) AML blasts incubated alone or with adipocytes or BMSC for 6 days and AML blasts counted using flow cytometry and Trypan blue exclusion (n = 12). The line through the data indicates the median. (F) Nonmalignant CD34+ cells cultured alone or in coculture with BMSC or adipocytes and CD34+ cell counted using flow cytometry and Trypan blue exclusion (n = 5). The line through the data indicates the median. (G) AML blasts from 3 different patients were cultured alone or with adipocytes or BMSC for 6 days and then placed in a CFC assay for 15 days. Colonies were then counted. Data are represented as mean ± standard deviation. (H) primary AML cells (2 × 106; 4 individual patient AML) cultured on BM adipocytes or cultured alone and then 2 × 106 viable cells were injected into NSG mice. Engraftment was measured using human CD33 and human CD45. Shown in the flow figure are the characteristics of AML#12 engraftment into BM and spleen. In the dot plot, each AML engraftment into NSG mice is shown for BM and spleen, and the engraftment of the AML cultured alone is shown by a shaded circle. The line through the data indicates the median. *P < .05. Ad, adipocytes; DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast; MFI, mean fluorescent intensity.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/10/10.1182_blood-2016-08-734798/4/m_blood734798f1.jpeg?Expires=1769095273&Signature=WgH7Jia8si~YqANyniADzE~iAreW088zvWtHqfmmx148yEqq0YoIYtc3R49G1KM5VgD-N4wZsqOUL6TprztpCXl6Orm-XuiEvsQcPeE5ZkhhQuHRwZN06Ila0oIyXG4cx7GQYHOgBEw-h1Yd5gYCjx9OnDorKWiHahOxa6Qo53nuARKb65AFXrzO20m9JHU-iZzOQ0C8VWX-m~M6RP6hRsEEBK7K8OmZwZSh7iPtIjG-CaQfQshqxCK~-KCv7n6QzlvpOCvqYzUzp5OVhgmiksEJEcRToiD7NQOveCHsyI6of9~XFCGBpZSsSxa8l0DXGNxN2cAD9C9W9feV1Td5BA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal